Abstract
We have recently shown that heterochromatin protein 1 (HP1) interacts with the nuclear envelope in an acetylation-dependent manner. Using purified components and in vitro assays, we now demonstrate that HP1 forms a quaternary complex with the inner nuclear membrane protein LBR and a sub-set of core histones. This complex involves histone H3/H4 oligomers, which mediate binding of LBR to HP1 and cross-link these two proteins that do not interact directly with each other. Consistent with previous observations, HP1 and LBR binding to core histones is strongly inhibited when H3/H4 are modified by recombinant CREB-binding protein, revealing a new mechanism for anchoring domains of under-acetylated chromatin to the inner nuclear membrane.
INTRODUCTION
Heterochromatin protein 1 (HP1) is a member of the Polycomb group and includes three distinct isotypes: HP1α, HP1β and HP1γ in humans, and mHP1α, M31 and M32 in mice (Cavalli and Paro, 1998; Jones et al., 2000). All HP1 variants are structurally similar and contain two sequence-related segments: the N-terminal chromodomain (CD) (Paro and Hogness, 1991; Singh et al., 1991) and the C-terminal chromoshadow domain (CSD) (Aasland and Stewart, 1995). These two domains consist of anti-parallel, three-stranded β-sheets packed against one or two α-helices and are separated from each other by a flexible hinge region (Ball et al., 1997; Brasher et al., 2000).
HP1 forms dimers via intermolecular interactions that involve the CSDs (Brasher et al., 2000), and binds to several chromatin remodelling factors and transcriptional regulators (Jones et al., 2000). In addition, specific interactions with the inner nuclear membrane protein LBR and histone H3 methylated at lysine 9 have been recently reported (Ye and Worman, 1996; Bannister et al., 2001; Lachner et al., 2001). The HP1–nuclear envelope associations (NE) are particularly intriguing: first, the anchorage of chromatin domains to the inner nuclear membrane is thought to affect transcriptional activity (Andrulis et al., 1998); secondly, transient binding involving components of condensed chromosomes and inner nuclear membrane proteins are thought to provide the basis for NE reassembly at the end of mitosis.
In previous studies (Kourmouli et al., 2000) we have shown that all three mouse HP1 proteins associate with the NE both in vivo and in vitro. To further investigate these interactions, we have now tried to identify specific NE sites to which these proteins attach. Biochemical data presented in this paper reveal that HP1 forms a tight complex with two of the core histones (H3/H4) and LBR. Consistent with our in vivo observations, these in vitro interactions are strongly inhibited by histone acetylation, suggesting a new mechanism for anchoring heterochromatic domains to the NE.
RESULTS
HP1 associates with LBR through the core histones
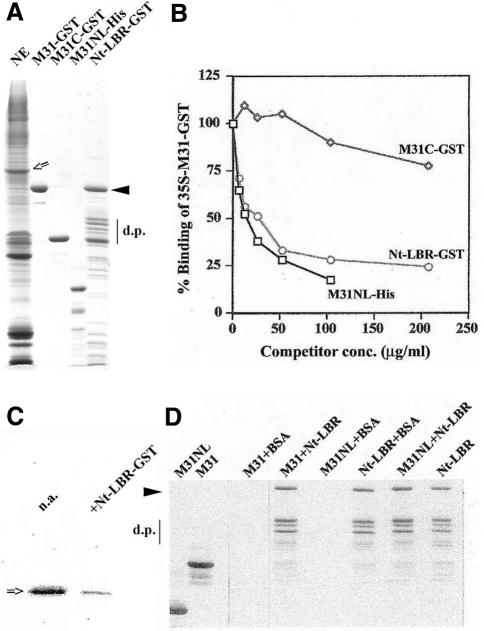
Previous studies have shown that all variants of mouse HP1 (mHP1α, M31 and M32) bind to salt–urea ‘stripped’ NEs (Kourmouli et al., 2000, 2001). Since the NEs primarily contain integral proteins and some tightly-adhering histones (see Figure 1A, lane NE; for additional information see Worman et al., 1988), we reasoned that LBR, a major constituent of the inner nuclear membrane, would be a good candidate for an HP1-docking site. To test this idea, we measured binding of unlabelled and metabolically labelled M31–GST (the mouse homologue of HP1β) to the NEs in the presence and absence of Nt-LBR, a water-soluble, N-terminal fragment, which contains ∼200 amino acids and represents the entire nucleoplasmic domain of this protein (for SDS–PAGE profiles of the proteins used see Figure 1A, lanes M31–GST and Nt-LBR–GST).
Fig. 1. The N-terminal domain of LBR inhibits binding of M31 to the NEs, but does not associate directly with M31. (A) SDS–PAGE profiles of the materials used in binding assays. Components are as indicated or explained in the text. An open arrow shows the position of native, full-length LBR. The closed arrowhead denotes the position of intact Nt-LBR–GST, while d.p. shows the characteristic proteolytic products of this recombinant protein that are produced after expression in bacteria (for relevant information see Nikolakaki et al., 1996; Ye and Worman, 1996). (B) Relative binding of [35S]M31–GST in the presence of Nt-LBR–GST, M31C–GST and M31NL-His, as detected by a co-pelleting assay and liquid scintillation counting. For technical details see Methods. (C) Experiment similar to the one shown in (B), using unlabelled M31–GST and western blotting with anti-M31 antibodies. n.a., sample in which Nt-LBR–GST was omitted; +Nt-LBR–GST, sample in which Nt-LBR–GST was added at a concentration of 200 µg/ml. (D) Pull-down assay with Nt-LBR–GST and M31-His (M31), M31NL-His (M31NL) or bovine serum albumin (BSA). The gel shows the material precipitated by glutathione beads after staining with Coomassie Blue.
As expected, Nt-LBR–GST inhibited M31–GST binding in a dose-dependent fashion (Figure 1B and C). In the same assay, M31–GST binding was also inhibited by M31NL-His, a recombinant protein that contains the NE-association site of M31, but not by M31C–GST, a C-terminal fragment of M31 that does not include the NE-binding region (for sequences and additional information see Kourmouli et al., 2000). However, neither M31-His nor M31NL-His bound directly to Nt-LBR–GST when purified proteins were tested in a pull-down assay (Figure 1D), suggesting that the HP1–LBR interactions are mediated by ‘linker’ proteins.
To identify such linker proteins, we extracted NEs with high salt/Triton X-100 and incubated the solubilized material with purified M31–GST. As shown in Figure 2, M31–GST precipitated specifically LBR and histones H3/H4. Detection of LBR required western blotting because the amount of this protein in the M31–GST precipitates was relatively small (Figure 2B; for a comment on this see Discussion). However, histones H3/H4 co-sedimenting with M31–GST were easily detectable by Coomassie Blue staining (Figure 2A). Neither LBR nor histones bound to the GST control (Figure 2, lane GST), while specific binding to mHP1α–GST and M32–GST, the two other variants of mouse HP1, could be readily documented using the same pull-down approach (H. Polioudaki and S.D. Georgatos, unpublished observations).
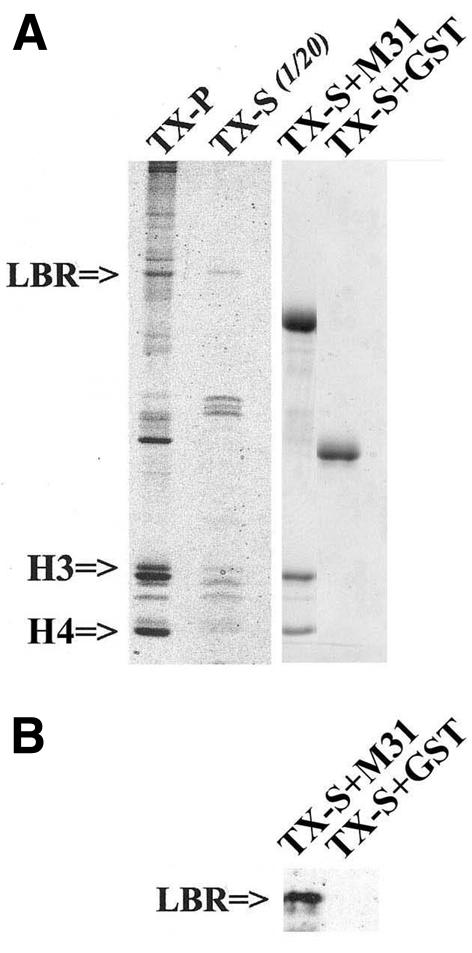
Fig. 2. Core histones and LBR are co-precipitated by M31–GST from NE extracts. NEs were extracted with 1 M NaCl and 1% Triton as described in Methods. The soluble fraction was adjusted to 500 mM NaCl and used in pull-down experiments with M31–GST or GST alone (control). The figure shows SDS–PAGE profiles of the following samples: TX-P, NE residue after high salt/Triton extraction; TX-S, high salt/Triton extract corresponding to 1/20 of the TX-P; TX-S+M31, material precipitated by M31–GST; TX-S+GST, material precipitated by GST. (A) Coomassie Blue-stained gel. (B) Part of a blot that was probed with affinity-purified anti-LBR antibodies.
The association of H3/H4 with LBR and M31 was not affected when the NE extracts were pre-digested with DNase I, indicating that DNA was not involved in these interactions (data not shown). However, to prove this point more rigorously, we repeated the binding assays using isolated histones. As shown in Figure 3, Nt-LBR–GST (Figure 3A) and M31–GST (Figure 3B) bound preferentially to histones H3 and H4, but not to histones H2A and H2B. The same type of association was detected when each core histone was tested alone, in combination with its closest partner in the core particle (e.g. H3/H4 and H2A/H2B) or in an ‘all histone’ mixture. Binding of M31 and Nt-LBR to histones H3/H4 was nearly stoichiometric and could still occur in the presence of 0.5–0.6 M salt.
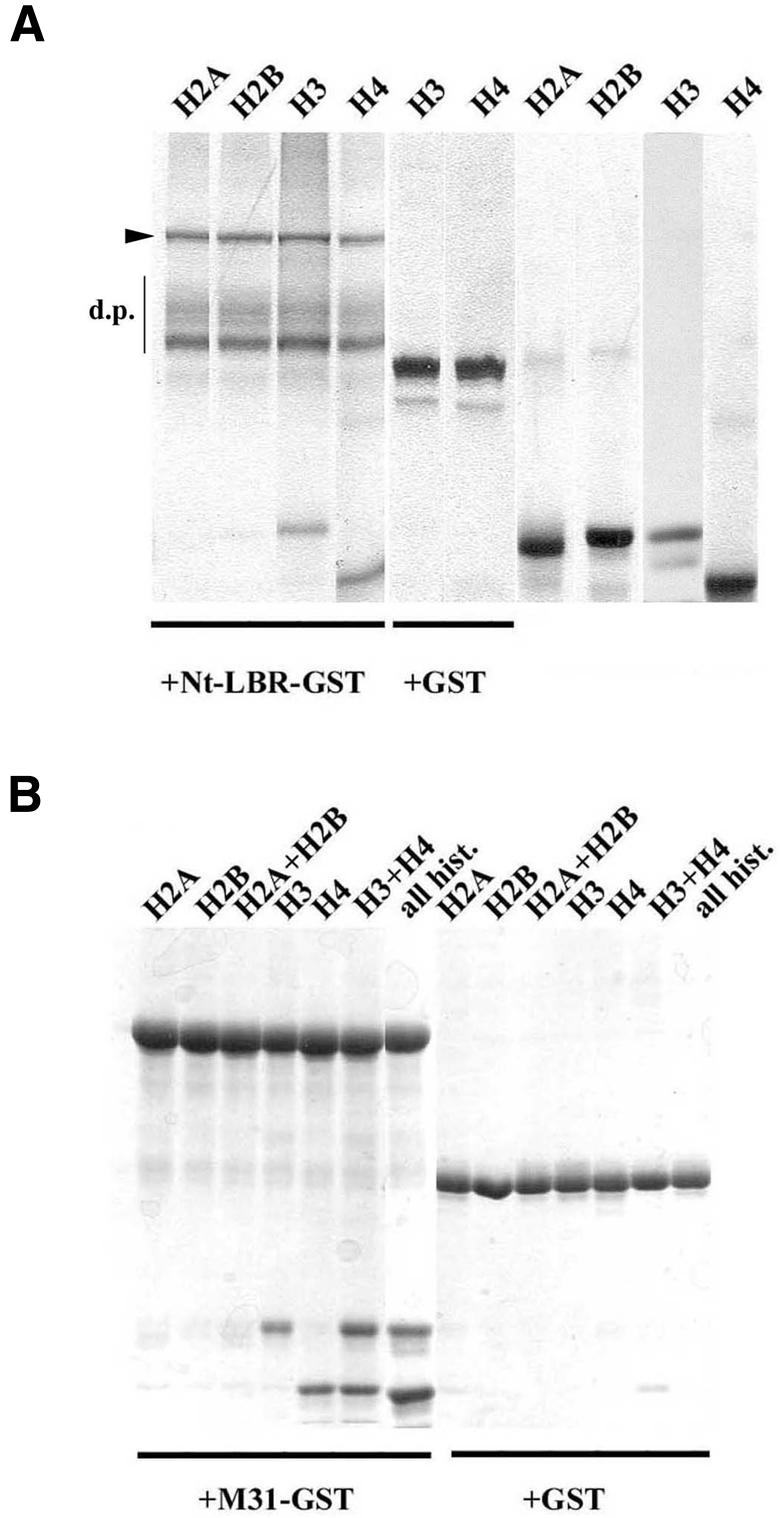
Fig. 3. Histones H3 and H4 bind to LBR and M31. Pull-down assays using purified core histones Nt-LBR–GST (A) and M31–GST (B). GST is also included in both cases as a control. Both panels show SDS–polyacrylamide gels. The samples analysed are as indicated. Lanes ‘all hist.’ correspond to a mixture of histones H2A, H2B, H3 and H4. The closed arrowhead denotes the position of intact Nt-LBR–GST, while d.p. signifies characteristic proteolytic products of this protein that are produced after expression in bacteria.
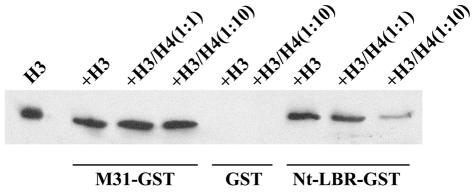
To find out whether H3 and H4 bound to distinct sites along the M31 and the LBR molecules, we proceeded with competition assays. In these experiments, a fixed quantity of histone H3 was incubated with either M31–GST or Nt-LBR–GST in the presence of increasing amounts of histone H4. H4 did not displace histone H3 from M31–GST, but did compete with H3 for binding to Nt-LBR–GST (Figure 4). From these results it can be inferred that the two core histones associate with the same site of LBR, but bind to distinct regions of the M31 molecule. In agreement with previously published observations (Bannister et al., 2001; Lachner et al., 2001), we could localize the H3-binding site of M31 in the CD; however, assignment of the H4-binding site could not be done with precision because fragments containing the N-terminal part of the molecule (amino acids 1–69) and the hinge region (residues 70–114) appeared to bind to H4 with comparable affinity (data not shown). More refined analysis needs to be done in order to characterize these binding sites and investigate in a more systematic fashion whether they accommodate differently modified forms of histone H4.
Fig. 4. Core histones bind to the same site of LBR, but associate with different sites of the M31 molecule. Competition assays using M31–GST, GST or Nt-LBR–GST, a constant amount of histone H3 and variable amounts of histone H4. The assay was executed as in Figure 3 and the H3 co-sedimenting with each GST fusion protein was detected by ECL/western blotting (for further details see text).
Reconstitution of the HP1–histone–LBR complex from recombinant proteins
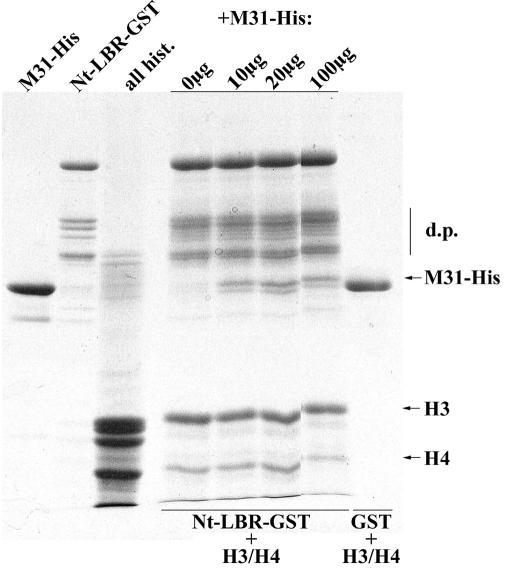
To examine whether the core histones cross-link LBR to M31 or bind to the two proteins in a mutually exclusive fashion, we co-incubated purified H3/H4 with Nt-LBR–GST and added to the reaction mixture increasing quantities of M31-His. As shown in Figure 5, M31-His co-precipitated with Nt-LBR and did not displace H3/H4 from the complex. This is in contrast to the data depicted in Figure 1D, which show that M31-His fails to bind Nt-LBR in the absence of histone proteins. From the sum of these observations we conclude that LBR, M31 and histones H3/H4 form a stable, quaternary complex.
Fig. 5. M31 forms a ternary complex with histones H3/H4 and Nt-LBR. Nt-LBR–GST was immobilized on glutathione beads and reacted sequentially with a standard amount of histones H3/H4 and increasing quantities of M31-His, as indicated. The formation of a ternary complex was assayed by pull-down experiments and SDS–PAGE. Designations are as in previous figures.
The interactions of core histones with HP1 and LBR are regulated by acetylation
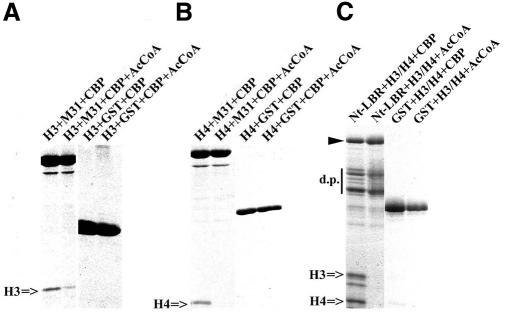
Employing in vivo approaches, we have previously shown that the HP1–NE association is abolished under hyper-acetylating conditions (e.g. when cells are treated with trichostatin A or sodium butyrate; see Kourmouli et al., 2000). Taking these and the current data into account, we set out to determine whether H3/H4 acetylation could account for this effect. Recombinant CREB-binding protein (CBP) was used to modify the core histones under controlled, in vitro conditions. The acetylated material, or mock-acetylated histones, was then incubated with M31–GST or Nt-LBR–GST and binding was assessed in a standard fashion. As shown in Figure 6, acetylated H3/H4 no longer bound to recombinant M31 or LBR. This inhibition was not observed when acetyl coenzyme A was omitted from the reactions, indicating a very specific effect.
Fig. 6. The M31–histone interaction is inhibited by CBP-mediated acetylation. Pull-down experiments with M31–GST and histone H3 (A), M31–GST and histone H4 (B), and Nt-LBR–GST and a mixture of histones H3/H4 (C). The histones were acetylated in vitro by recombinant CBP as explained in Methods. GST was included in all experiments as a control. AcCoA corresponds to acetyl coenzyme A.
DISCUSSION
Using in vitro assays, we have shown here that the inner nuclear membrane protein LBR associates with the heterochromatin-specific protein HP1 via histones H3/H4. These data differ from the results previously published by Worman and co-workers, who have claimed direct binding of HP1 proteins to LBR on the basis of two-hybrid assays and pull-down experiments with in vitro transcribed/translated proteins (Ye and Worman, 1996; Ye et al., 1997). The two-hybrid findings can be explained by indirect interactions involving the endogenous, yeast histones. However, the contradictory results obtained in biochemical assays cannot be easily understood unless we assume electrostatic, low-affinity binding between the basic (pI = 10), N-terminal domain of LBR and some acidic stretch of the HP1 molecule. Such electrostatic interactions, which could also account for the binding of Nt-LBR to naked DNA (Ye and Worman, 1996; Duband-Goulet and Courvalin, 2000), are unlikely to occur under more stringent conditions (e.g. in the presence of 0.5 M salt) and may not be physiologically meaningful.
The fact that histone–HP1 interactions are strongly affected by post-translational modifications is consistent with recent observations reported by Lachner et al. (2001) and Bannister et al. (2001), which document specific binding of HP1 to methylated histone H3. Since methylation of the critical lysine 9 in the N-terminal tail of H3 cannot occur when the same residue is acetylated (Rea et al., 2000), the two types of modification may constitute an ‘on/off’ switch that allows regulated binding of HP1 proteins to the core particle. However, it is also possible that modifications in the histone tails may also affect interactions of HP1 with the ‘histone fold’ part of the H3/H4 molecules, as suggested in a recently published report (Nielsen et al., 2001).
Agreeing in part with the report by Zhao et al. (2000), we have found that H4 (independently of H3) associates with both HP1 and LBR under in vitro conditions. However, HP1–H4 binding was not observed by Lachner et al. (2001), who have used as a ligand a synthetic peptide representing the N-terminal tail of histone H4. Moreover, this interaction was not detected by Bannister et al. (2001), who have employed intact H4 obtained from the same commercial source that we have used in the current study. One parameter that might account for these discrepancies is the modification state of the core histones. A non-modified synthetic peptide representing the tail of H4 is physically different from native, partially modified (e.g. phosphorylated) H4. Furthermore, commercial H4 preparations may differ in their acetylation state, depending on source and method of purification. With regards to this, it is interesting to note that of six commercially obtained H4 lots that we have tested, one contained an apparently inactive form of the protein, which consistently failed to bind HP1 under standard assay conditions.
Speculation
Since LBR and H3/H4 are co-precipitated by HP1–GST from salt/detergent extracts of NEs, it is likely that these proteins form a specific, multi-component complex. This complex may represent a ‘junctional’ assembly that anchors HP1-enriched heterochromatin to the NE.
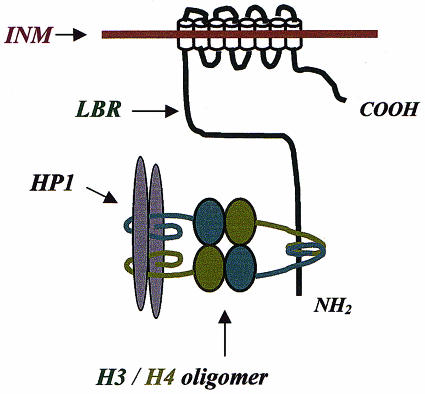
Considering the information currently available, it is likely that H3/H4 operate as a bivalent ligand (perhaps a tetrameric oligomer), cross-linking LBR and HP1, which do not bind directly to each other (for a hypothetical model see Figure 7). Depending on the stability of this oligomer, the LBR–histone–HP1 complex may occur in two different states: the fully assembled state (histone oligomer intact and LBR stably connected to HP1) and the partially dissociated state (histone oligomer disassembled and LBR disconnected from HP1). A hint favouring this interpretation is the fact that although H3/H4 bind stoichiometrically to M31–GST and Nt-LBR–GST, only a small amount of LBR is co-precipitated with M31–GST and histones under conditions that are expected to be sub-optimal for the stability of histone oligomers (e.g. 0.5 M salt).
Fig. 7. A provisional model for the LBR–histone–HP1 complex. LBR and HP1 are postulated to associate with one another via an H3/H4 oligomer (tetramer). Based on biochemical data presented in Figures 1–6, it is proposed that H3/H4 bind to the same site of the N-terminal domain of LBR, but interact with two separate regions of the HP1 molecule (depicted here as a dimer) through their tail regions (curved lines).
METHODS
Antibodies, plasmids and other materials. The polyclonal anti-LBR antibodies used have been described in previous studies (Meier and Georgatos, 1994), while the anti-histone H3 antibodies were obtained through commercial sources (Santa Cruz). M31 (full-length), or the C-terminal segment (M31C), and M32 were expressed as fusion proteins with GST using pGEX1. GST–mHP1α was cloned in pGEX-4T-1. Full-length M31 and M31NL were expressed as His6-tagged proteins employing pET-25b. Purified histones were purchased from Roche. CBP was a generous gift from Niki Kretsovali (Institute of Molecular Biology and Biotechnology, Heraklion, Crete, Greece).
Expression, purification and metabolic labelling of recombinant proteins. GST fusion proteins and His6-tagged polypeptides were expressed in BL21 (DE3) cells and purified from bacterial lysates according to standard procedures (Sambrook et al., 1993). For metabolic labelling, the cells were grown in methionine-free medium (M9-based) to an OD of 0.9. IPTG (0.1 mM) and [35S]-methionine (200–300 mCi) were added and incubation ensued for 3 h at 37°C. The bacteria were then harvested and the recombinant proteins purified as usual.
Isolation of NEs. Turkey erythrocyte NEs were isolated as specified by Georgatos and Blobel (1987). After isolation, the membranes were washed sequentially with 2 M KCl, 50 mM Tris–HCl pH 7.5, 1 mM DTT, 1 mM PMSF and protease inhibitors (leupeptin, pepstatin, aprotinin and antipain at 2 µg/ml), distilled water, 8 M urea, 10 mM Tris–HCl pH 8.0, 4 mM EDTA and 1 mM PMSF (when specified), and distilled water. Before use, NEs were washed and resuspended in assay buffer (see below) using mild sonication.
Preparation of NE extracts. NEs were extracted with 1 M NaCl, 20 mM Tris–HCl pH 8.0, 1 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 1 mM PMSF and 1% Triton X-100 at 4°C. The suspension was ultracentrifuged and the clarified supernatant was adjusted at 500 mM NaCl and used in pull-down assays.
NE binding assays. All reactions were carried out in Eppendorf tubes coated with 1% boiled/filtered fish skin gelatin. Ten to fifty micrograms of NEs, or the exact equivalent of proteolyzed membranes, were combined with increasing amounts of GST proteins dissolved in assay buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.4, 2 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 0.2 mM PMSF, 10% sucrose and 0.1% gelatin) and adjusted in volume to 100 µl. After a 45-min incubation at room temperature (mixing by rotation), the samples were spun in a Microfuge (12 000 g, 30 min) and the pellets washed with 300 µl of assay buffer. Following another centrifugation (15 min, in the same fashion), the supernatants were carefully aspirated and the walls of the tubes wiped with cotton swabs. The final pellets, representing nuclear membranes and associated material, were either solubilized in Laemmli buffer, or dissolved in scintillation fluid. Binding was detected by SDS–PAGE/western blotting (unlabelled proteins), or by liquid scintillation counting (35S-labelled probes). All quantitative experiments were repeated at least three times in duplicate or triplicate.
Pull-down assays. GST fusion proteins (15–30 µg) were mixed with NE extracts (∼200 µg of total protein) or purified histones (15–30 µg) at 500 mM NaCl buffer (see above) and incubated for 1 h at room temperature. After addition of glutathione beads (30 µl packed beads) and another 1 h incubation, the sediments were washed five times with the same buffer and analysed by SDS–PAGE and/or western blotting.
In vitro acetylation assays. Purified histones (15 µg) were mixed with GST–CBP and incubated with or without acetyl CoA (5 mM) in assay buffer (see above) for 20 min at room temperature. This mixture was adjusted to 500 mM NaCl and used in pull-down assays.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Niki Kretsovali (IMBB, Crete, Greece) and Thomais Papamarcaki (University of Ioannina, Greece) for materials and feedback. This work was supported by PENED 99 from the Greek Secretariat of Research and Technology (S.D.G.), a BBSRC Core Strategic Grant (P.B.S.) and a Royal Society (UK) grant (P.B.S. and S.D.G.).
REFERENCES
- Aasland R. and Stewart, A.F. (1995) The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res., 23, 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Neiman, A.M., Zappulla, D.C. and Sternglanz, R. (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature, 394, 592–595. [DOI] [PubMed] [Google Scholar]
- Ball L.J. et al. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C. and Kouzarides, T. (2001) Selective recognition of methylated lysine 9 on histone H3 by HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Brasher S.V. et al. (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J., 19, 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. and Paro, R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. [DOI] [PubMed] [Google Scholar]
- Duband-Goulet I. and Courvalin, J.-C. (2000) Inner nuclear membrane protein LBR preferentially interacts with DNA secondary structures and nucleosomal linker. Biochemistry, 39, 6483–6488. [DOI] [PubMed] [Google Scholar]
- Georgatos S.D. and Blobel, G. (1987) Two distinct attachment sites for vimentin along the plasma membrane and the nuclear envelope in avian erythrocytes: a basis for a vectorial assembly of intermediate filaments. J. Cell Biol., 105, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.O., Cowell, I.G. and Singh, P.B. (2000) Mammalian chromodomain proteins: their role in genome organisation and expression. BioEssays, 22, 124–137. [DOI] [PubMed] [Google Scholar]
- Kourmouli N., Theodoropoulos, P.A., Dialynas, G., Bakou, A., Politou, A.S., Cowell, I.G., Singh, P.B. and Georgatos, S.D. (2000) Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J., 19, 6558–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N., Dialynas, G., Petraki, C., Pyrpasopoulou, A., Singh, P.B., Georgatos, S.D. and Theodoropoulos, P.A. (2001) Binding of Heterochromatin Protein 1 to the nuclear envelope is regulated by a soluble form of tubulin. J. Biol. Chem., 276, 13007–13014. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll, D., Rea, S., Mechtler, K. and Jenuwein, T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Meier J. and Georgatos, S.D. (1994) Type B lamins remain associated with the integral membrane protein p58 during mitosis: implications for nuclear reassembly. EMBO J., 13, 1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani, M., Ortiz, J.A., Remboutsika, E., Chambon, P. and Losson, R. (2001) Heterochromatin formation in mammalian cells: interactions between histones and HP1 proteins. Mol. Cell, 4, 729–739. [DOI] [PubMed] [Google Scholar]
- Nikolakaki E., Simos, G., Georgatos, S.D. and Giannakouros, T. (1996) A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J. Biol. Chem., 271, 8365–8372. [DOI] [PubMed] [Google Scholar]
- Paro R. and Hogness, D.S. (1991) The polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F. and Maniatis, T. (1993) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Singh P.B., Miller, J.R., Pearce, J., Kothary, R., Burton, R.D., Paro, R., James, T.C. and Gaunt, S.J. (1991) A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res., 19, 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., Yuan, G., Blobel, G. and Georgatos, S.D. (1988) A lamin B receptor at the nuclear envelope. Proc. Natl Acad. Sci. USA, 85, 8531–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q. and Worman, H.J. (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem., 271, 14653–14656. [DOI] [PubMed] [Google Scholar]
- Ye Q., Callebaut, I., Pezhman, A., Courvalin, J.C. and Worman, H.J. (1997) Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem., 272, 14983–14989. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk, T., Allis, C.D. and Eissenberg, J.C. (2000) Heterochromatin protein 1 (HP1) binds to nucleosomes and DNA in vitro. J. Biol. Chem., 275, 28332–28338. [DOI] [PubMed] [Google Scholar]