Abstract
Dynactin has been proposed to link the microtubule-associated motor cytoplasmic dynein with membranous cargo; however, the mechanism by which dynactin–membrane interaction is regulated is unknown. Here we show that dynein and dynactin exist in discrete cytosolic and membrane-bound states in the filamentous fungus Neurospora crassa. Results from in vitro membrane-binding studies show that dynein and dynactin–membrane interaction is co-dependent. p150Glued of dynactin has been shown to interact with dynein intermediate chain and dynactin Arp1 filament; however, it is not known to play a direct role in membrane binding. In this report we describe our analysis of 43 p150Glued mutants, and we show that C-terminal deletions which remove the terminal coiled-coil (CC2) and basic domain (BD) result in constitutive dynactin–membrane binding. In vitro addition of recombinant p150Glued CC2+BD protein blocks dynactin–membrane binding. We propose that the C-terminal domains of p150Glued regulate dynactin–membrane binding through a steric mechanism that controls accessibility of the Arp1 filament of dynactin to membranous cargo.
INTRODUCTION
Cytoplasmic dynein is a multisubunit, microtubule-associated force-producing enzyme required for a wide range of cellular processes (Allan and Schroer, 1999; Karki and Holzbaur, 1999). Dynein-dependent movement of cargo requires a number of steps such as cargo attachment, motor activation, translocation of cargo, motor inactivation, cargo release, and then possibly motor reactivation for return transport. The mechanisms by which these steps are regulated are not understood. Dynactin, an additional multisubunit complex, has been proposed to link dynein with membranous organelles, and activate motor and regulate its processivity (Schroer et al., 1996; Holleran et al., 1998; King and Schroer, 2000; Kumar et al., 2000). The most abundant subunit of dynactin is actin-related protein 1 (Arp1), which forms a short (37 nm) filament that has been proposed to interact with a spectrin-like cytoskeleton associated with membranous cargo.
The largest subunit of dynactin, p150Glued, has been shown to mediate dynein/dynactin interaction; however, it has not been shown to function directly in membrane binding. p150Glued is predicted to contain five major structural domains, with three globular regions alternating with two α-helical coiled-coil domains (Schroer, 1996). A microtubule-binding domain is located in the first globular domain, and interaction of p150Glued with dynein intermediate chain occurs through the central region of p150Glued (Vaughan and Vallee, 1995). Residues within the second coiled-coil (CC2) domain have been shown to bind to Arp1 (Waterman-Storer et al., 1995). The function of the C-terminal globular basic domain (BD) is unknown.
As with vertebrates, conventional kinesin and cytoplasmic dynein of filamentous fungi have been shown to be required for anterograde and retrograde transport of membranous cargo, respectively (Seiler et al., 1999). In contrast to vertebrates, these transport systems are not essential for viability, and this makes filamentous fungi excellent genetic models for the study of microtubule-dependent, long-range transport of membranous organelles. We have developed a screen that allows the isolation of hundreds of Neurospora crassa mutants (referred to as ‘ropy’) that are defective in dynein/dynactin function (Bruno et al., 1996). Here, we examine dynein– and dynactin–membrane interaction in wild-type N. crassa and specific p150Glued mutants. Our results suggest that the C-terminal domains of p150Glued regulate the interaction of membranous cargo with the Arp1 filament of dynactin.
RESULTS AND DISCUSSION
Dynein/dynactin exist in soluble and membrane-bound states
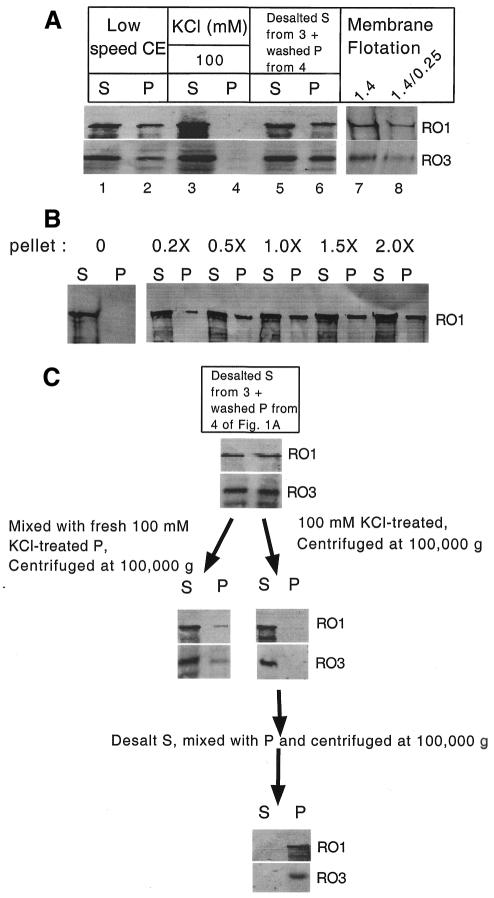
As a first step in the investigation of dynein and dynactin interaction with membranous organelles in N. crassa, we examined salt-dependent binding and release of dynein and dynactin from membrane by high-speed pelleting (100 000 g) and flotation in sucrose step gradients. Approximately 30% of dynein and dynactin was found in the P100 fraction (i.e. the 100 000 g pellet) following high-speed centrifugation of a low-speed extract (Figure 1A, lanes 1 and 2). Addition of KCl (100 mM) to cell extracts prior to high-speed centrifugation results in the release of all dynein and dynactin from P100 pellet (Figure 1A, lanes 3 and 4). When this S100 is desalted, mixed with a washed dynein/dynactin-free pellet, and then subjected to high-speed centrifugation, ∼40% of both dynein and dynactin was found in the P100 pellet (Figure 1A, lanes 5 and 6). The salt sensitivity of dynein and dynactin fractionation with the high-speed pellet suggests that these complexes are associated with membrane. To further analyze dynein/dynactin membrane association, we carried out a membrane flotation experiment by resuspending the P100 fraction in 2 M sucrose and overlaying with 1.4 and 0.25 M sucrose layers (Niclas et al., 1996). Results showed that ∼70% of dynein and dynactin floated upward into the 1.4 and 1.4/0.25 M interface layers, further supporting that dynein and dynactin contained within the P100 fraction represents complexes associated with membrane (Figure 1A, lanes 7 and 8). While the P100 fraction contains total cellular membranes, we recently showed that dynein/dynactin associate with a subset of membranes, suggesting that in vitro dynein/dynactin–membrane interaction is specific (Lee et al., 2001).
Fig. 1. Dynein and dynactin exist in two states: soluble and membrane bound. (A) Salt-dependent pelleting and flotation of dynein and dynactin with membrane from wild-type N. crassa. RO1 (dynein heavy chain) and RO3 (dynactin p150Glued) were detected by western analysis and are labeled on the right. Lanes 1 and 2 are supernatant and pellet, respectively, following 100 000 g centrifugation of a low-speed cell extract. Lanes 3 and 4 are supernatant and pellet, respectively, following 100 000 g centrifugation of 100 mM KCl-treated low-speed cell extract. Lanes 5 and 6 are supernatant and pellet, respectively, following 100 000 g centrifugation of recombined desalted supernatant and pellet from lanes 3 and 4. Lanes 7 and 8 are 1.4 M sucrose and 1.4/0.25 M sucrose interface layers following flotation of membrane pellet (resuspended in 2.0 M sucrose from lane 6; see Methods). (B) Membrane-dependent pelleting of dynein. S and P are supernatant and pellet, respectively, following 100 000 g centrifugation of recombined desalted supernatant with different amounts of salt-washed membrane pellet. (C) A schematic demonstrating the discrete nature of soluble and membrane-bound dynein and dynactin. The first two lanes are supernatant and pellet, respectively, followed by centrifugation at 100 000 g.
We consistently observed that 40–50% of dynein/dynactin was associated with membrane, suggesting that the soluble and membrane-bound pools represent discrete states. To test this hypothesis, we carried out a salt-dependent pelleting experiment in which different amounts of a washed dynein- and dynactin-free pellet were added to a salt-released, and then desalted dynein and dynactin S100 fraction (i.e. Figure 1A, lane 3). We observed that the dynein and dynactin pelleting is saturated to ∼40% even if double the amount of membrane pellet was added to the dynein and dynactin fraction (Figure 1B).
To further examine whether soluble and pelleted dynein/dynactin represent discrete states, we looked separately at the membrane-binding properties of each pool. We found that membrane-bound dynein and dynactin could be fully released by 100 mM KCl, and the released dynein and dynactin, upon desalting, were able to rebind efficiently with membrane (Figure 1C). In addition, >90% of the soluble dynein and dynactin remained in the supernatant upon mixing with washed pellet followed by centrifugation at 100 000 g (Figure 1C). These results indicate that dynein and dynactin exist in two discrete states: soluble and membrane bound. Both forms were found to have similar levels of dynein ATPase activity and identical sedimentation coefficients, making it unlikely that there is a gross structural difference between these pools or that one pool has been denatured (Kumar et al., 2000; data not shown).
Dynein and dynactin are co-dependent for membrane interaction
Dynein and dynactin do not generally co-immunoprecipitate with each other (Paschal et al., 1993), and it has been suggested that dynein and dynactin supercomplexes may represent the activated state of the motor that is capable of cargo interaction (Faulkner et al., 2000). To determine whether membrane association in N. crassa correlates with stabilized dynein–dynactin interaction, we immunoprecipitated dynein and dynactin from both soluble and membrane-bound pools. We found that dynein and dynactin do not show appreciable co-immunoprecipitation in the presence of 150 mM KCl (Figure 2A, lanes 2 and 3). However, in the absence of salt, dynein and dynactin efficiently co-immunoprecipitate regardless of whether they are from the soluble or membrane-bound pools (Figure 2A, lanes 4–7). These results suggest that the triggering event for membrane interaction is unlikely to be the stabilization of dynein and dynactin association.
Fig. 2. Dynein and dynactin are co-dependent for membrane interaction. (A) Salt-sensitive co-immunoprecipitation of dynein and dynactin from soluble and membrane-bound pools. RO1 (dynein heavy chain) and RO3 (dynactin p150Glued) were detected by western analysis and are labeled on the right. Lanes 1, 2 and 3 are immunoblots of immunoprecipitated proteins from the soluble (S100) fraction of wild type using pre-immune serum (pre-I), anti-RO1 and anti-RO3 antibodies, respectively, in the presence of 150 mM KCl. Lanes 4 and 5 are immunoprecipitations of dynein and dynactin from the desalted soluble pool using anti-RO1 and anti-RO3 antibodies, respectively. Lanes 6 and 7 are immunoprecipitations of dynein and dynactin from the salt-released membrane-bound (MB) pool using anti-RO1 and anti-RO3 antibodies, respectively. (B) Effects of dynein and dynactin mutations on salt-dependent pelleting and flotation of dynein and dynactin with membrane. Neurospora crassa strains used are labeled on the left. Lanes 1 and 2 are supernatant and pellet, respectively, following 100 000 g centrifugation of 100 mM KCl-treated low-speed cell extracts. Lanes 3 and 4 are supernatant and pellet, respectively, following 100 000 g centrifugation of desalted supernatants from lane 1 that were recombined with washed membrane pellet from lane 2. Lanes 5 and 6 are 1.4 M sucrose and 1.4/0.25 M sucrose interface layers following flotation of membrane pellet (lane 4 resuspended in 2.0 M sucrose).
Dynein alone has been shown to bind to membrane in vitro (Lacey and Haimo, 1994; Tai et al., 1999); therefore, we examined the dependency of dynein and dynactin interaction with membrane by using ro-1 (lacking dynein heavy chain) and Δro-3 mutants (lacking dynactin p150Glued). Consistent with dynactin’s proposed membrane-linker function, we find that dynein can not bind to membrane when dynactin is removed genetically (Figure 2B, lanes 3 and 4). Unexpectedly, we find that dynactin also can not bind to membrane when dynein is removed genetically (Figure 2B, lanes 3 and 4). We also mixed equal amounts of cell extracts from ro-1 and Δro-3 mutants followed by membrane-binding analysis. We found that in mixed extracts dynein and dynactin were capable of binding to membrane (Figure 2B, lanes 3 and 4). These data suggest that dynein– and dynactin–membrane interaction is co-dependent. Addition of a 3-fold excess of extract from either the ro-1 or the Δro-3 mutant did not result in increased membrane binding for the respective dynein or dynactin complexes (data not shown). The inability of excess dynactin to drive all dynein into the membrane pellet or excess dynein to drive all dynactin into the membrane pellet suggests that even in ro-1 and Δro-3 mutants the respective dynein and dynactin complexes exist in two discrete states: one that is capable of binding to membrane and one that is not.
C-terminal domains of p150Glued are involved in dynactin–membrane interaction
Previously it had been proposed that membranous organelles bind directly to dynactin by way of an organelle-associated spectrin binding to the dynactin Arp1 filament (Holleran et al., 1996; Muresan et al., 2001). A region within the CC2 domain of p150Glued has been shown to interact with Arp1 filament (Waterman-Storer et al., 1995); however, it is not known whether this domain is required for assembly of the dynactin complex or regulation of Arp1 filament–membrane interaction. To explore a possible role of p150Glued in dynactin–membrane interaction, we examined dynein– and dynactin–membrane binding in a large collection of ro-3 mutants. To identify ro-3 mutants that produce polypeptides which are defective in dynactin function, and not production of RO3 protein, we screened 43 independently isolated ro-3 mutants for the presence of RO3. We identified 10 ro-3 mutants that produced RO3 polypeptides which ranged in size from 90 to 145 kDa. The sites of the respective ro-3 mutations are presented in Figure 3A. Nine of the ro-3 mutations were nonsense, frameshift or deletion mutations that resulted in the production of truncated polypeptides. Only one of the mutants contained a missense mutation (near the N-terminal microtubule-binding domain). Interestingly, of the nine mutants producing truncated polypeptides, five remove residues contained within the CC2 domain that have been shown to interact with Arp1. In each of these mutants, the sedimentation value of dynactin was not altered. In addition, the truncated RO3 proteins co-immunoprecipitated with the dynactin subunits RO2 (p62), RO4 (Arp1) and RO7 (Arp11) (Figure 3B). These results indicate that the Arp1-binding site in CC2 is not required for incorporation of RO3 into the dynactin complex.
Fig. 3. C-terminal domains of p150Glued are involved in dynactin–membrane interaction. (A) Sites of mutations present in 10 independent alleles of ro-3. With the exception of the a7.3 missense mutation, the asterisks represent the sites of individual mutations that either introduce a nonsense or frameshift mutation, which results in the production of truncated RO3 protein. Mutant A25.3 possesses an internal deletion in the ro-3 gene. (B) Co-immunoprecipitation of dynactin subunits from wild type and C-terminal deletion mutants of ro-3. Proteins immunoblotted are labeled on the right of the blot, and cell extracts and anti-protein antibodies are labeled on top of the blot. (C) Salt-dependent pelleting of dynein and dynactin from wild type and C-terminal deletion mutants of ro-3. The relative lengths of RO3 (p150Glued) polypeptides from wild type and the C-terminal deletion mutants A8.2 and a1.8 are presented on the left. The proteins detected by western analysis are labeled on the right. The first two lanes represent supernatant (S) and pellet (P) fractions following centrifugation at 100 000 g of low-speed extracts in the presence of 100 mM KCl. The last four lanes are where the respective supernatants from the first lane were desalted, mixed with either 1× or 2× washed membrane pellet, and then centrifuged at 100 000 g. (D) Salt-dependent pelleting of membrane-bound dynein and dynactin from wild type and the A8.2 mutant in the absence and presence of recombinant p150Glued C-terminal domains. The methodology of the experiment is presented schematically on the left. Membrane-bound (MB) dynein and dynactin from wild type and the A8.2 mutant lacking p150Glued CC2+BD are labeled on the left. Membrane-bound dynein and dynactin were first released by 100 mM KCl, desalted by gel filtration, and then incubated with either 100 µg of CC2 or CC2+BD for 20 min followed by incubation with washed membrane pellet for 1 h and centrifugation at 100 000 g. Control represents the addition of 100 µg of bovine serum albumin in place of the recombinant proteins.
To determine whether the mutations removing the RO3 (p150Glued) CC2 and BD or only the BD affect dynactin–membrane interaction, we conducted salt-dependent pelleting and flotation experiments. The results from two representative mutants, A8.2 and a1.8, which remove CC2+BD or the BD alone, respectively, are presented (Figure 3C). In the A8.2 mutant, nearly all the dynactin binds to membrane in a salt-sensitive fashion, while in the a1.8 mutant there is only a slight increase in the proportion of dynactin associated with membrane relative to the wild-type control. However, if double the amount of membrane pellet is used in the pelleting assay of the a1.8 mutant, almost all the dynactin is present in the membrane fraction. As before, increasing the amount of membrane pellet used in the assay of wild type does not affect the amount of dynein and dynactin present in the membrane-bound state (Figures 1B and 3C). The results indicate that the removal of CC2+BD results in constitutive dynactin–membrane binding, while removal of only the BD domain results in a dynactin complex that can be completely driven into the membrane-bound state by the addition of excess membrane fraction. Interestingly, dynein–membrane interaction is relatively unaffected by these ro-3 mutations. This result is consistent with our extract mixing experiments (described above), which show that in the absence of dynactin, dynein still exists in two discrete states: one that is capable of binding to membrane and one that is not.
The results suggest that p150Glued CC2 and BD may regulate dynactin–membrane interaction by sterically affecting the accessibility of the Arp1 filament to membranous cargo. To test this hypothesis, we expressed in Escherichia coli His-tagged p150Glued recombinant polypeptides containing either CC2+BD or only CC2 and then determined whether these polypeptides could block dynactin–membrane interaction in vitro. As shown in Figure 3D, the addition of the recombinant CC2+BD protein to salt-released dynactin from wild type or the A8.2 mutant was able to effectively block the re-association of dynactin with membrane. The His-tagged polypeptide containing only the CC2 domain had only a slight effect on dynactin–membrane rebinding. To determine whether these recombinant proteins are likely to affect dynactin–membrane interaction through a direct physical interaction with dynactin, we examined whether a column containing the His-tagged polypeptides could retain dynactin from extracts of wild type and the A8.2 mutant. Our results showed that columns containing either CC2+BD or CC2 bound dynactin from wild type and the A8.2 mutant (data not shown). Our results suggest that the C-terminal domains of p150Glued function in regulating dynactin–membrane interaction through controlling the accessibility of the Arp1 filament to membranous cargo (Figure 4).
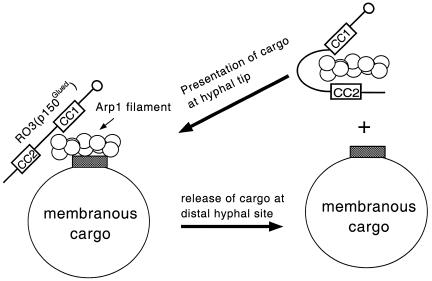
Fig. 4. Model for the regulation of dynactin–membrane interaction in N. crassa. For simplicity, only the dynactin Arp1 filament and a single p150Glued subunit are shown. The C-terminal CC2+BD of p150Glued interacts with Arp1 filament and prevents dynactin–membrane binding (i.e. dynactin is in the soluble state). Upon interaction with membranous cargo, the C-terminal domains of p150Glued release from the Arp1 filament to allow cargo to bind to the Arp1 filament. Following translocation to the cargo release site, the membranous cargo dissociates from the Arp1 filament, the C-terminal domains of p150Glued reassociate with the Arp1 filament, and dynein and dynactin return to the hyphal tip to initiate another round of transport.
We propose that soluble and membrane-bound states of dynactin result directly from two different conformational states of the C-terminal domains of p150Glued (Figure 4). In the soluble state, the CC2+BD domains of p150Glued bind to the Arp1 filament and sterically block its interaction with membranous cargoes. (Our calculation of the length of CC2+BD is ∼30 nm, sufficient to cover the 37 nm Arp1 filament and thereby prevent interaction with membranous cargo.) In the membrane-bound state, these domains fold-back (i.e. either bind to N-terminal segments of p150Glued or the p24/p50 dynamitin subunits contained within the shoulder/sidearm subcomplex of dynactin) and allow Arp1–membrane interaction. Alternatively, in both states the CC2 domain of p150Glued may remain associated with the Arp1 filament and regulate Arp1 filament–membrane interaction in a manner similar to tropomyosin-based regulation of myosin–actin interaction in muscle tissue (Huxley, 1972). A further understanding of dynactin–membrane interaction will require identification of the switch that regulates the conformation state of the C-terminal domains of p150Glued.
METHODS
Strains, growth conditions and genetic techniques. The N. crassa wild-type (74-OR23-1A; FGSC 987) and ro-1 (B15) (FGSC 4352) strains were obtained from the Fungal Genetic Stock Center (FGSC), Department of Microbiology, University of Kansas Medical Center, Kansas City, KS. Strains deleted for the ro-3 gene have been described (Tinsley et al., 1996). Media, growth conditions and sexual crosses were as described (Davis and de Serres, 1970).
Sequence analysis of ro-3 mutants. Genomic DNA from ro-3 mutants was isolated using the Dneasy Plant Mini Kit (QIAGEN Inc., Santa Clarita, CA). The ro-3 gene was amplified from the mutants by PCR using pfu Turbo DNA polymerase from Stratagene (La Jolla, CA). The entire DNA sequence of the ro-3 gene was determined for each mutant ro-3 allele.
Expression of RO3 recombinant proteins. DNA fragments encoding ro-3 CC2+BD and CC2 domains were amplified by PCR using a wild-type copy of the ro-3 gene. The PCR-generated fragments were inserted into a modified pQE41 vector to produce RO3-His6-tagged constructs (Qiagen). The DNA sequence of both constructs was determined to ensure maintenance of the proper reading frame and the absence of any mutations. The constructs were transformed into E. coli, high-level expression was induced, and recombinant His-tagged CC2+BD and CC2 proteins were purified using the QIA Expression Kit (Qiagen).
Immunoprecipitation and immunoblotting. Anti-RO1, anti-RO2, anti-RO3, anti-RO4 and anti-RO7 antibodies were produced as described (Minke et al., 1999, 2000; Lee et al., 2001). Immunoprecipitation was performed as described (Beckwith et al., 1998; Kumar et al., 2000). For immunoblotting, proteins resolved by SDS–PAGE were electroblotted onto nitrocellulose membrane (Schleicher & Schuell, Keene, NH) and then probed with anti-RO1 and anti-RO3 antibodies at 1:1000 dilution followed by goat anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase at 1:15 000 dilution (Promega, Madison, WI). Western blot processing was performed as described (Promega).
Dynein/dynactin–membrane binding assay. Frozen mycelia (0.5 g each) were suspended in 1.5 ml of extraction buffer (EB; 50 mM PIPES pH 7.0, 50 mM HEPES, 2 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol and protease inhibitors: 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 10 µg/ml TAME, 1 µg/ml pepstatin A and 10 µg/ml soybean trypsin inhibitor) and the hyphae were ground with zirconium beads using a mortar and pestle. Debris were removed by centrifugation at 5000 g for 10 min. Extracts were cleared by centrifugation at 28 000 g for 10 min. Supernatant (0.5 ml) was incubated with 100 or 200 mM KCl (to remove loosely bound membrane-associated proteins) for 60 min, overlaid over a 7.5% sucrose cushion (0.2 ml in EB), and then centrifuged at 100 000 g in a Beckman Ti 100.3 rotor for 45 min to pellet down membranous organelles and membrane-associated proteins. The supernatant was desalted using Sephadex gel filtration column NAP (Amersham Pharmacia Biotech, Piscataway, NJ). The pellet was washed in 0.3 ml of 7.5% sucrose in EB and centrifuged at 100 000 g for 45 min. The pellet was then resuspended with desalted supernatant (0.5 ml), incubated for 60 min, overlaid over a 7.5% sucrose cushion (0.2 ml in EB), and recentrifuged at 100 000 g for 45 min. The pellet obtained from high-speed centrifugation was resuspended to the same volume as supernatant. Samples (60 µl) from each supernatant and pellet were analyzed by SDS–PAGE followed by western blotting. Membrane flotation experiments were performed as described (Niclas et al., 1996).
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Seiler for providing helpful comments. This work was supported by grant GM51217 from the National Institutes of Health.
REFERENCES
- Allan V.J. and Schroer, T.A. (1999) Membrane motors. Curr. Opin. Cell Biol. 11, 476–482. [DOI] [PubMed] [Google Scholar]
- Beckwith S.M., Roghi, C.H., Liu, B. and Morris, N.R. (1998) The 8-kD cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J. Cell Biol., 143, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno K.S., Tinsley, J.H., Minke, P.F. and Plamann, M. (1996) Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc. Natl Acad. Sci. USA, 93, 4775–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.H. and de Serres, F.J. (1970) Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol., 27A, 79–143. [Google Scholar]
- Faulkner N.E., Dujardin, D.L., Tai, C.Y., Vaughan, K.T., O’Connell, C.B. and Vallee, R.B. (2000) A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nature Cell Biol., 2, 784–791. [DOI] [PubMed] [Google Scholar]
- Holleran E.A., Tokio, M.K., Karki, S. and Holzbaur, E.L.F. (1996) Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J. Cell Biol., 135, 1815–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran E.A., Karki, S. and Holzbaur, E.L.F. (1998) The role of the dynactin complex in intracellular motility. Int. Rev. Cytol., 182, 69–109. [DOI] [PubMed] [Google Scholar]
- Huxley H.E. (1972) Structural changes in the actin- and myosin-containing filaments during contraction. Cold Spring Harb. Symp. Quant. Biol., 37, 361–376. [Google Scholar]
- Karki S. and Holzbaur, E.L.F. (1999) Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol., 11, 45–53. [DOI] [PubMed] [Google Scholar]
- King S.J. and Schroer, T.A. (2000) Dynactin increases the processivity of the cytoplasmic dynein motor. Nature Cell Biol., 2, 20–24. [DOI] [PubMed] [Google Scholar]
- Kumar S., Lee, I.H. and Plamann, M. (2000) Cytoplasmic dynein ATPase is regulated by dynactin-dependent phosphorylation. J. Biol. Chem., 275, 31798–31804. [DOI] [PubMed] [Google Scholar]
- Lacey M.L. and Haimo, L.T. (1994) Cytoplasmic dynein binds to phospholipid vesicles. Cell Motil. Cytoskeleton, 28, 205–212. [DOI] [PubMed] [Google Scholar]
- Lee I.H., Kumar, S. and Plamann, M. (2001) Null mutants of the Neurospora Arp1 pointed-end complex show distinct phenotypes. Mol. Biol. Cell, 12, 2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke, P.F., Lee, I.H., Tinsley, J.H., Bruno, K.S. and Plamann, M. (1999) Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol., 32, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Minke P.F., Lee, I.H., Tinsley, J.T. and Plamann, M. (2000) A Neurospora crassa Arp1 mutation affecting cytoplasmic dynein and dynactin localization. Mol. Gen. Genet., 264, 433–440. [DOI] [PubMed] [Google Scholar]
- Muresan V., Stankewich, M.C., Steffen, W., Morrow, J.S., Holzbaur, E.L.F. and Schnapp, B.J. (2001) Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins: a role for spectrin and acidic phospholipids. Mol. Cell, 7, 173–183. [DOI] [PubMed] [Google Scholar]
- Niclas J., Allan, V.J. and Vale, R.D. (1996) Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J. Cell Biol., 133, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M., Holzbaur, E.L.F., Pfister, K.K., Clark, S., Meyer, D.I. and Vallee, R.B. (1993) Characterization of a 50-kDa polypeptide in cytoplasmic dynein preparations reveals a complex with p150Glued and a novel actin. J. Biol. Chem., 268, 15318–15323. [PubMed] [Google Scholar]
- Schroer T.A. (1996) Structure and function of dynactin. Semin. Cell Biol., 7, 321–328. [Google Scholar]
- Schroer T.A., Bingham, J.B. and Gill, S.T. (1996) Actin-related protein 1 and cytoplasmic dynein-based motility—what’s the connection? Trends Cell Biol., 6, 212–215. [DOI] [PubMed] [Google Scholar]
- Seiler S., Plamann, M. and Schliwa, M. (1999) Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol., 9, 779–785. [DOI] [PubMed] [Google Scholar]
- Tai A.W., Chuang, J.Z., Bode, C., Wolfrum, U. and Sung, C.H. (1999) Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell, 97, 877–887. [DOI] [PubMed] [Google Scholar]
- Tinsley J.H., Minke, P.F., Bruno, K.S. and Plamann, M. (1996) p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol. Biol. Cell, 7, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K.T. and Vallee, R.B. (1995) Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol., 131, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Karki, S. and Holzbaur, E.L.F. (1995) The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp1). Proc. Natl Acad. Sci. USA, 92, 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]