Abstract
Rad23 is a DNA repair protein that promotes the assembly of the nucleotide excision repair complex. Rad23 can interact with the 26S proteasome through an N-terminal ubiquitin-like domain, and inhibits the assembly of substrate-linked multi-ubiquitin (multi-Ub) chains in vitro and in vivo. Significantly, Rad23 can bind a proteolytic substrate that is conjugated to a few ubiquitin (Ub) moieties. We report here that two ubiquitin-associated (UBA) domains in Rad23 form non-covalent interactions with Ub. A mutant that lacked either UBA sequence was capable of blocking the assembly of substrate-linked multi-Ub chains, although a mutant that lacked both UBA domains was significantly impaired. These studies suggest that the interaction with Ub is required for Rad23 activity, and that other UBA-containing proteins may have a similar function.
INTRODUCTION
Nucleotide excision repair (NER) involves the sequential assembly of proteins at the site of DNA lesions (Guzder et al., 1996). The yeast Rad23 protein plays an important role in recruiting Rad14 and TFIIH to damaged DNA (Guzder et al., 1995a). Yeast and human Rad23 proteins interact with Rad4 (XPC in humans) (Masutani et al., 1994; Guzder et al., 1995b), and the complex preferentially binds damaged DNA (Guzder et al., 1998; Jansen et al., 1998). The loss of most NER genes causes a severe decrease in survival (Prakash et al., 1993; de Laat et al., 1999). In contrast, deletion of yeast RAD23 results in intermediate sensitivity to UV light (Watkins et al., 1993), suggesting that it plays a regulatory role. Rad23 proteins contain N-terminal ubiquitin-like (UbL) domains (Watkins et al., 1993), which bind the 26S proteasome (Schauber et al., 1998). However, the significance of Rad23/proteasome interaction is poorly understood.
We reported that Rad23 inhibited the assembly of substrate-linked multi-ubiquitin (multi-Ub) chains (Ortolan et al., 2000), and proposed that it could control protein stability by binding and preventing the expansion of a nascent multi-Ub chain. In agreement with this conjecture, we found that Rad23 could be purified in a complex with a proteolytic substrate that was already ligated to one or two ubiquitin moieties (Ortolan et al., 2000). Rad23 does not possess Ub-specific isopeptidase activity because multi-Ub chains that were pre-assembled on histone H2B were unaffected by Rad23. Although the mechanism of inhibition of multi-Ub chain formation was not determined, we found that Rad23 did not prevent E1 and E2 enzymes from forming thioester intermediates with Ub and, furthermore, did not bind and inactivate the E2 protein (Ubc2/Rad6).
Ubiquitin-associated (UBA) domains are present in many unrelated proteins, and were proposed to bind ubiquitin (Hofman and Bucher, 1996). Rad23 contains two UBA domains (van der Spek et al., 1996) whose role in DNA repair, cell cycle progression and proteolysis has not been well characterized. The N-terminal UBA domain (UBA1) has no known effectors. In contrast, the C-terminal UBA domain (UBA2) plays an overlapping genetic role with Ddi1, another UBA-containing protein that regulates a DNA replication-specific checkpoint (Clarke et al., 2001). UBA2 also interacts with Vpr (an HIV-1-encoded accessory protein; Withers-Ward et al., 1997), MPG protein (a 3-methyladenine DNA glycosylase; Miao et al., 2000) and possibly with Png1 (a deglycosylating enzyme; Suzuki et al., 2001), as well as the transcription regulator p300/CBP (Zhu et al., 2001). However, none of these interactions has revealed the biochemical function of the UBA sequence, or its effect on Rad23 function. We report here that both UBA domains in Rad23 form non-covalent interactions with Ub. Significantly, a Rad23 mutant that lacked both UBA domains was unable to block the assembly of substrate-linked multi-Ub chains efficiently. Bertolaet et al. (2001) have also reported recently that the UBA domains in both Rad23 and Ddi1 can bind Ub, indicating that the interaction with Ub may be an evolutionarily conserved property of UBA domains. Although free Ub is unlikely to represent the physiological target of Rad23, these results are consistent with the ability of Rad23 to bind a substrate that is ligated to a short multi-Ub chain in vivo (Ortolan et al., 2000). Taken together, our studies suggest that the interaction between Rad23 and Ub will be important for its biochemical activities (Ortolan et al., 2000) in DNA repair (Guzder et al., 1995a), cell cycle progression (Lambertson et al., 1999; Clarke et al., 2001) and the stress response (Lambertson et al., 1999).
RESULTS
Sequence alignment revealed significant similarity between UBA1 and a UBA sequence in certain Ub-isopeptidases (Figure 1). Because Ub-isopeptidases target polymeric Ub, and Rad23 binds ubiquitylated substrates (Ortolan et al., 2000), we investigated whether Rad23 could bind Ub.
Fig. 1. Sequence similarity between UBA domains in Rad23 and certain Ub-isopeptidases (identical residues are indicated in bold). The numbers on the right indicate the amino acid residue at the C-terminus of the conserved UBA sequence in each respective protein. The upper two lines represent the sequence of the two UBA domains in yeast Rad23 protein. Only UBA1 sequences from human and mouse Rad23 are shown.
To examine Rad23/Ub interaction, we developed a filtration assay using Centricon-30, which can retain proteins larger than ∼30 000 mol. wt. We prepared a ubiquitylation reaction containing E1, E2, [32P]Ub and histone H2B (Ortolan et al., 2000), and following incubation at 30°C for 60 min the reaction was applied to a Centricon-30 filter to remove free [32P]Ub (8500 mol. wt), E2 (19 700 mol. wt), unconjugated H2B (14 000 mol. wt) and ATP. The retentate was separated in an SDS-containing 12% polyacrylamide gel and exposed to X-ray film and, as expected, free [32P]Ub was entirely removed following centrifugation (data not shown). However, high levels of [32P]Ub were retained if the reactions contained Rad23 (Figure 2). To ensure that [32P]Ub was not bound non-specifically to components in the reaction, we determined the effect of an unrelated protein, bovine serum albumin (BSA). While the addition of increasing amounts of Rad23 (Figure 2A, lanes 1–3) led to the recovery of higher levels of [32P]Ub (Figure 2B, lanes 1–3), high amounts of BSA did not cause any retention of Ub (Figure 2B, lanes 4–6), indicating that Rad23 specifically caused the effect.
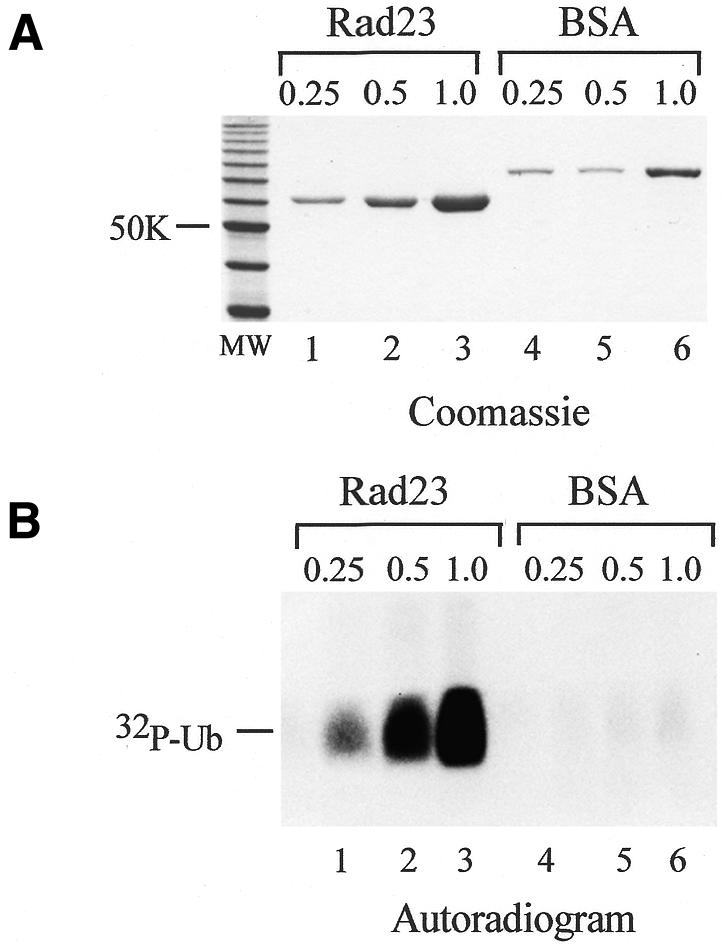
Fig. 2. Rad23 prevents the passage of Ub (∼8500 mol. wt) through a 30 000 mol. wt cut-off filter. (A) A Coomassie Blue stained gel showing the amounts of Rad23 and BSA (µg) that were used in a Ub-binding assay using Centricon-30. (B) Rad23 and BSA were added to ubiquitylation reactions that contained [32P]Ub, and subjected to ultrafiltration in Centricon-30 (autoradiogram). The retentate was separated on a 12% polyacrylamide gel and exposed to X-ray film. The retention of [32P]Ub increased in the presence of higher levels of Rad23 (lanes 1–3), while only a trace amount of [32P]Ub was recovered in reactions that contained BSA (lanes 4–6).
We resolved Rad23 and Ub, separately and combined, in a Superdex-200/PC-3.2/30 analytical gel-filtration column to address the concern that Rad23 might have caused aggregation of Ub, thus preventing its passage through the filter. Fractions were collected, precipitated with 10% trichloroacetic acid and separated by SDS–PAGE. Immunoblotting revealed that the major fraction of Ub migrated as a monomeric protein, demonstrating that it was not aggregated (data not shown). However, a low level of Ub and higher amounts of di-ubiquitin (di-Ub) were detected in fractions that contained Rad23, indicating a direct interaction. It is possible that larger quantities of Ub were not detected in the Rad23-containing fractions because dilution of the protein sample in the column might have caused dissociation.
Rad23 contains an N-terminal ubiquitin-like (UbLR23) domain, which interacts with the 26S proteasome (Schauber et al., 1998). However, UbLR23 is not required for inhibiting the assembly of substrate-linked multi-Ub chains, or for interacting with a ubiquitylated substrate in vivo (Ortolan et al., 2000). Consistent with these results, we determined that Rad23 lacking UbLR23 could still retain [32P]Ub in Centricon-30 (data not shown). Although UbLR23 is required for complete resistance to DNA damage and stress (Lambertson et al., 1999), our results support the hypothesis that it functions primarily as a proteasome-targeting signal.
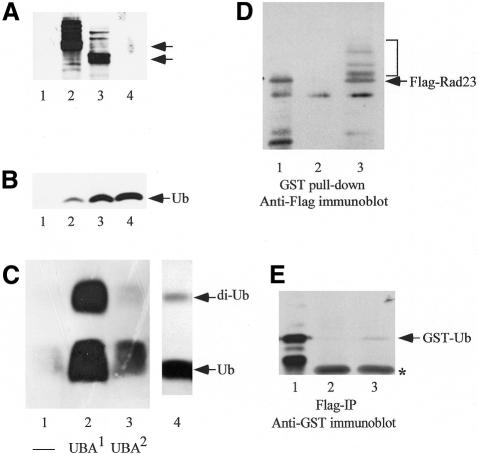
To examine directly the requirement of UbLR23 in Rad23/Ub binding, we purified GST, GST–Rad23 and GST–ΔUbLrad23 (Ortolan et al., 2000) from yeast cells (Figure 3A, lanes 1–3), and incubated the proteins with Ub. We determined that both GST–Rad23 and GST–ΔUbLrad23 could bind Ub (Figure 3B, lanes 2 and 3). We then examined the interaction between Ub and GST–UBA1 and GST–UBA2 fusion proteins that were purified from Escherichia coli. A low level of non-specific interaction between Ub and GST was observed (Figure 3C, lane 1). However, a significant interaction was detected with GST–UBA1 (Figure 3C, lane 2), and weaker binding was noted with GST–UBA2 (Figure 3C, lane 3). The commercial preparation of Ub contained detectable levels of di-Ub (indicated in lane 4), and both Ub and di-Ub bound the UBA domains. Since equal amounts of the GST fusion proteins were incubated with Ub, our findings indicate that UBA1 may form a stronger interaction with Ub than UBA2. In agreement with our binding studies, a previous report showed that loss of UBA1 reduced Rad23/Ub interaction to a greater degree than loss of UBA2 function (Bertolaet et al., 2001).
Fig. 3. Direct interaction between Rad23 and Ub. (A) GST, GST–Rad23 and GST–ΔUbLrad23 were purified to homogeneity from yeast, and equal amounts of protein (0.1 µg) were adsorbed to glutathione–Sepharose (lanes 1–3). The adsorbed proteins were incubated with unlabelled Ub (2.5 µg), and the affinity beads were washed and suspended in SDS–electrophoresis buffer. Proteins were separated by SDS–PAGE, transferred to nitrocellulose, and the upper half of the filter was incubated with anti-GST antibodies. Arrows to the right indicate the positions of GST–Rad23 (∼85 000 mol. wt) and GST–ΔUbLrad23 (∼75 000 mol. wt). The higher molecular weight species in lanes 2 and 3 are believed to be ubiquitin-conjugated derivatives of the GST fusion proteins. GST (∼26 000 mol. wt) is not visible. (B) The lower half of the nitrocellulose filter from (A) was incubated with antibodies against Ub. We detected an interaction between Ub and both GST–Rad23 and GST–ΔUbLrad23 (lanes 2 and 3), but not with GST (lane 1). Lane 4 contained 25 ng of purified Ub. (C) The UBA domains in Rad23 were expressed as fusions to GST (GST–UBA1 and GST–UBA2) and purified from E. coli. Equal amounts of the purified proteins were adsorbed to glutathione–Sepharose and incubated in binding buffer containing unlabelled Ub. A control reaction contained only GST protein (lane 1). The affinity beads were washed in buffer containing 0.5% Triton X-100 and the bound proteins examined in an immunoblot. We found that GST–UBA1 formed a strong interaction with Ub and di-Ub (lane 2), while GST–UBA2 formed a weaker interaction. Lane 4 contains a sample of the commercial Ub used in this study, and the positions of Ub and di-Ub are indicated. (D) Extracts that were prepared from a yeast strain that expressed Flag-Rad23 and GST–Ub were applied to glutathione–Sepharose beads. Following incubation at 4°C, the beads were washed extensively in buffer containing 0.5% Triton X-100, suspended in SDS-containing gel-loading buffer and resolved by SDS–PAGE. The separated proteins were transferred to a nitrocellulose filter and incubated with antibodies against the Flag epitope. Lane 1 contains an aliquot of the cell extract, and the arrow on the right indicates the position of Flag-Rad23 (∼60 000 mol. wt). Flag-Rad23 was precipitated only from extracts that contained GST–Ub (lane 3), but not if the strain did not express GST–Ub (lane 2). The bracket to the right indicates the presence of high molecular weight derivatives of Flag-Rad23, which could represent conjugation to Ub (also see A). (E) In a reciprocal experiment, we incubated yeast cell extracts with antibodies against the Flag epitope and examined the precipitated proteins for the presence of GST–Ub. Lane 1 shows the position of GST–Ub in total extracts. A low amount of GST–Ub (∼35 000 mol. wt) was non-specifically precipitated from an extract that lacked Flag-Rad23 (lane 2), although higher amounts were detected if the extract contained Flag-Rad23. An asterisk indicates an antibody cross-reaction against the IgG light chain (∼22 000 mol. wt).
To obtain evidence for interaction between Rad23 and Ub in vivo, we expressed Flag-tagged Rad23 and GST–Ub in yeast cells. The C-terminal glycine residue of Ub (in GST–Ub) was changed to alanine to prevent its conjugation to cellular proteins. Yeast cell extracts were incubated with either glutathione–Sepharose, or anti-Flag antibodies, and the precipitated proteins were separated on an SDS–10% polyacrylamide gel. The immunoblots were incubated with antibodies against Flag (Figure 3D) or GST (Figure 3E). Flag-Rad23 was recovered on glutathione Sepharose only when it was co-expressed with GST–Ub (Figure 3D, lane 3), and not from extracts that did not contain GST–Ub (Figure 3D, lane 2). Previous studies reported that a small fraction of yeast and human Rad23 proteins are conjugated to ubiquitin in vivo (Watkins et al., 1993; Kumar et al., 1999). Interestingly, high molecular weight derivatives of Flag-Rad23 were precipitated with GST–Ub, although they were undetectable in total cell extracts (Figure 3D, lane 1). An immunoblot containing proteins that were precipitated with Flag antibodies was incubated with anti-GST serum, and GST–Ub was detected in extracts that contained Flag-Rad23 (Figure 3E, lane 3). In a control reaction, extracts that did not contain Flag-Rad23 showed only a low level of non-specific precipitation of GST–Ub (Figure 3E, lane 2). The positions of Flag-Rad23 (Figure 3D, lane 1) and GST–Ub (Figure 3E, lane 1) in total extracts are shown.
A specific amino acid motif that is present in certain Ub-isopeptidases is highly conserved in UBA1 of Rad23 proteins (Figure 1). However, this sequence is not readily identifiable in UBA2 of yeast, mouse and human Rad23 proteins (van der Spek et al., 1996), suggesting that UBA sequences may be functionally distinct. We purified Rad23 mutants that lacked the UBA domains to examine the role of each UBA domain in inhibiting the assembly of substrate-linked multi-Ub chains. The ∼40 amino acid residues that encompass UBA2 in human and yeast Rad23 form a stable and independently folded domain (Dieckmann et al., 1998). We used this structural information to generate a set of Rad23 mutants that lacked UBA1 (rad23ΔUBA1), UBA2 (rad23ΔUBA2), and both UBA1 and UBA2 (rad23ΔUBA1,2). We added the mutant proteins to a ubiquitylation reaction and found that the single mutants could efficiently inhibit multi-Ub chain formation (Figure 4B, lanes 3 and 4). However, the double mutant rad23ΔUBA1,2 was significantly less efficient at preventing the assembly of multi-Ub chains, as observed by the formation of highly ubiquitylated histone H2B (Figure 4B, lane 5). In a control reaction, recombinant Rad23 was added to a ubiquitylation reaction and inhibition of multi-Ub chain assembly was observed (Figure 4A, lane 2). As we reported previously, Rad23 is itself conjugated to Ub in these reactions (see arrow), although it is not known whether the Ub is ligated to the same lysine residue in vitro and in vivo. It is also unclear whether the conjugated Ub serves as a template for multi-Ub chain formation, or whether it affects the biochemical activities of Rad23. Interestingly, all three Rad23 mutants also became conjugated to Ub (arrows in Figure 4B). These results demonstrate that both UBA domains contribute to Rad23 function, and suggest that they are either redundant, or cooperate in binding Ub. However, it is possible that the loss of single UBA domains only causes minor physiological defects in NER, cell cycle control and stress response, because the deficiency can be compensated by the second UBA domain, or by other UBA-containing proteins (Clarke et al., 2001). While these studies would predict that Rad23 lacking both UBA sequences should not bind Ub, preliminary studies revealed a low-level interaction between rad23ΔUBA1,2 and Ub. Since wild-type and mutant Rad23 proteins (lacking UBA domains) became conjugated to Ub in a ubiquitylation reaction (Figure 4), we speculate that this modification might also promote an interaction with Ub in vivo.
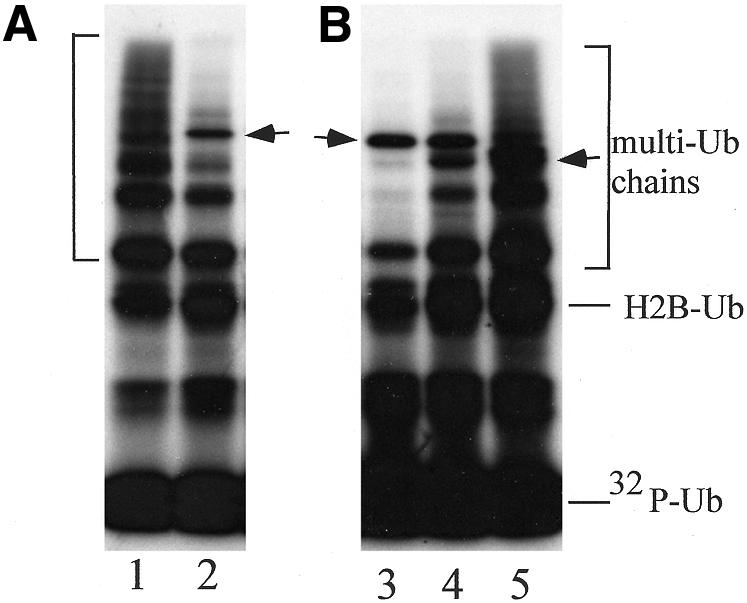
Fig. 4. The UBA domains in Rad23 are required for efficient inhibition of multi-Ub chain assembly. (A) Substrate-linked multi-Ub chains were assembled efficiently in a ubiquitylation reaction that lacked Rad23 protein (see bracket adjacent to lane 1). However, in the presence of Rad23 the assembly of multi-Ub chains on H2B was significantly reduced (lane 2). As we reported previously, Rad23 itself becomes mono-ubiquitylated in these reactions (arrow adjacent to lane 2). (B) Lanes 3–5 represent ubiquitylation reactions that contained rad23ΔUBA1, rad23ΔUBA2 and rad23ΔUBA1,2. Rad23 single mutants, lacking either UBA1 (lane 3) or UBA2 (lane 4), could efficiently inhibit the formation of multi-Ub chains. In contrast, a Rad23 mutant that lacked both UBA domains (lane 5) was a poor inhibitor of substrate-linked multi-Ub chain assembly (see bracket adjacent to lane 5). Similar to the mono-ubiquitylation of Rad23, all three UBA mutants (lanes 3–5) were also ligated to a single [32P]Ub in these reactions (arrows adjacent to lanes 3 and 5). (A) and (B) represent the same experiment and gel, but the exposure of (B) was 4-fold longer to illustrate the defect in multi-Ub chain inhibition by rad23ΔUBA1,2.
Rad23 binds Rad4 and promotes the assembly of the nucleotide excision–repair complex on damaged DNA (Guzder et al., 1995a; Jansen et al., 1998), and can stimulate DNA incision activity (Sugasawa et al., 1996). Rad23 can also bind the 26S proteasome and has an overlapping genetic role with Rpn10 (Lambertson et al., 1999), a proteasome subunit that binds multi-ubiquitylated substrates (van Nocker et al., 1996). We reported that Rad23 could inhibit the assembly of substrate-linked multi-Ub chains, and interacted with a ubiquitylated protein (Ortolan et al., 2000). Collectively, these studies provide compelling support for the idea that the various functions of Rad23 in DNA repair, cell cycle progression and stress response involve a role in proteolysis. In a recent report, deletion of both UBA domains failed to elicit a noticeable defect in sensitivity to UV light (Bertolaet et al., 2001). Since a plating assay may require ∼3 days of recovery following exposure to UV light, this long incubation might not reveal regulatory defects. In addition, we note that the proteolytic role of Rad23 is likely to be a post-incision step that involves Rad23/proteasome interaction. It was previously shown that loss of UbLR23, which prevents proteasome interaction (Schauber et al., 1998), causes only intermediate sensitivity to UV light (Watkins et al., 1993). Therefore, it will be important to compare the UV sensitivity of Rad23 that lacks UbLR23 (ΔUbLrad23) with mutants lacking UBA domains (rad23ΔUBA1,2). We speculate that the UBA domains in Rad23 interact with ubiquitylated substrates to inhibit the expansion of a nascent multi-Ub chain. Subsequent translocation of the tethered substrate to the proteasome, by the ubiquitin-like domain in Rad23 (UbLR23), could facilitate degradation by proteasome-associated E2 and E3 factors (Tongaonkar et al., 2000; Xie and Varshavsky, 2000). Because UbLR23 is not required for Rad23/Ub binding, or for inhibiting multi-Ub chain formation (Ortolan et al., 2000), we propose that it functions primarily as a proteasome localization signal.
We report here an interaction between UBA domains in Rad23 and ubiquitin. Other investigators have recently reported similar findings (Bertolaet et al., 2001). The three-dimensional structure of the ∼40 amino acid residues encompassing UBA2 in human and yeast Rad23 was determined by NMR spectroscopy, and found to consist of a compact three helix bundle with the conserved residues participating in helix–helix interactions (Dieckmann et al., 1998). The surface of the UBA domain contains hydrophobic residues whose accessibility could promote interaction with effectors (Withers-Ward et al., 2000). Significantly, Bertolaet et al. (2001) showed that the interaction between Ub and the UBA domains in Rad23 and Ddi1 required a highly conserved leucine residue, whose conversion to alanine inhibited interaction with Ub. Our results suggest that UBA1 may form a stronger interaction with Ub and di-Ub, in agreement with previous studies (Bertolaet et al., 2001).
We propose that Rad23 can interact with physiological substrates that are already ligated to Ub, as we showed previously for a test substrate in vivo (Ortolan et al., 2000). We speculate that by specifically binding only those proteins that are already ubiquitylated, non-specific and potentially deleterious interactions between Rad23 and other cellular proteins are avoided.
METHODS
Ubiquitylation assays. The purification of E1, E2 (Ubc2), Rad23, ΔUbLrad23 and [32P]Ub was previously described Tongaonkar and Madura, 1998, Ortolan et al., 2000. Rad23 mutants lacking UBA domains (rad23ΔUBA1, rad23ΔUBA2 and rad23ΔUBA1,2) were generated by PCR and expressed in E. coli using the T7-expression system (pET11d; Novogen). The DNA fragments encoding these mutants contained 5′ NcoI and 3′ BamHI DNA restriction sites. The expression, purification and radiolabelling of GST–Ub with [γ-32P]ATP, and cleavage with thrombin, was as previously described (Tongaonkar and Madura, 1998). GST–UBA1 and GST–UBA2 were expressed in pGEX2TK, and purified to homogeneity from E. coli.
Ubiquitin-binding assays. ubiquitylation reactions (30 µl) were diluted into 2 ml of 50 mM Tris–HCl pH 8.0 and centrifuged in Centricon-30 (Amicon) at 4°C, until the volume was reduced to ∼0.05 ml. The retentate was washed with 50 mM Tris–HCl pH 8.0 to ensure that all unbound [32P]Ub was removed. After the volume was reduced to ∼0.05 ml, 4× SDS-loading buffer was added and the sample was resolved in an SDS–12% polyacrylamide gel and examined by autoradiography.
Oligonucleotides. DNA fragments were cloned using 5′ NcoI and 3′ BamHI DNA restriction sites, and subjected to DNA sequencing analysis to confirm the accuracy of the constructs. To generate rad23ΔUBA1 we amplified a 5′ DNA fragment that encoded amino acid residues 1–140, and a 3′ fragment encoding amino acid residues 191–398 of the RAD23 gene. The two DNA fragments were treated with KpnI restriction enzyme and ligated. The resulting DNA fragment, which contained the RAD23 gene lacking UBA1, was digested with NcoI and BamHI and ligated into pET11d. To generate rad23ΔUBA2, we amplified a DNA fragment using a forward primer that annealed at the beginning of the gene, and a reverse primer containing a termination codon, that annealed at a position corresponding to amino acid 350. The double mutant rad23ΔUBA1,2 was generated by amplifying the DNA encoding amino acid residues 1–140 and 191–350. The DNA fragments were treated with KpnI restriction enzyme and ligated. The resulting product was digested with NcoI and BamHI and ligated into pET11d. Plasmids encoding GST–UBA1 and GST–UBA2 expressed residues 141–190 and 351–398 of the Rad23 protein, and were cloned into pGEX2TK using BamHI and EcoRI DNA restriction sites.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank J. Dutta for experimental assistance performed during a laboratory rotation. Members of the laboratory are thanked for discussion and criticism of the manuscript. This work was supported by NIH grant CA83875 to K.M.
REFERENCES
- Bertolaet B.L., Clarke, D.J., Wolff, M., Watson, M.H., Henze, M., Divita, G. and Reed, S.I. (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nature Struct. Biol., 8, 417–422. [DOI] [PubMed] [Google Scholar]
- Clarke D.J., Mondesert, G., Segal, M., Bertolaet, B.L., Jensen, S., Wolff, M., Henze, M. and Reed, S.I. (2001) Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol., 21, 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers, N.G.J. and Hoeijmakers, J.H.J. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Dieckmann T., Withers-Ward, E.S., Jarosinski, M.A., Liu, C.-F., Chen, I.S.Y. and Feigon, J. (1998) Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nature Struct. Biol., 5, 1042–1046. [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Bailly, V., Sung, P., Prakash, L. and Prakash, S. (1995a) Yeast DNA repair protein RAD23 promotes complex formation between transcription factor TFIIH and DNA damage recognition factor RAD14. J. Biol. Chem., 270, 8385–8388. [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Habraken, Y., Sung, P., Prakash, L. and Prakash, S. (1995b) Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem., 270, 12973–12976. [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Sung, P., Prakash, L. and Prakash, S. (1996) Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem., 271, 8903–8910. [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Sung, P., Prakash, L. and Prakash, S. (1998) Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem., 273, 31541–31546. [DOI] [PubMed] [Google Scholar]
- Hofman K. and Bucher, P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitylation pathway. Trends Biol. Sci., 21, 172–173. [PubMed] [Google Scholar]
- Jansen L.E.T., Verhage, R.A. and Brouwer, J. (1998) Preferential binding of yeast Rad4–Rad23 complex to damaged DNA. J. Biol. Chem., 273, 33111–33114. [DOI] [PubMed] [Google Scholar]
- Kumar S., Talis, A.L. and Howley, P.M. (1999) Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitylation. J. Biol. Chem., 274, 18785–18792. [DOI] [PubMed] [Google Scholar]
- Lambertson D., Chen, L. and Madura, K. (1999) Pleiotropic growth and proteolytic defects caused by the loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics, 153, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C. et al. (1994) Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J., 13, 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F., Bouziane, M., Dammann, R., Masutani, C., Hanaoka, F., Pfeifer, G. and O’Connor, T.R. (2000) 3-methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem., 275, 28433–28438. [DOI] [PubMed] [Google Scholar]
- Ortolan T.G., Tongaonkar, P., Lambertson, D., Chen, L. and Madura, K. (2000) The Rad23 DNA repair protein is a negative regulator of substrate-linked multi-ubiquitin chain assembly. Nature Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- Prakash S., Sung, P. and Prakash, L. (1993) DNA repair genes and proteins of Saccharomyces cerevisiae.Annu. Rev. Genet., 27, 33–70. [DOI] [PubMed] [Google Scholar]
- Schauber C., Chen, L., Tongaonkar, P., Vega, I., Lambertson, D., Potts, W. and Madura, K. (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature, 391, 715–718. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Masutani, C., Uchida, A., Maekawa, T., van der Spek, P.J., Bootsma, D., Hoeijmakers, J.H.J. and Hanaoka, F. (1996) HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol., 16, 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Park, H., Kwofie, M.A. and Lennarz, W.J. (2001) Rad23 provides a link between the Png1 deglycosylating enzyme and the 26S proteasome in yeast. J. Biol. Chem., 276, 21601–21607. [DOI] [PubMed] [Google Scholar]
- Tongaonkar P., and Madura, K. (1998) Reconstituting ubiquitination reactions with affinity purified components and 32P-ubuitin. Anal. Biochem, 260, 135–141. [DOI] [PubMed] [Google Scholar]
- Tongaonkar P., Chen, L., Lambertson, D., Ko, B. and Madura, K. (2000) Evidence for an interaction between ubiquitin-conjugating enzymes and the 26S proteasome. Mol. Cell. Biol., 20, 4691–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spek P.J., Visser, C.C., Hanaoka, F., Smit, B., Hagemeijer, A., Bootsma, D. and Hoeijmakers, J.H.J. (1996) Cloning, comparative mapping, and RNA expression of the mouse homologues of the Saccharomyces cerevisiae nucleotide excision repair gene RAD23. Genomics, 31, 20–27. [DOI] [PubMed] [Google Scholar]
- van Nocker S., Sadis, S., Rubin, D.M., Glickman, M., Fu, H., Coux, O., Wefes, I., Finley, D. and Vierstra, R.D. (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol., 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins J.F., Sung, P., Prakash, L. and Prakash, S. (1993) The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol., 13, 7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Ward E.S. et al. (1997) Human immunodeficiency virus Type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J. Virol., 71, 9732–9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Ward E.S., Mueller, T.D., Chen, I.S.Y. and Feigon, J. (2000) Biochemical and structural analysis of the interaction between the UBA(2) domain on the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry, 39, 14103–14122. [DOI] [PubMed] [Google Scholar]
- Xie Y. and Varshavsky, A. (2000) Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl Acad. Sci. USA, 97, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Wani, G., Wani, M.A. and Wani, A.A. (2001) Human homologue of yeast Rad23 protein A interacts with p300/Cyclic AMP-responsive element binding (CREB)-binding protein to down-regulate transcriptional activity of p53. Cancer Res., 61, 64–70. [PubMed] [Google Scholar]