Abstract
Molecular chaperones are known to facilitate cellular protein folding. They bind non-native proteins and orchestrate the folding process in conjunction with regulatory cofactors that modulate the affinity of the chaperone for its substrate. However, not every attempt to fold a protein is successful and chaperones can direct misfolded proteins to the cellular degradation machinery for destruction. Protein quality control thus appears to involve close cooperation between molecular chaperones and energy-dependent proteases. Molecular mechanisms underlying this interplay have been largely enigmatic so far. Here we present a novel concept for the regulation of the eukaryotic Hsp70 and Hsp90 chaperone systems during protein folding and protein degradation.
Introduction
The function of a protein is determined by the three-dimensional structure of its amino acid chain. The native structure is attained following translation or translocation and is under constant threat of unfolding as a consequence of chemical equilibrium and cellular stress. Systems that actively maintain and control protein structure are thus a prerequisite for cell survival, and involve both molecular chaperones and energy-dependent proteases (Wickner et al., 1999). Molecular chaperones bind non-native proteins, inhibit protein aggregation and promote folding to the native state (Hartl, 1996; Bukau and Horwich, 1998). Energy-dependent proteases, on the other hand, eliminate proteins that fail to attain their native conformation (Wickner et al., 1999). Thus, chaperones and proteases appear to form a cellular surveillance system that monitors protein quality. Recent findings provide insight into molecular mechanisms that underlie the interplay of chaperones and proteases.
The Hsp70 and Hsp90 chaperones
Major chaperones in the mammalian cytosol and nucleus are the 70 and 90 kDa heat shock proteins (Hsp70 and Hsp90). Hsp70 participates in the folding of newly synthesized proteins, the protection of proteins during cellular stress and intracellular protein trafficking (Hartl, 1996; Frydman and Höhfeld, 1997; Bukau and Horwich, 1998). Hsp90 function appears to be more restricted but, again, a role in stress protection has been demonstrated (Buchner, 1999; Caplan, 1999). Both classes of chaperones seem to associate with non-native protein substrates through recognition of hydrophobic patches ultimately buried in the native structure. In addition, they mediate the conformational regulation of a wide range of client proteins involved in signal transduction, cell proliferation and apoptosis (Frydman and Höhfeld, 1997; Caplan, 1999; Pandey et al, 2000; Beere and Green, 2001). Interaction with non-native protein substrates and client proteins is highly dynamic and coupled to cycles of ATP binding and ATP hydrolysis by the chaperones (Frydman and Höhfeld, 1997; Prodromou et al, 1999; Young and Hartl, 2000). Upon release, further protein folding or biogenesis can occur. This may involve rebinding to the same or another chaperone, or transfer to other cellular machines. Hsp70 and Hsp90 have been shown to cooperate with the degradation machinery (Schneider et al., 1996; Bercovich et al., 1997; Dul et al., 2001). Of particular interest is the pharmacological shifting of Hsp90 function from protein folding to protein degradation, induced by anti-tumor agents like geldanamycin (Schneider et al., 1996; Whitesell and Cook, 1996). However, the underlying molecular mechanisms remain largely elusive.
It is now widely recognized that Hsp70 and Hsp90 do not act on their own, but co-operate with several ancillary proteins, so-called chaperone cofactors or co-chaperones (Frydman and Höhfeld, 1997; Buchner, 1999; Caplan, 1999). In principle, chaperone cofactors have two options for modulating chaperone function. They can either regulate the ATPase cycle of the chaperone to influence its affinity for protein substrates, or recruit the chaperones to specific proteins, protein complexes and subcellular compartments. Many cofactors combine both activities and, consequently, exhibit a modular structure comprising chaperone-binding/chaperone-regulating motifs plus other functional domains. A chaperone-binding motif found in several Hsp70 and Hsp90 cofactors is characterized by a tandem arrangement of three degenerate 34 amino acid repeats (tetratricopeptide repeats, TPRs; Frydman and Höhfeld, 1997). The Hsp70/Hsp90-organizing protein Hop possesses multiple TPR domains, enabling it to simultaneously bind Hsp70 and Hsp90, and to promote chaperone cooperation during the regulation of signal transduction pathways (Frydman and Höhfeld, 1997; Buchner, 1999; Caplan, 1999). In each domain, three tandem TPRs align with an adjacent α-helix to form a groove that accommodates conserved peptide motifs present at the C-terminus of Hsp70 and Hsp90 (Scheufler et al., 2000). Although the presence of TPRs is not restricted to chaperone cofactors, they appear to form a stably folded adaptor well suited to mediate chaperone/cofactor contacts, even under conditions of cellular stress.
CHIP links chaperones to the degradation machinery
CHIP was initially identified in a screen for human TPR-containing proteins (Ballinger et al., 1999). At its N-terminus three tandem TPRs are located, which, together with an adjacent, highly charged α-helix, form a chaperone adaptor (Figure 1). CHIP utilizes this single adaptor to contact either Hsp70 or Hsp90 (Ballinger et al., 1999; Connell et al., 2001). Association with Hsp70 blocks the ATPase cycle of the chaperone and inhibits its ability to refold non-native proteins in vitro (Ballinger et al., 1999). Similarly, binding of CHIP to Hsp90 prevents the cooperation of the chaperone with other cofactors required for productive chaperone function (Connell et al., 2001). Apparently, CHIP abrogates the ability of both chaperones, Hsp70 and Hsp90, to assist cellular protein folding.
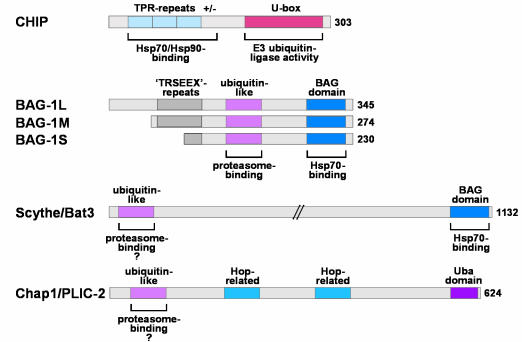
Fig. 1. Cofactors that appear to link molecular chaperones to the ubiquitin/proteasome system. CHIP possesses an N-terminal chaperone binding motif formed by three TPRs and an adjacent highly charged region. A U-box required for ubiquitin ligase activity is present at the C-terminus. The BAG-1 isoforms share a ubiquitin-like domain involved in proteasome binding and a BAG domain that mediates interaction with Hsp70. Like the BAG-1 proteins, Scythe/Bat3 possesses a ubiquitin-like domain that may be used for proteasome association and a BAG domain used for binding and regulation of Hsp70. Chap1/PLIC-2 combines a ubiquitin-like domain and a Uba domain, the latter of which is found in several proteins involved in ubiquitin conjugation. In addition, regions structurally related to the chaperone cofactor Hop are present in Chap1/PLIC-2.
Interestingly, the C-terminus of CHIP displays structural similarities to components of the ubiquitin/proteasome system, a major protein degradation pathway in eukaryotic cells (Figure 1). Proteins destined for degradation are labelled with a multiubiquitin chain and then targeted to a large heterooligomeric protease, the 26S proteasome (Varshavsky, 1997; Baumeister et al., 1998). Ubiquitylation is mediated by a complex cellular machinery comprising a ubiquitin activator (the E1 enzyme), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3). E2 and E3 enzymes are recruited from large protein families and the broad repertoire of distinct E2/E3 pairs is likely to ensure specific recognition of diverse substrate proteins. Additional proteins, including the yeast ubiquitylation factor Ufd2, cooperate with the E2/E3 machinery (Koegl et al., 1999). Interestingly, Ufd2 and CHIP both possess a U-box domain shown to participate in ubiquitin conjugation. The modular structure of CHIP may thus enable the cofactor to directly link molecular chaperones to the degradation machinery.
In support of this notion, CHIP was recently shown to promote the degradation of chaperone substrates by the ubiquitin/proteasome system (Connell et al., 2001; Meacham et al., 2001). Connell et al. (2001) investigated the effects of CHIP on the activation and degradation of the glucocorticoid hormone receptor (GR). Like several other steroid hormone receptors, GR undergoes transient interactions with the Hsp70 and Hsp90 chaperone systems during its activation (Frydman and Höhfeld, 1997). The high-affinity hormone binding state is finally reached in association with a Hsp90 heterocomplex that includes cofactors such as p23. In vitro experiments revealed that incorporation of CHIP into Hsp90 heterocomplexes leads to a dissociation of p23 and, consequently, Hsp90-associated GR no longer reaches its steroid-binding conformation (Connell et al., 2001). In addition, remodelling of the Hsp90 chaperone complex by CHIP is accompanied by induced ubiquitylation of the hormone receptor. Apparently, the cofactor not only blocks GR activation but also diverts the receptor to the degradation machinery; increasing the cellular level of CHIP is indeed sufficient to induce degradation of GR by the proteasome.
Similar findings were obtained for the cystic fibrosis transmembrane conductance regulator (CFTR), a plasma membrane chloride-ion channel. Folding and maturation of CFTR is rather inefficient, and most nascent CFTR is retained in the endoplasmic reticulum (ER) and degraded by the ubiquitin/proteasome system (Kopito, 1999). Hsp70 and Hsp90 were shown to interact with cytoplasmic domains of CFTR and to facilitate the folding process (Loo et al., 1998; Meacham et al., 1999). Consequently, CFTR maturation is sensitive to expression of the CHIP cofactor. Elevation of CHIP induced the ubiquitylation and degradation of immature, ER-localized forms of CFTR (Meacham et al., 2001). CHIP apparently targets diverse chaperone substrates to the ubiquitin/proteasome system and in this way modulates the balance of protein folding and protein degradation during protein quality control.
CHIP is a chaperone-associated ubiquitin ligase
It was conceivable that CHIP-induced degradation was simply a consequence of the cofactor’s ability to inhibit chaperone function. However, structure/function analysis of CHIP suggests an active role in the ubiquitylation of chaperone substrates. A deletion fragment that lacks the C-terminal U-box retains the ability to contact Hsp70 and to inhibit the ATPase cycle of the chaperone (Ballinger et al., 1999; Meacham et al., 2001). Moreover, it is sufficient to induce remodelling of Hsp90 heterocomplexes (Connell et al., 2001). Nevertheless, the fragment does not induce the ubiquitylation and degradation of GR and CFTR in transient transfection assays (Connell et al., 2001; Meacham et al., 2001). In fact, it acts as a dominant-negative factor that blocks ubiquitylation and degradation. These findings point to a critical role for the U-box in CHIP-induced targeting of chaperone substrates for proteasomal degradation.
Another U-box-containing protein is Ufd2, which cooperates with certain E2 and E3 enzymes to promote the formation of multiubiquitin chains, and was therefore termed E4 (Koegl et al., 1999). The enzymatic function of Ufd2 remains enigmatic. However, the U-box was shown to bind to ubiquitin moieties of the modified substrate protein. This may stabilize the association of the substrate with the conjugation machinery and, as a consequence, may increase the efficiency of the conjugation reaction. The presence of the U-box within CHIP suggested an E4-like activity, but it was also conceivable that CHIP fulfils an E3-like function, as the U-box is structurally related to RING-finger domains found in several E3 ubiquitin ligases (Aravind and Koonin, 2000). In the E3s, the RING-finger mediates the binding and activation of E2 enzymes (Jackson et al., 2000). In fact, CHIP utilizes its U-box for cooperation with E2 enzymes of the Ubc4/5 family, and thus displays E3 ubiquitin ligase activity (Demand et al., 2001; Hatakeyama et al., 2001; Jiang et al., 2001). E3 and E4 enzymes appear to promote ubiquitylation in a similar fashion, by facilitating the interaction between the conjugation machinery and the substrate protein.
CHIP is the first ubiquitin ligase known to directly associate with molecular chaperones. Indeed, the chaperone itself can be seen as an integral part of the ubiquitin ligase complex (Figure 2). Through its ability to recognize non-native proteins and specific client proteins, the chaperone apparently selects substrates for CHIP-mediated ubiquitylation. The chaperone–CHIP complex thus resembles other multisubunit ubiquitin ligases, such as the Skp1–cullin–F-box protein (SCF) complex. In SCF, an E3 teams up with diverse F-box proteins that act as adaptors to specifically recognize a broad range of substrate proteins (Jackson et al., 2000). Similarly, CHIP association with Hsp70 and Hsp90 appears to promote the ubiquitylation of diverse chaperone substrates.
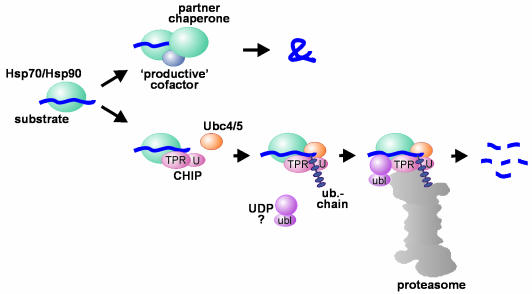
Fig. 2. Hsp70 and Hsp90 in protein folding and degradation. An initial decision to fold or degrade a Hsp70- or Hsp90-associated substrate protein may be reached through competition between a ‘productive’ cofactor such as Hop, and the ubiquitin ligase CHIP. During the folding process, Hsp70 and Hsp90 may co-operate with other chaperone proteins (termed partner chaperones). On the degradation pathway, CHIP associates with Hsp70 or Hsp90 via its TPR chaperone adaptor (TPR), and at the same time recruits E2 ubiquitin conjugating enzymes of the Ubc4/5 family to the chaperone complex. This may involve binding of the E2 to the U-box of the cofactor (U). In conjunction with E2, CHIP mediates ubiquitin attachment to the chaperone substrate and induces its targeting to the proteasome for degradation. The targeting process may be facilitated by a ubiquitin domain protein (UDP), such as BAG-1, which binds to Hsp70 and utilizes its ubiquitin-like domain (ubl) for proteasomal association.
Chaperone machines: folding versus degradation
Previously, kinetic partitioning of non-native proteins between molecular chaperones and proteases has been proposed to underlie protein quality control (Wickner et al., 1999). This model infers a competition between chaperones and components of the degradation machinery. The characterization of the CHIP cofactor now points to a different concept. Through association with CHIP, the molecular chaperones Hsp70 and Hsp90 are directly turned into protein degradation factors (Figure 2). This suggests an elegant solution to the problem of how the degradation machinery recognizes aberrant proteins: the factors that bind non-native proteins in the course of protein folding also appear to be employed to select substrates for protein degradation. According to this model, a balance of protein folding and protein degradation would be achieved through regulation of chaperone function. As CHIP occupies commonly used cofactor binding sites on Hsp70 and Hsp90, competition with other chaperone cofactors may provide the means to modulate quality control decisions. The cofactor Hop most likely fulfils a role as a CHIP antagonist. Like CHIP, Hop associates with the C-terminus of Hsp70 via a TPR chaperone adaptor. Similarly, the two cofactors appear to compete for a common TPR acceptor site on Hsp90 (Demand et al., 1998; Ballinger et al., 1999; Connell et al., 2001). In contrast to CHIP, Hop has been shown to assist chaperone-mediated protein folding (Buchner et al., 1999; Caplan et al, 1999). Competing cofactors may thus determine whether a given chaperone machine is involved in protein folding or protein degradation (Figures 2 and 3).
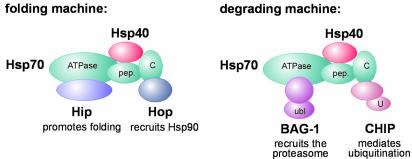
Fig. 3. Model for Hsp70 chaperone machines. Binding of distinct chaperone cofactors to the N-terminal ATPase domain of Hsp70 (ATPase) and to its C-terminus (C) may give rise to chaperone machines involved in protein folding and protein degradation, respectively. The cofactors Hip and BAG-1 compete for binding to the ATPase domain, while Hop and CHIP associate with the C-terminus in a competitive manner. During folding and degradation, Hsp70 appears to co-operate with cofactors of the Hsp40 protein family. pep., peptide binding domain of Hsp70; ubl, ubiquitin-like domain of BAG-1; U, U-box of CHIP.
The Hsp70 cofactor BAG-1 has also been proposed to act as a link between molecular chaperones and the ubiquitin/proteasome system (Lüders et al., 2000). BAG-1 exists in multiple isoforms in mammalian cells. These share a C-terminal BAG domain required for Hsp70 interaction, and an N-terminal ubiquitin-like domain (Figure 1). BAG-1 thus belongs to a family of ubiquitin domain proteins (UDPs) (Jentsch and Pyrowolakis, 2000). The chaperone cofactor utilizes its ubiquitin domain at least in part for binding to the proteasome (Lüders et al., 2000). In this way, BAG-1 promotes an association between Hsp70 and the proteolytic complex. As BAG-1 also acts as a nucleotide exchange factor that induces the release of substrate from Hsp70, a role in assisting substrate transfer to the proteasome is conceivable (Höhfeld and Jentsch, 1997; Lüders et al., 1998; Sondermann et al., 2001). However, experimental evidence in support of this notion is still missing. In fact, overexpression of BAG-1 was shown to promote the activation of the raf-1 protein kinase and to stimulate the anti-apoptotic function of Bcl-2 (Takayama et al., 1995; Song et al., 2001). These observations are difficult to reconcile with an exclusive role of BAG-1 in chaperone–proteasome coupling. Only a subpopulation of cellular BAG-1 may thus fulfil such a coupling function, or coupling may affect only a subset of BAG-1 and Hsp70 substrate proteins. Alternatively, BAG-1 may closely cooperate with other, possibly rate-limiting cofactors during the sorting of chaperone substrates to the proteasome. As a consequence, elevation of BAG-1 levels alone may be insufficient to promote substrate degradation.
A likely candidate for a rate-limiting cofactor is CHIP. In fact, the ubiquitin ligase was recently shown to team up with the BAG-1 ubiquitin domain protein to regulate proteasomal sorting of chaperone substrates (Demand et al., 2001; Figure 2). This apparently involves the formation of a chaperone complex that comprises both cofactors (Demand et al., 2001; Figure 3). Notably, BAG-1 binds to the amino-terminal ATPase domain of Hsp70, whereas CHIP occupies the carboxyl terminus of the chaperone (Ballinger et al., 1999; Sondermann et al., 2001). Intriguingly, BAG-1 competes with the folding-stimulating cofactor Hip in binding to the ATPase domain of Hsp70 (Höhfeld and Jentsch, 1997). This further points to a role of cooperating and competing cofactors in determining the function of molecular chaperones in protein folding and protein degradation (Figure 3).
BAG-1 was previously shown to associate with another RING-finger type ubiquitin ligase, the Siah protein (Matsuzawa et al., 1998). The functional consequences of this association remain unclear. However, it could provide a means for Siah to gain access to chaperone substrates and to promote their ubiquitylation and degradation. BAG-1/Siah cooperation would provide an additional pathway for protein sorting between Hsp70 and the proteasome. The existence of additional pathways is also implied by the fact that BAG-1 is not the only ubiquitin domain protein that cooperates with molecular chaperones (Figure 1). The human UDPs Chap1 and Chap2 were recently identified in a screen for proteins which, like BAG-1, bind to the ATPase domain of a Hsp70 family member (Kaye et al., 2000). Chap1 (also termed PLIC-2) and its relative PLIC-1 represent human homologues of the yeast Dsk2 protein which is required for cell-cycle regulation. Both PLIC proteins were shown to associate with the proteasome and with ubiquitin ligases, and may thus coordinate the activities of the ubiquitin conjugation machinery and the proteolytic complex (Kleijnen et al., 2000). The observed association with molecular chaperones suggests that this coordinating function may involve chaperone regulation. Affected chaperone substrates, however, remain to be identified. Similarly, a role for Chap2 in the degradation of chaperone substrates awaits experimental confirmation. Chap2 is also known as Bat3 and human Scythe. Like BAG-1, Scythe combines an N-terminal ubiquitin domain with a BAG domain that is used for binding to Hsp70 and for inhibition of Hsp70 chaperone function (Thress et al., 2001; Figure 1). Thus, Chap2/Scythe most likely acts as an additional coupling factor between the molecular chaperone and the proteasome.
Perspectives
Interactions of Hsp70 and Hsp90 with a variety of cofactors enable these chaperones to accompany substrate proteins from the cradle to the grave. The emergence and characterization of multiple cofactor families now paves the way to elucidate the roles of chaperones in protein folding and degradation. The modulation of quality control decisions during signal transduction and apoptosis becomes feasible. Nevertheless, our knowledge of cellular targets that are affected by diverse chaperone cofactors is still limited, and further work is required to understand the mechanisms underlying substrate selection. In any case, molecular chaperones and their ancillary cofactors have taken centre stage in protein quality control.

Jörg Höhfeld, Douglas M. Cyr & Cam Patterson


Acknowledgments
Acknowledgements
Work in the authors’ laboratories described here was supported by grants from the Deutsche Forschungsgemeinschaft, the Bonner Forum Biomedizin and the NIH.
REFERENCES
- Aravind L. and Koonin, E.V. (2000) The U box is a modified RING finger — a common domain in ubiquitylation. Curr. Biol., 10, R132–R134. [DOI] [PubMed] [Google Scholar]
- Ballinger C.A., Connell, P., Wu, Y., Hu, Z., Thompson, L.J., Yin, L.-Y. and Patterson, C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol., 19, 4535–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Walz, J., Zühl, F. and Seemüller, E. (1998) The proteasome: a paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Beere H.M. and Green, D.R. (2001) Stress management — heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol., 11, 6–10. [DOI] [PubMed] [Google Scholar]
- Bercovich B., Stancovski, I., Mayer, A., Blumenfeld, N., Laszlo, A., Schwartz, A.L. and Ciechanover, A. (1997) Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem., 272, 9002–9010. [DOI] [PubMed] [Google Scholar]
- Buchner J. (1999) Hsp90 & co. — a holding for folding. Trends Biochem. Sci., 24, 136–141. [DOI] [PubMed] [Google Scholar]
- Bukau B. and Horwich, A.L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Caplan A.J. (1999) Hsp90’s secrets unfold: new insights from structural and functional studies. Trends Cell Biol., 9, 262–268. [DOI] [PubMed] [Google Scholar]
- Connell P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L.J., Höhfeld, J. and Patterson, C. (2001) Regulation of heat shock protein-mediated protein triage decisions by the co-chaperone CHIP. Nature Cell Biol., 3, 93–96. [DOI] [PubMed] [Google Scholar]
- Demand J., Lüders, J. and Höhfeld, J. (1998) The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol., 18, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demand, , Alberti, S., Patterson, C. and Höhfeld, J. (2001) Cooperation of an ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol, in press. [DOI] [PubMed] [Google Scholar]
- Dul J.L., Davis, D.P., Williamson, E.K., Stevens, F.J. and Argon, Y. (2001) Hsp70 and antifibrillogenic peptides promote degradation and inhibit intracellular aggregation of amyloidogenic light chains. J. Cell Biol., 152, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J. and Höhfeld, J. (1997) Chaperones get in touch: the Hip-Hop connection. Trends Biochem. Sci., 22, 87–92. [DOI] [PubMed] [Google Scholar]
- Hartl F.-U. (1996) Molecular chaperones in protein folding. Nature, 381, 571–580. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Yada, M., Matsumoto, M., Ishida, N. and Nakayama, K.-I. (2001) U-box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem., 276, 33111–33120. [DOI] [PubMed] [Google Scholar]
- Höhfeld J. and Jentsch, S. (1997) GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J., 16, 6209–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P.K., Eldridge, A.G., Freed, E., Furstenthal, L., Hsu, J.Y., Kaiser, B.K. and Reimann, J.D.R. (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol., 10, 429–439. [DOI] [PubMed] [Google Scholar]
- Jentsch S. and Pyrowolakis, G. (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol., 10, 335–342. [DOI] [PubMed] [Google Scholar]
- Jiang J., Ballinger, C.A., Cyr, D.M., Höhfeld, J. and Patterson, C. (2001) CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Kaye F.J., Modi, S., Ivanovska, I., Koonin, E.V., Thress, K., Kubo, A., Kornbluth, S. and Rose, M.D. (2000) A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett., 467, 348–352. [DOI] [PubMed] [Google Scholar]
- Kleijnen M.F., Shih, A.H., Zhou, P., Kumar, S., Soccio, R.E., Kedersha, N.L., Gill, G. and Howley, P.M. (2000) The hPLIC proteins may provide a link between the ubiquitylation machinery and the proteasome. Mol. Cell, 6, 409–419. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U. and Jentsch, S. (1999) A novel ubiquitylation factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. (1999) Biosynthesis and degradation of CFTR. Physiol. Rev., 79, S167–S173. [DOI] [PubMed] [Google Scholar]
- Loo M.A., Jensen, T.J., Cui, L., Hou, Y., Chang, X.B. and Riordan, J.R. (1998) Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J., 17, 6879–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Demand, J., Schönfelder, S., Frien, M., Zimmermann, R. and Höhfeld, J. (1998) Cofactor-induced modulation of the functional specificity of the molecular chaperone Hsc70. Biol. Chem., 379, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Lüders J., Demand, J. and Höhfeld, J. (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem., 275, 4613–4617. [DOI] [PubMed] [Google Scholar]
- Matsuzawa S., Takayama, S., Froesch, B.A., Zapata, J.M. and Reed, J.C. (1998) p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J., 17, 2736–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G.C., Lu, Z., King, S., Sorscher, E., Tousson, A. and Cyr, D.M. (1999) The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J., 18, 1492–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G.C., Patterson, C., Zhang, W., Younger, J.M. and Cyr, D. (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nature Cell Biol., 3, 100–105. [DOI] [PubMed] [Google Scholar]
- Pandey P. et al. (2000) Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. EMBO J., 19, 4310–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C. et al. (1999) Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J., 18, 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C., Brinker, A., Bourenkov, G., Pegoraro, S., Moroder, L., Bartunik, H., Hartl, F.U. and Moarefi, I. (2000) Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell, 101, 199–210. [DOI] [PubMed] [Google Scholar]
- Schneider C., Sepp-Lorenzino, L., Nimmesgern, E., Ouerfelli, O., Danishefsky, S., Rosen, N. and Hartl, F.-U. (1996) Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl Acad. Sci. USA, 93, 14536–14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H., Scheufler, C., Schneider, C., Höhfeld, J., Hartl, F.-U. and Moarefi, I. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science, 291, 1553–1557. [DOI] [PubMed] [Google Scholar]
- Song J., Takeda, M. and Morimoto, R.I. (2001) Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nature Cell Biol., 3, 276–282. [DOI] [PubMed] [Google Scholar]
- Takayama S., Sato, T., Krajewski, S., Kochel, K., Irie, S., Millan, J.A. and Reed, J.C. (1995) Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell, 80, 279–284. [DOI] [PubMed] [Google Scholar]
- Thress K., Song, J., Morimoto, R.I. and Kornbluth, S. (2001) Reversible inhibition of Hsp70 chaperone function by Sythe and Reaper. EMBO J., 20, 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (1997) The ubiquitin system. Trends Biochem. Sci., 22, 383–387. [DOI] [PubMed] [Google Scholar]
- Whitesell L. and Cook, P. (1996) Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol. Endocrinol., 10, 705–712. [DOI] [PubMed] [Google Scholar]
- Wickner S., Maurizi, M.R. and Gottesman, S. (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science, 286, 1888–1893. [DOI] [PubMed] [Google Scholar]
- Young J.C. and Hartl, F.U. (2000) Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J., 19, 5930–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]