Abstract
The Pharmacogene Variation Consortium (PharmVar) provides nomenclature for the highly polymorphic human CYP2D6 gene locus and a comprehensive summary of structural variation. CYP2D6 contributes to the metabolism of numerous drugs and thus, genetic variation in its gene impacts drug efficacy and safety. To accurately predict a patient’s CYP2D6 phenotype, testing must include structural variants including gene deletions, duplications, hybrid genes, and combinations thereof. This tutorial offers a comprehensive overview of CYP2D6 structural variation, terms and definitions, a review of methods suitable for their detection and characterization, and practical examples to address the lack of standards to describe CYP2D6 structural variants or any other pharmacogene. This PharmVar tutorial offers practical guidance on how to detect the many, often complex, structural variants, as well as recommends terms and definitions for clinical and research reporting. Uniform reporting is not only essential for electronic health record-keeping but also for accurate translation of a patient’s genotype into phenotype which is typically utilized to guide drug therapy.
1. Introduction
The Cytochrome P450 2D6 gene (CYP2D6) is one of the most complex and clinically relevant pharmacogenes (1). The gene encodes CYP2D6, a membrane-bound drug-metabolizing enzyme that contributes to the catabolism or bioactivation of over 20% of clinically used drugs (2). The Clinical Pharmacogenetics Implementation Consortium (CPIC) classifies individuals with increased, normal, decreased, and no CYP2D6 activity as ultrarapid (UM), normal (NM), intermediate (IM), and poor (PM) metabolizers, respectively (3). Due to the high degree of genetic variability, CYP2D6 genetic testing also presents diverse analytical challenges for research and diagnostic laboratories. The Pharmacogenomics Knowledgebase (PharmGKB) provides information for CYP2D6, including reference tables, drug label information, and clinical annotations, among many others (4).
In addition to harboring numerous single nucleotide polymorphisms and small insertions/deletions (collectively referred to as single nucleotide variants (SNVs), or variants) that can impact gene function, the CYP2D6 gene locus also exhibits an array of structural variants (SVs) or copy number variants (CNVs), including gene deletions, duplications/multiplications, and hybrid genes that consist of various portions of CYP2D6 and the homologous CYP2D7 pseudogene (5). The term “SV” appears to be preferentially used in the literature when describing rearranged gene structures such as hybrid genes, and “CNV” appears to be the term of choice within the context of gene copy number testing. Since all known CYP2D6 SVs are CNVs, we have elected to refer to these collectively as SV/CNVs throughout this tutorial to reflect that either term is appropriate. Known CYP2D6 haplotypes with SVs/CNVs can be nonfunctional or have decreased, normal, increased, or unknown function.
A large body of evidence supports the clinical relevance of gene variation within the CYP2D6 gene locus, including SVs/CNVs (1, 6). It is therefore important to not only interrogate SNVs, but also determine whether an SV/CNV is present to predict a patient’s CYP2D6 metabolizer status accurately. Clinical guidelines published by CPIC, and the Dutch Pharmacogenetics Working Group (DPWG) support these observations. Examples include specific recommendations for CYP2D6 UMs, in which more than two copies of a normal function CYP2D6 gene copy are present, for several drugs, among them antidepressants (7, 8), opioids (9), tamoxifen (10), atomoxetine (11), and ondansetron (12). Recommendations may also be found in FDA prescribing labels as exemplified by eliglustat (13). Alternately, SVs/CNVs may cause loss of CYP2D6 function and contribute to IM or PM status, for which several guidelines also recommend modified therapy. Moreover, a maximum daily dose may be recommended for an IM or PM patient. For example, the FDA prescribing labels for tetrabenazine (14) and pimozide (15) limit dosages for CYP2D6 PM individuals. These guidelines highlight the importance of testing for CYP2D6 SVs/CNVs to reduce the risk of adverse events or treatment failure.
Additionally, variants in regulatory regions within the extended 22q13 locus and a variant in NFIB have also been suggested to impact CYP2D6 function (16–21). While those within CYP2D6 may directly impact metabolic capacity, the clinical utility of variants indirectly acting on function via regulatory mechanisms has, however, not yet been demonstrated. A patient may also have a rare variant(s) in the CYP2D6 gene which may not be interrogated but could conceivably impact function or variants in other genes that contribute to drug metabolism and disposition.
2. CYP2D Gene Locus
The CYP2D locus on chromosome 22 harbors CYP2D6 and two pseudogenes with high homology, CYP2D7 and CYP2D8 (also annotated as CYP2D7P and CYP2D8P, respectively, as per the HUGO Gene Nomenclature Committee) (Figure 1a). Chromosome 22 is known for its instability and numerous genetic aberrations that can alter CYP2D6 gene copy number status, such as duplication (22) or the 22q13.2 microdeletion (23) and 22q13.3 microdeletion (Phelan-McDermid) syndromes (24, 25). Large copy number gains of 1.1 Mb and 8.7 Mb are also annotated in the ClinVar database (entries 147621 and 154688, respectively).
Figure 1. Overview of the CYP2D6 gene locus and structural variants.
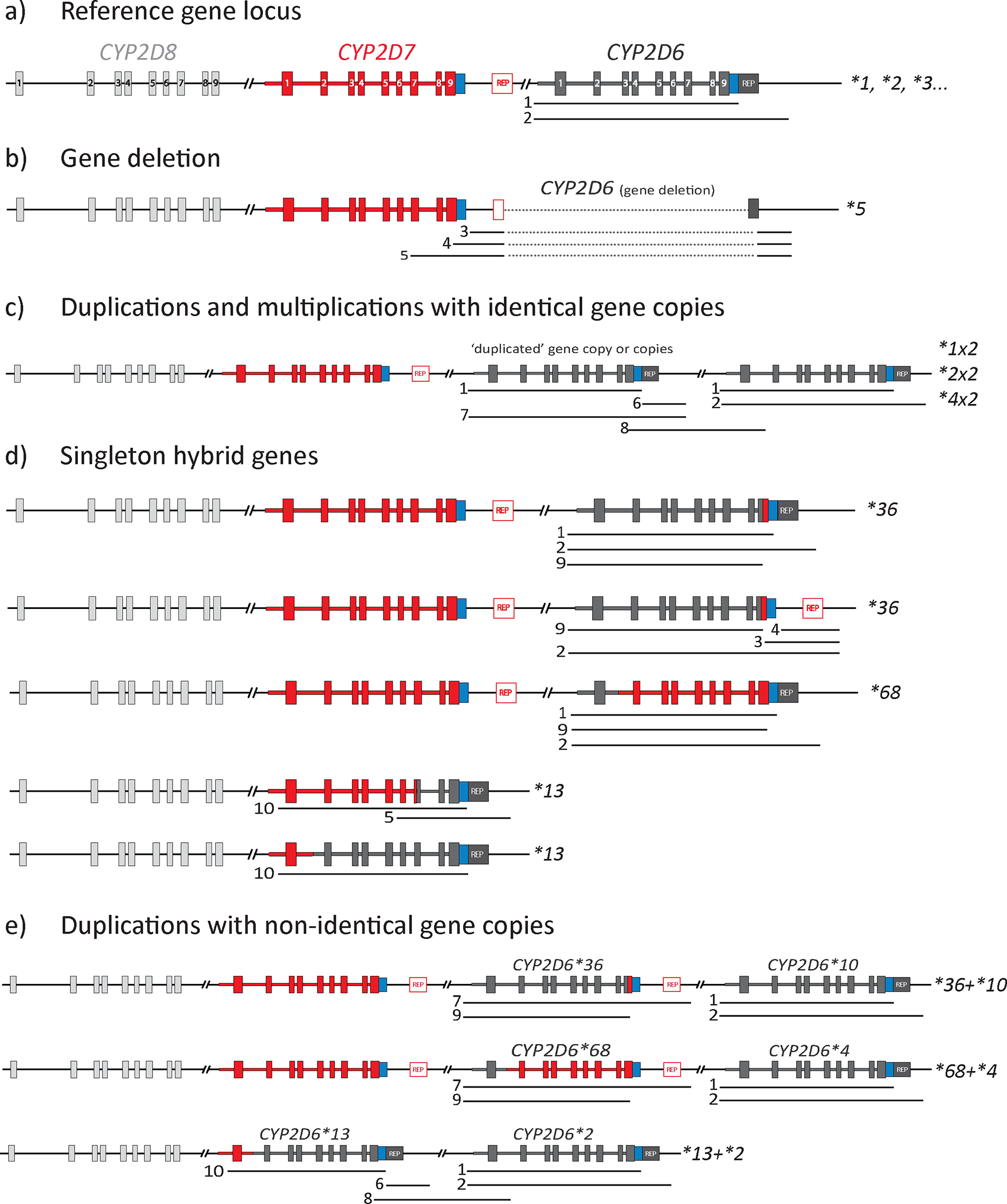
Each structural variant (SV) can also be described as a copy number variant (CNV) (Table 1). The three genes within the locus are shown in dark gray (CYP2D6), red (CYP2D7), and light gray (CYP2D8); boxes within each gene represent exons (numbered in panel a). Regions that can be amplified by XL-PCR to detect and characterize SVs/CNVs are shown by numbered lines underneath allele representations (see Table 3 for detailed descriptions). Repetitive sequences (REP) are shown in red (1.6-kb spacer is present) or dark gray (spacer is absent). Panel a depicts the reference gene locus containing CYP2D6, CYP2D7, and CYP2D8; sequence variants within the CYP2D6 gene define numerous star alleles. Panel b depicts the CYP2D6*5 gene deletion. Panel c depicts SV/CNV alleles with two or more identical gene copies (identity being on the core allele level); examples include *1×2, *2×2, and *4×2). Panel d depicts alleles having a hybrid gene consisting of CYP2D6 and CYP2D7 sequences; these alleles may or may not have a full CYP2D7 gene copy in addition to the hybrid gene. Lastly, panel e provides a selection of alleles with two or more non-identical gene copies, one of which is a full CYP2D6 gene copy (normal, decreased, or no function), and one is a hybrid gene.
The CYP2D gene locus is encoded on the negative strand of the genome in reverse orientation. The Pharmacogene Variation Consortium (PharmVar), however, displays the gene locus in the forward orientation (i.e., relative to the direction of transcription and translation of the CYP2D6 gene) (Figure 1a) and, therefore, illustrations in this tutorial and those provided on the PharmVar CYP2D6 gene page (26) and in the CYP2D6 PharmVar GeneFocus review (5) are shown in the forward direction (5’-CYP2D8-CYP2D7-CYP2D6-3’).
Historically, CYP2D6 variant coordinates have been annotated and reported using the genomic reference sequence (RefSeq) rather than the transcript (5). For consistency, PharmVar and this tutorial annotate variants using the NG_008376.4 RefSeq with the “A” of the ATG translation start codon being +1 (positions are not marked with a “g.” to avoid potential confusion with chromosomal positions). The outdated M33388 sequence should no longer be used for any purpose, and any laboratories or vendors still using M33388 are strongly encouraged to update annotations to NG_008376.4 to avoid confusion and errors. For example, rs3892097 corresponds to 1847G>A on the genomic RefSeq NG_008376.4 but 1846G>A on M33388; the corresponding cDNA position on the transcript NM_000106.6 is c.506–1G>A. The PharmVar CYP2D6 gene page enables easy conversion of positions among different genome builds and count modes.
It is also important to realize that genome build GRCh37 contains a sequence that matches CYP2D6*2, while for GRCh38, the genomic NG_008376.4 and transcript NM_000106.6 reference sequences correspond to CYP2D6*1. Additional information regarding differences among reference sequences can be found in the Read Me document on the PharmVar CYP2D6 gene page (26). CYP2D6 variant positions listed in this tutorial are according to NG_008376.4 or GRCh38.
2.1. CYP2D6 Gene Structure
The CYP2D6 gene has nine exons and is relatively small at 4311 bp (chr22:42126499 to 42130810) (Figure 1a). It is located 8.7 kb downstream of the homologous CYP2D7 pseudogene. CYP2D7 contains several key differences, one of which is a T-insertion in exon 1 that causes a frameshift and premature stop codon that renders it nonfunctional (27).
Repetitive sequences have enabled numerous unequal cross-over events by non-allelic homologous recombination between CYP2D6 and CYP2D7, leading to the formation of stable duplications, multiplications, deletions, and hybrids (5, 28–34). Two repeat elements lie downstream of CYP2D6, a 0.5-kb element that directly follows the coding sequence and a 2.8-kb repeat. In CYP2D7, these two elements are interrupted by a 1.6-kb sequence known as a “spacer”. These downstream elements likely facilitated the formation of the stable, heritable recombination events (35, 36) characteristic of the various SV structures found in CYP2D6. To our knowledge, there are no reports in the literature of de novo SVs/CNVs. The 2.8-kb repetitive elements, commonly referred to as REP6 and REP7 to indicate their respective CYP2D6 and CYP2D7 origins, also contain an Alu element and tandem 10-bp repeats that may promote homologous recombination. REP elements correspond to chr22:42123192–42125972 (NG_008376.4: 9839–12619 (REP6) and chr22:42135344–42138124 (REP7) (27,28) and are not believed to impact function. In some reports, REP elements may be denoted as REP-DEL (found in SVs/CNVs with a gene deletion) and REP-DUP (found in SVs/CNVs with a gene duplication). Due to the lack of systematic characterization of the REP elements within various SVs/CNVs, their sequence variability in these elements remains unclear. Therefore, graphical displays in this tutorial do not differentiate REP6, REP7, REP-DUP, and REP-DEL, while illustrations in the PharmVar Structural Variation document do (37).
The CYP2D7 downstream region is highly similar to the CYP2D6 downstream region, with a major difference being the aforementioned 1.6-kb insertion known as the spacer, which maps to GRCh38 chr22:42138118 to 42139679) (34, 38, 39). The corresponding insertion site downstream of CYP2D6 maps to NG_008376.4 position 9845_9846 or 9848_9849 depending on alignment parameters corresponding to GRCh38 positions chr22:42125963_42125962 and chr22: 42125960_42125959, respectively. A graphical depiction of the spacer and CYP2D6 insertion site is available in the PharmVar Structural Variation document (37). Several CYP2D6::CYP2D7 hybrid alleles have the spacer in their downstream region, including CYP2D6*36.001 (note that *36 can also occur as a singleton with a CYP2D6-like downstream region without the spacer) (Figure 1d). The unique spacer sequence has been exploited for primer design to specifically amplify alleles with or without this structural feature.
Variants in CYP2D7 can also cause interference with CYP2D6 genetic analyses. For example, a reported SNV in CYP2D7 reverted the sequence to match CYP2D6, allowing primer binding and amplification that caused false-positive heterozygous calls for 137insT and 31G>A, interfering with CYP2D6*15 and *35 genotyping assay results (40). Therefore, CYP2D7 variants may interfere with assays and methods interrogating CYP2D6 SV/CNV status.
PharmVar currently lists CYP2D6*1 through *172 for a total of 163 distinct star alleles (nine star numbers are retired), not counting suballeles, many of which harbor numerous SNVs. Star allele designations have also been issued to describe gene deletions (CYP2D6*5) and hybrid alleles (CYP2D7::CYP2D6 hybrids: CYP2D6*13; CYP2D6::CYP2D7 hybrids: CYP2D6*36, *61, *63, *68, *83). Unlike CYP2D7::CYP2D6 hybrids, which differ in switch region but are nonetheless all nonfunctional and collectively referred to as CYP2D6*13, there is no single star allele designation for CYP2D6::CYP2D7 hybrid genes, as it has not been conclusively shown that the exon 9 conversion, a shared feature, renders these nonfunctional. SVs/CNVs with duplications or multiplications do not receive individual star designations and are described in detail below.
3. CYP2D6 Structural Variants
Recombination events have created an array of SVs, all of which are CNVs. An overview of SVs/CNVs is shown in Figure 1 (see the PharmVar Structural Variation document (37) for a comprehensive summary).
Population-based frequency data are available for the commonly tested CYP2D6*5 gene deletion. Although some frequency data have been published for other SVs/CNVs, their accuracy depends on how testing was performed. For example, diplotypes with a CYP2D6*2 allele and a duplication may have been reported as having a CYP2D6*2 duplication by default. In the absence of additional testing, this defaulting strategy can lead to the misclassification of other duplication alleles such as CYP2D6*1×2 or *4×2 as *2×2. This practice, which is now discouraged, likely led to an overestimation of the frequency of CYP2D6*2×2, especially in older publications. More recent studies have shown that approximately 3.5–8.4% of the United States population is predicted to have a CYP2D6 SV/CNV, depending on ancestry (41, 42). Frequencies may even exceed 30% in some Asian populations (42–46), indicating that SVs/CNVs are not rare in individuals of non-European descent. Therefore, their inclusion in routine testing is important for both comprehensive and equitable patient care.
CYP2D6 allele frequencies are provided in the PharmGKB allele frequency table (47). However, due to study limitations, the tabulated frequencies are likely to be incomplete and, including those for SVs/CNVs, should be treated as estimates.
3.1. Definitions and Terms
Describing CYP2D6 structural variants in a standardized manner is challenging, especially because there is no guidance from the Human Genome Variation Society (HGVS) on how best to describe SVs/CNVs. Table 1 provides definitions for SV/CNV-related terms (48, 49) and those used specifically for pharmacogenes, as illustrated for CYP2D6. PharmVar recommends utilizing these terms and the activity score (AS) system (3, 50) to facilitate standardized descriptions and reporting of CYP2D6 structural variants and translating SV/CNV-containing diplotypes into phenotypes.
Table 1.
Terms used to describe structural variants in pharmacogenes, including CYP2D6
| Terms (abbreviation) | What the term means for CYP2D6 |
|---|---|
| Star allele and haplotype | A star allele (*) describes a haplotype using a defined gene region (e.g., CYP2D6*1, *2). A star allele can have any number of variants and contain an SV/CNV. The region used to define CYP2D6 star alleles encompasses 6066 bp (NG_008376.4 positions 3436 through 9501 counting from the sequence start or -1584 through 4482 counting from the translation start). The core allele for each star number represents all variants that i) result in an amino acid or ii) have been shown to alter function and are present in all suballeles. The term allele is often used to describe all variants present in a gene copy on the nucleotide level (e.g., a C to G change); CYP2D6 star allele nomenclature should not be used to refer to a single variant unless that star allele only has a single variant (e.g., *18). For example, 100C>T (rs1065852) itself should not be described as a CYP2D6*10 allele or reported as CYP2D6*10 allele frequency because the variant also occurs in many other star alleles or haplotypes, including the common CYP2D6*4 allele. |
| Diplotype and genotype | A combination of two haplotypes (star alleles) constitutes the diplotype. For example, CYP2D6*2/*10 indicates the presence of one *2 allele and one *10 allele. The term genotype is often used instead of diplotype, although the latter is more accurate. |
| Structural variation (SVs) and gene copy number variation (CNVs) | An SV is a rearrangement involving a region of DNA ≥1 kb. SVs include copy-neutral inversions and balanced translocations, as well as rearrangements resulting in genomic imbalances, such as insertions and deletions. These genomic imbalances are also commonly referred to as copy number variants (CNVs). All CNVs are SVs, but not all SVs are CNVs. |
| Copy number variation (CNV) | All CNVs are SVs. CNVs include gene deletions (copy number loss) and duplications or multiplications, with the latter representing copy number gains. A CNV may include one or several exons or full genes and exceeds 1 kb. There are no known examples of intragenic duplications or deletions of only one or several exons of CYP2D6. The terms SV and CNV are often used interchangeably. |
| Multiallelic CNV (MCNV) | This term indicates that there are multiple forms of CNVs for a gene. CYP2D6 can be described as multiallelic CNV because there are alleles with deletions, duplications, or multiple gene copies. This term, however, is rarely used to describe CYP2D6 CNVs. |
| Insertion–deletion variants (indels and delins) | Insertion–deletion variants (indels) are DNA insertions, deletions, or combined insertion-deletion variants referred to as delins. These are typically less than 1 kb. Although the term indel could be used to describe small CYP2D7-derived regions within CYP2D6, these are typically referred to as “conversions” (see below). |
| Identical gene duplications | At least two identical or near-identical copies of the entire CYP2D6 gene on the same allele, e.g., *1x2, *2x3, *4xN. Over 90% of CYP2D6 SVs involve identical copies of the gene. NCBI refers to a duplication of two identical, adjacent DNA regions as a tandem duplication. Gene duplications may be referred to in the literature or reports as copy number gain. |
| Non-identical gene duplications | At least two non-identical gene copies on the same allele. Each gene copy has nine exons. One of the gene copies may be a hybrid with one or more exons corresponding to CYP2D7. Non-identical gene copies are defined as having different core star allele designations. Examples include *36+*10, *68+*4, *1+*90, and *164+*4. Non-identical gene copies should be listed in order of appearance, with the 5’-most gene copy listed first. Non-identical gene duplications have also been referred to as “tandem” duplications; however, to avoid conflict with NCBI’s definition, PharmVar does not recommend this terminology. |
| Full/entire gene | Full or entire gene (or gene copy) indicates that all nine exons are from CYP2D6 or CYP2D7. |
| Gene deletion | Deletion of the entire CYP2D6 gene; this SV is designated as CYP2D6*5. Gene deletions may be referred to in the literature or in reports as copy number loss. |
| Hybrid gene | A gene copy consisting of nine exons with portions of CYP2D6 and CYP2D7; these have also been referred to as “fusion” genes in the literature. Designations of CYP2D7::CYP2D6 and CYP2D6::CYP2D7 are used to differentiate which gene comprises the 5’ portion of the hybrid. Examples include *13 (CYP2D7::CYP2D6 hybrid) and *68 (CYP2D6::CYP2D7 hybrid). PharmVar defines two categories of CYP2D6::CYP2D7 hybrid genes. Category A hybrid genes have one or multiple CYP2D7-derived exon(s), including exon 9, but switch back to CYP2D6 either late within exon 9 or the immediate 3’UTR; these hybrids have a CYP2D6-like downstream region and lack the CYP2D7 spacer. In category B hybrid genes, the CYP2D7 portion extends beyond exon 9; therefore, these entities have a CYP2D7-like downstream region. A *36 in a *36+*10 arrangement is typically a category B hybrid gene featuring a CYP2D7-like downstream region, while the singleton *36 in a *1/*36 represents a category A hybrid gene with a CYP2D6-like downstream region. Category A and B hybrid genes are typically not distinguished or specified in clinical reports. |
| CYP2D7 conversions | A small region of CYP2D7-derived sequence embedded within the CYP2D6 gene. CYP2D7 conversions in intron 1 and exon 9 are often referred to as the “intron 1” and “exon 9” conversions. A singleton CYP2D6*36 (category A hybrid) is an example of an allele with an embedded exon 9 conversion. The intron 1 conversion is found on several star alleles, including many *2 alleles. Conversions are typically not detailed in clinical test reports, where they may not be differentiated from hybrids. |
| Downstream gene copy | The 3’-most of two or more gene copies on the same chromosome. This gene copy may also be referred to as the non-duplicated gene copy. For example, CYP2D6*10 is the downstream (3’) gene copy of a *36+*10. Of note, the “non-duplicated copy” may also refer to the allele on the opposite chromosome, e.g., the CYP2D6*1 in a *1/*36+*10. |
| Upstream gene copy or copies | The 5’-most gene copy (or copies) located between the CYP2D7 gene and the downstream (3’) gene copy. This gene copy may also be referred to as the duplicated gene copy or copies. For example, the CYP2D6*36 and one of the *10 gene copies are the upstream (5’) gene copies of a *36+*10×2. |
| Copy Number (CN) assays | CN assays provide quantitative measurement of copy number at a given genomic location. For CYP2D6, commonly targeted regions for CN assays include the 5’UTR, introns 2 and 6, and exon 9. CN assay calls at these loci are used to identify CNVs and derive the most likely gene structure. |
| Singleton | The term singleton is often used to indicate that a gene copy, specifically a hybrid gene such as CYP2D6*36 or *68, is not in a duplication arrangement but rather the only gene copy present. |
3.2. Categories of CYP2D6 Structural Variants
Structural variants of CYP2D6 can be classified by several common features when compared to the reference gene locus.
3.2.1. Gene Deletions
A large deletion encompassing the entire CYP2D6 gene was first described in 1991 in a subset of individuals presenting as PMs (28), and subsequently designated as the CYP2D6*5 allele (Figure 1b). This allele (activity value for AS calculation = 0) is the most interrogated SV/CNV due to its relatively high frequency across populations and ease of detection.
CYP2D6*5 allele frequencies vary considerably, with the highest reported in the Xhosa population (up to 16%). In contrast, it is rare or absent in some American and East Asian populations.
3.2.2. Gene Duplications and Multiplications
The gain of a complete gene copy or copies (unit(s) with nine exons) is referred to as a gene duplication (when an allele has gained one complete gene copy) or multiplication (when an allele has gained two or more complete gene copies) (Figure 1c and 1e). Gene duplications were first described in the early 1990s (51–53) and identified as the underpinning cause of ultrarapid metabolism. The most common allele with a copy number gain is the CYP2D6*2×2 duplication, with frequencies ranging from 0% to 3.3% across the major population groups, but considerably higher frequencies in Ethiopians (CYP2D6*2×2, x3, x4, and x5, 16%) (54) and Saudi Arabians (CYP2D6*2xN, 10.4%) (55). Since testing was limited when the latter studies were conducted, these reported frequencies should be viewed cautiously. Other reported CYP2D6 gene duplications include CYP2D6*1×2, *3×2, *4×2, *6×2, *9×2, *10×2, *17×2, *28×2, *29×2, *35×2, *41×2, *43×2, *45×2, and *146×2. Depending on the activity conveyed by each gene copy, per CPIC clinical allele function assignments (3), duplications can confer increased function (e.g., *1×2, *2×2; activity value = 2), no function (e.g., *3×2, *4×2; activity value = 0), decreased function (e.g., *10×2, *41×2 [CPIC downgraded the value assigned to CYP2D6*41 to 0.25 in March 2023 to reflect the function of this allele more accurately] activity value = 0.5), normal function (e.g., *17×2, activity value = 1), or uncertain function (e.g., *146×2). Therefore, it is of utmost importance to know the nature of the gene duplication to accurately predict phenotype. Diplotypes that confer a predicted UM status include CYP2D6*1/*2×2 and *1/*35×2, while *2×2/*4 and *2/*4×2 predict NM and IM phenotypes, respectively. For a complete list of diplotypes with activity scores, see the PharmGKB CYP2D6 Diplotype to Phenotype table (47).
3.2.2. Hybrid genes
In addition to CYP2D6 gene duplications and deletions, unequal recombination events also generated gene copies containing a combination of CYP2D6 and CYP2D7 sequences. PharmVar refers to these genes as hybrids, but they have also been described as “fusion” genes. The first of these alleles was reported in 1995 (56).
There are two recognized types of hybrid genes. CYP2D7::CYP2D6 hybrids have 5’ portions of CYP2D7 (including at least exon 1) and 3’ portions of CYP2D6 (31, 32, 34, 42). Such hybrid genes were consolidated under a single star number, CYP2D6*13, since they are all nonfunctional (activity value = 0) due to a T-insertion in their CYP2D7-derived exon 1 (57). Figure 1d illustrates two such alleles, one switching in exon 7 and one in intron 1. CYP2D6*13 alleles occur as singletons and in duplication arrangements (e.g., with a CYP2D6*2 gene copy, Figure 1e), neither of which carry a full copy of the CYP2D7 gene.
CYP2D6::CYP2D7 hybrids have 5’ portions of CYP2D6 and 3’ portions of CYP2D7, along with a full, upstream copy of CYP2D7. Although all such hybrids have a CYP2D7-derived exon 9, they are not consolidated under a single star designation due to uncertainty regarding whether the variants comprising the CYP2D7 exon 9 conversion on their own render the protein nonfunctional. CYP2D6::CYP2D7 hybrid genes differ in the location of the switch to CYP2D7 and whether they have a CYP2D6 (category A) or CYP2D7-derived (category B) downstream region (an overview of hybrid classification categories is provided in the PharmVar SV document) (37). The two most common CYP2D6::CYP2D7 hybrid genes are CYP2D6*36 and *68 (both have an activity value of 0) (Figure 1d). Both can occur as singletons, or, more commonly, in duplication arrangements. CYP2D6*36 and *68 are most often found upstream of a *10 and a *4, respectively, in CYP2D6*36+*10 and *68+*4 duplication structures (Figure 1e). The former is relatively common in East Asians (45) but rare in Europeans (41). In European populations, 15% to 25% of CYP2D6*4 alleles are in a *68+*4 configurations (unpublished author observations).
4. PharmVar-recommended Nomenclature for CYP2D6 Structural Variants
An array of naming approaches has been used throughout the literature and clinical reporting to describe CYP2D6 diplotypes (commonly referred to as genotypes but often used interchangeably), including those with SVs, creating a significant risk of misinterpretation. For example, genotypes with duplications are often reported as CYP2D6*2/*4 (dup) or (*2/*4)xN if the nature of the duplication is unknown; the allele with the duplication is listed first or second, e.g., *1×2/*1 or *1/*1×2; lower or upper case “x” denotes the copy number, e.g., *2×2 or *2X2; non-identical gene copies are displayed in different orders separated by a “+” or “-“, e.g., *10-*36 or *10+*36, and the list goes on. In addition, due to limitations of certain electronic health records (EHR) or other (automated) report delivery software, the star (*) may not be included in some reports (e.g., CYP2D6 1×2/1).
The lack of conformity may be driven by individual preferences in the absence of directives, as well as limitations of software or fixed database requirements. Due to requirements and limitations of laboratory information systems, EHRs, or interfaces between laboratories and client sites, a result of CYP2D6*68+*4/*5 may ultimately be reported as 68+4/5, *4+*68/*5, or other formats that do not conform to the PharmVar recommendations.
Likewise, hybrids have been described using a hyphen or forward slash, e.g., CYP2D6/2D7, CYP2D6-CYP2D7, CYP2D6-D7, or similar. The Hugo Gene Nomenclature Committee (HUGO) has recently introduced a unique double-colon separator to describe hybrid genes (58). To standardize reporting, PharmVar has adopted the HUGO recommendation to represent hybrid genes as CYP2D6::CYP2D7 and CYP2D7::CYP2D6.
PharmVar recommends annotating and reporting SV/CNV alleles as summarized in Table 2 and encourages all stakeholders to adopt these recommendations to facilitate standardized reporting of SVs/CNVs.
Table 2.
PharmVar-recommended annotations and reporting of structural variants. The table provides a selection of SV/CNV alleles to exemplify the recommendations.
| Recommendation | Examples | Interpretation |
|---|---|---|
| The allele with the lower star number is written first. | *1/*4 | A CYP2D6*1 gene copy on one allele and a *4 gene copy on the other allele. |
| *5/*10 | A full deletion on one allele and a CYP2D6*10 gene copy on the other allele. | |
| For identical gene copies on the same allele (in cis), the star allele is followed by an “×” and the number of gene copies. Gene copies may vary on the suballele level. If the number of gene copies is unknown, write the star number followed by “×N”. |
*1×2 *2×2 *4×2, etc. |
Two CYP2D6 gene copies on the same allele. |
|
*2×3
*41×3 |
Three CYP2D6 gene copies on the same allele. | |
|
*2×N
*36×N |
Two or more CYP2D6 gene copies on the same allele. | |
| When an SV/CNV is present, but the duplicated allele cannot be discriminated, write both possible diplotypes; i) if the number of gene copies is unknown, write the star number followed by “×N”; ii) alternatively, genotypes may also be reported in brackets to denote ambiguity. | *2×2/*4 or *2/*4×2 | Two CYP2D6*2 gene copies on one allele, and one *4 gene copy on the other allele, or one *2 gene copy on one allele, and two *4 gene copies on the other allele. This may introduce phenotypic ambiguity. |
| *2×N/*4 or *2/*4×N | The number of gene copies is unknown, and it was not determined which allele has the SV/CNV. | |
|
(*2/*4)×3
(*2/*4)×N |
Provision of the genotype in brackets denotes that it was not determined which allele has the SV/CNV; ×3 indicates a total number of gene copies; ×N denotes that the total number of gene copies is unknown. | |
| For non-identical gene copies on the same allele (in cis), or multiple SVs/CNVs on the same allele, the upstream gene copy is written first, followed by a “+” and the downstream gene copy. Although the order of the gene copies may not be experimentally determined in routine clinical testing, they should be displayed in their most likely order for consistency. |
*1+*90 | One CYP2D6*1 gene copy is upstream of one *90 gene copy on the same allele. |
| *68+*4 | One CYP2D6*68 hybrid gene copy is upstream of one *4 gene copy on the same allele. | |
| *13+*2 | One CYP2D6*13 hybrid gene copy is upstream of one *2 gene copy on the same allele. | |
| *36+*10 | One CYP2D6*36 hybrid gene copy is upstream of one *10 gene copy on the same allele. | |
|
*36×2+*10×2 *36+*10×2
*36×2+*10 *36×N+*10×N |
One or two CYP2D6*36 hybrid gene copies are upstream of one or two *10 gene copies on the same allele; higher copy number states likely also exist. | |
| *13+68×2+*4 | One CYP2D6*13 hybrid gene copy is upstream of two CYP2D6*68 hybrid gene copies upstream of one *4 gene copy on the same allele. | |
| For diplotypes containing an SV/CNV, if both alleles have the same star number, write the allele without the SV first. For SV alleles with two nonidentical gene copies, the star number of the downstream (3’) gene copy determines whether the allele is displayed first or second in a diplotype. |
*1/*1×2 | One CYP2D6*1 gene copy on one allele and two *1 gene copies on the other allele. |
| *2×2/*4 | Two CYP2D6*2 gene copies on one allele and one *4 gene copy on the other allele. | |
| *68+*4/*10 | One CYP2D6*68 hybrid gene copy upstream of one *4 gene copy on one allele, and one *10 gene copy on the other allele; the *68+*4 allele is written first due to *4 being the 3’ gene with the lower star number. | |
| *2/*36+*10 | One CYP2D6*2 gene copy on one allele and a *36 hybrid gene copy upstream of one *10 gene copy on the other allele; the *36+*10 is written second due to *10 being the 3’ gene with the higher star number. | |
| *1/*13+*2 | One CYP2D6*1 gene copy on one allele, and a *13 hybrid gene copy upstream of one *2 gene copy on the other allele; the *13+*2 is written second due to *2 being the 3’ gene with the higher star number. | |
| For diplotypes that contain SVs/CNVs on both alleles, the downstream gene with the lowest star number is written first. | *2×2/*41×2 | Two *2 gene copies on one allele and two *41 gene copies on the other. |
| *2×2/*68+*4 | Two CYP2D6*2 gene copies on one allele, and one CYP2D6*68 hybrid gene copy upstream of one *4 gene copy on the other allele. | |
| *13+*2/*36+*10 | One CYP2D6*13 hybrid gene copy upstream of one *2 gene copy on one allele, and one *36 hybrid gene copy upstream of one *10 gene copy on the other allele. | |
| *68+*4/*1+*90 | One CYP2D6*68 hybrid gene copy upstream of one *4 gene copy on one allele and one *1 gene copy upstream of one *90 gene copy on the other allele. | |
| When an SV/CNV is present, but its structure has not been further validated, i) a default assignment may be used or ii) all possible diplotypes written. |
*13+*2/*4 |
The CYP2D6*13 gene copy is defaulted to being upstream of the *2 gene copy as *13+*4 has not been described. Other possible diplotypes include i) *2+*4/*13, ii) *4+*2/*13, or ii) *2, *4, and *13 on one allele and a *5 gene deletion on the other, although (i), (ii), and (iii) have not been described and are less likely. |
| For diplotypes with SV/CNV(s) and SNV(s) that cannot be reconciled with known star alleles and SV/CNV structures. | n/a | Samples that cannot be interpreted should be reported as “indeterminate”. In such cases, it would be appropriate to inform the ordering provider that additional testing would be necessary to elucidate the SV/CNV structure(s) and determine the nature of the CYP2D6 gene copies present. |
| Gene symbols for hybrids are separated by a double colon. | CYP2D6::CYP2D7 or CYP2D7::CYP2D6 | Gene symbols are no longer separated by a hyphen or forward slash but by a double semicolon per HUGO recommendation (58). Adapting this recommendation for CYP2D6 facilitates instant recognition of such gene structures. |
5. Methods for Characterizing CYP2D6 Structural Variants
To accurately predict a patient’s CYP2D6 activity (metabolizer status), testing must include SV/CNV analysis (59) at more than one location within CYP2D6 to differentiate identical gene duplications (e.g., *2×2/*4) and hybrid genes (e.g., *13+*2/*4). Many approaches and platforms are available to characterize SVs/CNVs, each with inherent limitations. These approaches are often combined with genotyping to make genotype (or diplotype) calls. Following are summaries of some common methods used to interrogate SVs/CNVs, though other approaches have been utilized (60–62). Most methods perform well with standard DNA preparation procedures (e.g., column- or bead-based) while others may require high-molecular-weight DNA. While a review of DNA preparation methods is beyond the scope of this tutorial, some limitations and challenges related to DNA quality and/or purity are highlighted. It is recommended to use reference materials (see section 6.3 below) to establish and validate methods and to properly follow protocols and/or manufacturer’s recommendations.
5.1. Qualitative Testing Using Long-Range PCR (XL-PCR)
The presence of SVs/CNVs can be detected qualitatively by long-range PCR (commonly abbreviated as XL-PCR, although LR-PCR is also used), in which various combinations of CYP2D6- and CYP2D7-specific primers are employed (30–34, 63–65). Table 3 provides a selection of XL-PCR reactions that can be utilized to amplify SV/CNV-specific amplicons, and Figure 1b–e provides a graphical overview of the amplified region(s). For example, the presence of two or more gene copies on a single allele can be assessed with a CYP2D6-specific forward primer and a reverse primer that binds within the region between the duplicated gene copies (parts of this intergenic region are unique to duplication structures). Amplicons may encompass the entire upstream (5’) gene copy (previously referred to as the “duplicated” gene). A duplication structure can also be identified by other reactions targeting a specific region. XL-PCR has also been utilized to amplify a fragment that includes exon 9 of the 5’ gene copy, the intergenic region, and part of—or the full—3’ gene copy. Amplification of these products supports the presence of one or more additional gene copies. Amplicon size differs by 1.6 kb depending on whether the 5’ gene copy has a CYP2D6-like (no spacer) or CYP2D7-like (with spacer) downstream region. For example, amplicon 7 generated from the upstream CYP2D6*68 gene copy of a *68+*4 has the spacer, so it is 1.6 kb longer than the amplicon generated from the 5’ gene copy of a *2×2 (Figure 1). As shown for the CYP2D6*5 gene deletion and singleton CYP2D6::CYP2D7 hybrid genes, certain primer pairs amplify these SVs/CNVs if they have a CYP2D7-derived exon 9 and downstream region (Figure 1b, XL-PCRs 3 and 4) but fail to do so if their downstream regions are CYP2D6-derived. While most alleles generating this XL-PCR product are indeed CYP2D6*5, there are exceptions such as some CYP2D6*36 alleles.
Table 3.
Long-range PCR amplicons can be used to detect and characterize structural variants.
| XL-PCR | Description |
|---|---|
| 1 | • CYP2D6-specific forward (F) and reverse (R) primers. • The R-primer spans the spacer insertion site and can be paired with different F primers to amplify longer or shorter amplicons. • This XL-PCR amplifies each CYP2D6 gene copy in duplications and multiplications. |
| 2 | • The R primer binds downstream of the CYP2D locus (i.e., the most 3’ gene) and can be paired with different F primers to amplify longer or shorter amplicons. • In cases of duplications or multiplications, this XL-PCR only amplifies the 3’ gene copy. |
| 3, 4, 5 | • F primers binding within the CYP2D7-specific spacer (XL-PCR 3), CYP2D7 exon 9 (XL-PCR 4), or CYP2D7 intron 6 (XL-PCR 5) and paired with an R primer that binds downstream of the CYP2D6 locus will produce amplicons from the *5 gene deletion allele (F primer binds to the remaining CYP2D7 gene copy). • Since the R primer for these XL-PCRs is neither CYP2D6- nor CYP2D7-specific, it will bind regardless of the nature of the downstream region of the most 3’ gene in the allele. • XL-PCRs 3 and 4 also amplify singleton *36 if they have a CYP2D7 downstream region, and XL-PCR 5 amplifies singleton *13 hybrids if they switch to CYP2D6 upstream of intron 6. |
| 6 | • The F primer is specific for a CYP2D6-derived downstream region, and the R primer binds to the duplication-specific intergenic region. • This XL-PCR amplifies if the 5’ gene copy or copies in a duplication or multiplication arrangement have a CYP2D6-derived downstream region such as *2x2 and *41x3, but does not amplify when the 5’ gene copy has a CYP2D7-derived downstream region, e.g., the *36 of a *36+*10. |
| 7 | • The F primer is CYP2D6-specific, and the R primer binds to the duplication-specific intergenic region. • This XL-PCR amplifies the entire 5’ gene copy or copies regardless of whether they have a CYP2D6 or CYP2D7-derived downstream region. • Amplicons generated from the latter are 1.6 kb longer due to having the spacer. |
| 8 | • The F primer is specific to CYP2D6 exon 9, and the R primer is specific to intron 1. • This XL-PCR amplifies the intergenic region and small parts of the 5’ gene copy (exon 9) down through the 3’ region of the downstream gene copy (exon 1 and parts of intron 1). • This amplicon is 1.6 kb longer if the 5’ gene has a CYP2D7-derived downstream region with the spacer. |
| 9 | • The F primer is CYP2D6-specific, and the R primer is specific for CYP2D7 exon 9. • This XL-PCR amplifies CYP2D6::CYP2D7 hybrid genes. |
| 10 | • The F primer is CYP2D7-specific, and the R primer (same as XL-PCR 1) is specific for CYP2D6; this XL-PCR amplifies CYP2D7::CYP2D6 hybrid genes. • These hybrid genes can also be amplified with a CYP2D6-specific primer binding to exon 9. |
F and R denote forward and reverse primers; XL-PCR, long-range PCR.
The listed XL-PCR reactions represent a selection and are not intended to include the numerous primers and primer combinations that have been published. The selected reactions illustrate the concept of pairing primers that bind to CYP2D6 or CYP2D7 sequences to amplify specific SVs and highlight how certain primer combinations can amplify from multiple loci within an allele or amplify from various SVs. See references for primers, primer pairs, amplification lengths, and other details (30–34, 63–65).
This table accompanies the XL-PCR amplifications detailed in Figure 1.
Hybrid genes can be amplified using CYP2D6- and CYP2D7-specific forward and reverse primers regardless of their location within the structure. Specifically, XL-PCR 9 only amplifies CYP2D6::CYP2D7 hybrid genes, whereas XL-PCR 10 only amplifies CYP2D6*13, and XL-PCR 2 is designed to amplify only the 3’ gene copy.
There are limitations and caveats to XL-PCR analysis. The absence of a PCR product may be difficult to interpret. For example, it could mean that the interrogated structure (duplication, hybrid, deletion, etc.) is indeed absent, but could also be the result of failure to amplify due to inferior DNA quality, inhibitors in the DNA preparation, and/or the presence of interfering rare/novel SNV(s). In rare cases a novel SV/CNV (e.g., a previously unknown hybrid or deletion) may also escape detection because the primer(s) used in a reaction may not be able to bind or may be too distant to generate a PCR product. Also, some amplicons may be generated from multiple locations. For example, XL-PCR 1 amplifies all copies in SVs/CNVs with two or more identical CYP2D6 gene copies, and XL-PCR 3 and 4 amplify CYP2D6*5 gene deletions and certain singleton *13 hybrid genes (Figure 1e). Thus, it is important to carefully consider where the primers bind, and which structure(s) can be amplified. Lastly, although long fragments of up to 40 kb have been generated, XL-PCR products over 15 kb may be difficult to generate reproducibly.
Amplicons may be visualized by agarose gel or micro-capillary electrophoresis for size discrimination and then sequenced in full to determine their exact nucleotide identity, partially sequenced, or genotyped to determine the nature of a gene duplication.
5.2. Quantitative Testing
Quantitative testing measures how many copies are present at an interrogated gene region, resulting in a copy number (CN) determination. Different methods and assays are available, the most common of which are discussed here.
The overall performance of any CYP2D6 CN test method depends on how many targets within the CYP2D locus are interrogated (Table 4). Clinical recommendations for CYP2D6 genotyping suggest that at least two positions (ideally one targeting the 5′ untranslated region or exon 1 and another targeting exon 9) should be interrogated to detect gene duplications and deletions accurately and to identify and differentiate hybrid genes from full gene copies (59).
Table 4.
Quantitative copy number calls for four commonly targeted gene regions and qualitative testing by long-range PCR
| Diplotype | Activity Score (AS) | Coriell ID | 5’UTR | Intron 2 | Intron 6 | Exon 9 | XL-PCR1 |
|---|---|---|---|---|---|---|---|
| Copy number (CN) calls at the targeted region | Amplicons generated | ||||||
| *2×2/*4 2 | 2 | NA23296 | 3 | 3 | 3 | 3 | 1, 2, 6, 7, 8 |
| *2/*4×2 | 1 | NA19819 | 3 | 3 | 3 | 3 | 1, 2, 6, 7, 8 |
| *2/*41×3 | 1.75 | NA24217 | 4 | 4 | 4 | 4 | 1, 2, 6, 7, 8 |
| *1/*36+*10 2 | 1.25 | NA23093 | 3 | 3 | 3 | 2 | 1, 2, 7, 9 |
| *1/*13+*2 2 | 2 | NA19785 | 2 | 3 | 3 | 3 | 1, 2, 6, 8, 10 |
| *1/*13+*2 3 | 2 | NA19790 | 2 | 2 | 3 | 3 | 1, 2, 6, 8, 10 |
| *1/*4.013+*4 4 | 1 | NA10860 | 3 | 3 | 3 | 2 | 1, 2, 7, 9 |
| *1/*36×2+*10 | 1.25 | NA18526 | 4 | 4 | 4 | 2 | 1, 2, 7, 9 |
| *1+*90/*36+*10 | n/a5 | NA18642 | 4 | 4 | 4 | 3 | 1, 2, 6, 7, 8, 9 |
| *5/*43 | n/a6 | HG03246 | 1 | 1 | 1 | 1 | 1, 2, 3, 4, 5 |
| *68+*4/*5 2 | 0 | HG01190 | 2 | 1 | 1 | 1 | 1, 2, 3, 4, 5, 7, 9 |
| *5/*36×2+*10×2 | 0.5 | NA18545 | 4 | 4 | 4 | 2 | 1, 2, 3, 4, 5, 6, 7, 9 |
| *36×2+*10/*52 | 0.75 | NA18632 | 4 | 4 | 4 | 2 | 1, 2, 7, 9 |
| *13+*2/*68+*4 | 1 | n/a | 3 | 3 | 3 | 3 | 1, 2, 6, 7, 8, 9, 10 |
Details for long-range (XL-PCR) are provided in Table 3 and Figure 1. Amplicons expected to amplify with the various primer combinations are listed.
The CYP2D6*13 gene copy in NA19785 switches in intron 1, while the *13 in NA19790 switches in intron 4, thus accounting for the differing CN calls for introns 2 and 6.
The upstream CYP2D6*4 gene copy in NA10860 has an exon 9 conversion and thus produced a CN call of 2 for this region; this suballele is designated *4.013 (note that the *4N legacy designation does not denote “duplication”). Diplotypes with a CYP2D6*4 duplication containing this suballele can be reported as *4×2.
Since the function of CYP2D6*90 is uncertain, its phenotype is indeterminate (AS = not available); however, based on the presence of one CYP2D6*1 and one *10 gene copy, the AS is at least 1.25; some laboratories may provide a range of AS reflecting *90 having no, decreased, normal, or increased activity.
Since the function of *43 is uncertain, its phenotype is indeterminate (AS = unavailable); some laboratories may provide a range of AS reflecting *43 having no, decreased, normal, or increased activity.
5.2.1. Quantitative Real-Time PCR (qPCR)
In this relative quantitation method, CN assays specific for CYP2D6 and a reference gene are run as a duplex real-time PCR. The reference assay detects a sequence virtually devoid of CN variation, such as the housekeeping genes RNaseP, TERT, or GADPH. For CYP2D6, CN assays are designed to target specific regions throughout the gene, including but not limited to the 5’UTR, intron 2, intron 6, and exon 9. Experiments include unknown samples and enough “known” or “normal” 2-copy samples to ensure statistically sound analyses.
After amplification, the cycle threshold (CT) values for the target and reference genes in each reaction are analyzed by the comparative CT (ΔΔCT) relative quantitation method to determine the number of copies of the target sequence (66). First, the ΔCT is calculated for the target and reference gene in each well and averaged across sample replicates (multiple replicate reactions are required for each sample). Then, the ΔCT is compared to the ΔCT for the calibrator samples or against the average of the “normal” 2-copy samples.
5.2.2. Digital PCR (dPCR)
Absolute quantification of gene copies can be achieved with dPCR, in which the genomic DNA sample and reaction are divided into many partitions (typically thousands) so that within each partition, target DNA may or may not be present, and therefore amplification may or may not take place. Similar to relative quantification using qPCR, CYP2D6 and reference gene-specific CN assays are used, but normalization to a reference gene is optional. After thermal cycling, the number of amplification-positive and -negative partitions are measured, and a Poisson distribution-based algorithm quantifies the targeted gene region in the sample.
Although qPCR and dPCR allow quantitative detection of nucleic acids, dPCR has several advantages, including detection of small fold-changes, higher tolerance to interfering inhibitors, and independence from standard curves. Another advantage of dPCR is that two or more targets may be interrogated in the same reaction to save costs (67), as assays targeting the CYP2D6 5’UTR and exon 9 regions can be easily duplexed (46, 65).
5.2.3. Limitations and Caveats of qPCR and dPCR
qPCR is a powerful method that maintains statistical significance in samples with sufficient DNA concentration (≥100 copies of target DNA), allowing reliable quantification of up to five or more copies. However, DNA samples with suboptimal concentration, low quality (degradation, fragmentation), purity issues (presence of interfering substances), or a combination thereof may result in inaccurate CYP2D6 CN calls as normalized against a reference gene. The quality of DNA isolated from formalin-fixed paraffin-embedded (FFPE) tissues is typically compromised and unsuitable for CYP2D6 CN testing using qPCR ((68) and unpublished observations by authors); to our knowledge, there are no studies demonstrating that dPCR reliably measures CN in FFPE sample-derived DNA. Thus, for accurate and consistent results, DNA samples should undergo strict pre-analytical quality control before entering downstream analyses. Spectrophotometric assessment (i.e., A260/A280) and electrophoresis-based methods are two easy and inexpensive ways to determine the gross purity and relative state of sample degradation, respectively. In contrast, dPCR is more tolerant of inhibitors and suboptimal sample quality, allowing high-precision quantification of up to 8–11 copies (69, 70).
Both qPCR and dPCR enable a full multi-point analysis of hybrid gene copies, but they may be unable to detect (i) deletions in the presence of duplications or multiplications (i.e., CYP2D6*2×2/*5 vs. *2/*2); (ii) samples containing both CYP2D6::CYP2D7 and CYP2D7::CYP2D6 hybrids (these may also be referred to as “balanced hybrids”) may yield uniform CN calls across all loci, erroneously suggesting the presence of full gene copies (e.g., CYP2D6*13+*2/*68+*4 may produce CN calls of 3 depending on the assays used and where the *13 switches); (iii) CYP2D7::CYP2D6 hybrid genes switching after the exon 9 region (targeted by many CN assays) may escape detection (these could also be described as “CYP2D7 with a CYP2D6 downstream region”); (iv) variants in some star alleles may interfere with CN assays, inhibiting or preventing amplification of the interrogated region, causing allele drop-out and yielding results that are difficult to interpret (71); and (v) commonly used reference (control) genes including RNAseP and TERT may in rare instances have a CN or variant that leads to decreased amplification efficiency and thus, affect the accuracy of CYP2D6 CN calls (72).
5.3. Determination of Which Allele Is Duplicated
5.3.1. Utility of TaqMan Genotyping Assays and Other Methods
Real-time PCR genotyping assays, including TaqMan, may inform which allele carries the duplication by examining the amplification ratios of variants that differ between the two alleles (e.g., heterozygous calls). For example, heterozygosity at positions 100C>T (rs1065852), 1847G>A (rs3892097), and 2851C>T (rs16947) may be utilized to discriminate whether a sample has a duplication on the CYP2D6*2 or *4 allele.
As illustrated in Figures 2a and b for 1847G>A and 2851C>T, signal ratios are rarely clear-cut; samples with a duplication trend away from the heterozygous cluster towards one of the homozygous clusters. Therefore, this approach should be used with caution, particularly when only one informative (heterozygous) variant is present. As illustrated in Figures 2c and d, utilizing the Genotyper software (Thermo Fisher Scientific) “traces” feature can facilitate the identification of subclusters (IntelliQube software also enables real-time amplification trace viewing). Specifically, samples with a CYP2D6*4×2 duplication (Figure 2c, light blue) are traced between the heterozygous (1847G/A) and homozygous variant (1847A/A) clusters, while samples with other duplications, including CYP2D6*2×2, are between the heterozygous (1847G/A) and homozygous reference (1847G/G) clusters. Likewise, as shown in Figure 2d, samples with CYP2D6*2×2 duplications trace between the heterozygous (2851C/T) and homozygous variant (2851T/T) clusters while those with other duplications, including CYP2D6*4×2, are between the heterozygous (2851C/T) and homozygous reference (2851C/C) clusters. Although results for other variants may be similar to those shown in Figure 2, results may vary and must be individually assessed. Using the “classification scheme” of the TaqMan Genotyper Software can also be helpful. The “classification scheme” is a set of defined homozygous reference, heterozygous, and homozygous variant zones established by running samples with known genotypes; these can be used for comparison in clinical runs where the “n” may be smaller (73). Sometimes, samples with three copies may fall between zones or trend toward one side of the zone. Two or more variants should be considered for accurate duplication allele assignments. Not all samples that are off cluster necessarily have an SV/CNV; a sample may be off cluster due to DNA quality/purity or other reasons. Lastly, having a larger number of samples in a dataset (or the same run) facilitates subcluster identification, but this is not always possible in lower-throughput clinical testing scenarios.
Figure 2. Real-time PCR genotyping assays can inform SV/CNV state.
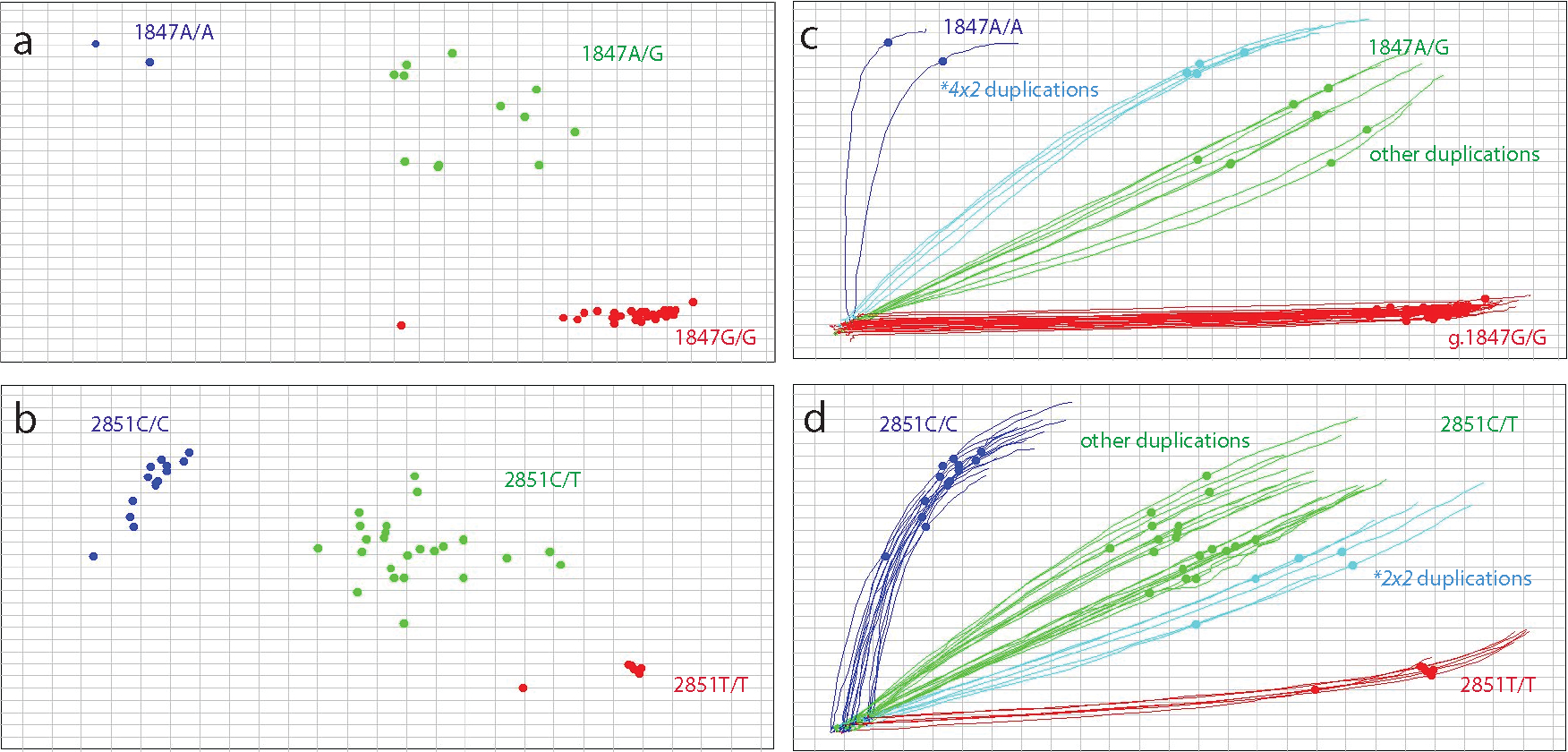
TaqMan assay results for the CYP2D6*4 core variant (1847G>A, rs3892097, panels a and c) and 2851C>T (rs16947, panels b and d) which is a core variant of many star alleles, including CYP2D6*2, are shown to illustrate how signal ratios and clustering of real-time genotyping assays can inform copy number (CN) state at the variant site. The “traces” feature in panels c and d allows the identification of clusters representing samples with copy number variants. Individuals with a CYP2D6*4×2 duplication are highlighted by the light blue traces in panel c. These samples represent diplotypes such as CYP2D6*1/*4×2, *2/*4×2 or *4×2/*41) and form a distinct cluster between the homozygous 1847A/A and heterozygous 1847G/A clusters suggesting that the sample has a CYP2D6*4 duplication. As shown in panel d, samples with a CYP2D6*2 duplication such as CYP2D6*1/*2×2, *2×2/*4 or *2×2/*10), highlighted by light blue traces, cluster between the homozygous 2851T/T and heterozygous 2851C/T clusters. The TaqMan assay for 1847G>A produces more distinct clusters than those observed for 2851C>T. Clustering also improves with higher sample numbers, including samples with CN variants.
Pyrosequencing (60, 74) and mass spectrometry-based methods such as the VeriDose CYP2D6 CNV panel (75) can also be employed to determine which allele has a duplication using variant signal ratios.
5.3.2. Genotyping or Sequencing Duplication-Specific XL-PCR Fragments
To determine whether a sample genotyped as CYP2D6*2×2/*4 or *2/*4×2 has a *2 or *4 duplication, the upstream gene copy can be specifically amplified by XL-PCR 7, for example (Figure 1c, Table 3), and genotyped for 100C>T, 1847G>A, and 2851C>T, or partially sequenced. If there is a CYP2D6*2 duplication, the XL-PCR amplicon will have 100C, 1847G, and 2851T, while a *4 duplication would yield 100T, 1847A, and 2851C. Since the amplicon is generated from one gene copy in this case, genotype results and sequence traces are expected to be homozygous. If a sample has a duplication on both alleles, e.g., CYP2D6*2×2/*4×2, XL-PCR will amplify both duplicated alleles and yield heterozygous genotype or sequencing results.
5.4. Other Methods and Platforms for Detecting CYP2D6 Structural Variants
5.4.1. Panel-Based Tests
CYP2D6 SVs/CNVs can be determined with commercially available platforms, including the VeriDose CYP2D6 CNV Panel (64, 75, 76) and Multiplex Ligation-dependent Probe Amplification (MLPA) panel (77), which are typically run along with SNV panels to determine CYP2D6 genotype. In contrast, Luminex xTAG (78), the PharmacoScan (79), and the Infinium Global Diversity Array with Enhanced PGx (80) include both variant and CN testing.
5.4.2. Sequencing-Based Approaches
PacBio has developed a specialized workflow that subjects XL-PCR amplicons to PacBio HiFi single molecule real-time (SMRT) sequencing to characterize SVs/CNVs (81). Long-read SMRT sequencing has been used to determine the phase of SNVs but can also inform SVs/CNVs (82–84). Lastly, Nanopore sequencing has been employed for genotyping and allele characterization, including SVs/CNVs (85).
Whole genome sequencing (WGS) data coupled with star allele calling software tools may be utilized to identify SVs/CNVs. While tools such as Aldy (86), StellarPGx (87), Stargazer (88), PyPGx (89), and Cyrius (38) appear to reliably call gene deletions, their variable fidelity in calling duplications with identical gene copies and some of the hybrid genes, especially complex SVs/CNVs, can make them unreliable in these scenarios (84, 90). Therefore, verification of SVs/CNVs with qPCR, dPCR, XL-PCR, or other methods should be considered. Whole exome sequencing (WES) has been evaluated for CYP2D6 SV/CNV detection, but there are inherent technical challenges, including difficulties with coverage and CNV/SV calling that preclude clinical implementation of WES-based CYP2D6 determination (91–95). In addition, CYP2D6 calling from WES data is inconsistent among star-allele calling tools, indicating limited clinical application for WES (96). Specialized software has been developed and successfully used for CYP2D6 calling from targeted NGS captures that include baits for CYP2D6 and CYP2D7 (97, 98); however, the software requires customization to the specific capture and significant adjustment to apply to other captures.
5.4.3. Cas9-Based Approaches
A recent report described Cas9-targeted nanopore DNA sequencing to detect CYP2D6::CYP2D7 hybrid alleles (99). This method uses Cas9-targeted cleavage of native genomic DNA at key sites within or flanking regions of interest and the addition of sequencing adapters at these sites. “Adapted” molecules are sequenced on a Nanopore instrument (the process is also called adaptive sequencing), which allows for long reads without the need for PCR. Although SNVs and CYP2D6::CYP2D7 hybrid genes were detected, low read depths were a limitation. This method, while promising, needs to be optimized for routine application.
Another Cas9-based approach targets the entire gene locus (CYP2D6, CYP2D7, and CYP2D8). The assay utilizes guide RNAs that target downstream of CYP2D6 and upstream of CYP2D8 and enriches continuous fragments greater than 52 kb. These fragments contain all three gene loci and any SV/CNV present. Long-read single-molecule sequencing then generates reads that allow for completely phased genotype and diplotype assignment, with read depths of 50–350× depending on sample source and quality (39). This approach removes the need for multiple assays to determine the SNV-level diplotypes, as well as quantitative CN testing and does not require knowledge about which SVs/CNVs may be present, which is often needed when selecting primers for XL-PCR.
5.4.4. Inheritance-Based Approaches (Duos/Trios)
Information regarding the presence or nature of an SV/CNV may also be obtained using mother/father/child trios or parent/child duos. For example, it can be deduced that the child has a CYP2D6*2×2/*4 diplotype with 3 gene copies if the mother is genotyped as CYP2D6*1×2/*2 or *1/*2×2 and the father is *4/*5. In this case, the child’s genotype also informs the mother’s genotype (CYP2D6*1/*2×2), as the child did not have a *1 allele, and the duplication was inherited from the mother. The knowledge of related individuals can also help resolve or fully characterize complex diplotypes having SVs/CNVs on both alleles. As shown by Gaedigk et al. (100), trio analysis was integral for the characterization of balanced hybrids—one allele had a CYP2D7::CYP2D6 hybrid in a duplication arrangement and the other a CYP2D7::CYP2D6 hybrid also in a duplication, thus producing 3-copy CN calls.
5.4.4. Genome-Wide Arrays
While genome-wide genotyping arrays may be able to identify SVs/CNVs, this method is neither sensitive nor accurate in determining the exact nature of the CYP2D6 SVs/CNVs. Given the polymorphic nature and high frequency of these events, findings are typically not reported clinically. One exception is the Illumina Infinium Global Diversity Array with Enhanced PGx (80), which interrogates 172 CYP2D6 SNVs and detects SVs/CNVs. However, there are no published data regarding the accuracy of CYP2D6 genotype or SV/CNV calls using this array.
6. Resources to Standardize Clinical Testing and Reporting
6.1. Test Recommendations
In collaboration with other organizations, the Association for Molecular Pathology (AMP) has published consensus recommendations for CYP2D6 allele selection (59). They utilized information from CPIC, PharmGKB, and the scientific literature to create a minimal panel (Tier 1 or “must test”) and an extended panel (Tier 2) of CYP2D6 variant alleles. The Tier 1 panel currently comprises 11 variant alleles, including gene deletion (CYP2D6*5) and duplication/multiplications (×N). CNV testing a minimum of one position (e.g., commonly exon 9) is sufficient to satisfy the Tier 1 recommendation. Also, Tier 1 does not require specification of which allele is duplicated. For Tier 2, testing at least two positions within the gene is recommended to differentiate hybrid alleles from full gene duplication and multiplication events. The Tier 2 panel adds 12 variant alleles, including hybrids; however, laboratories are not expected to characterize which hybrid is present but only to differentiate them from a full gene duplication/multiplication. Even when complying with Tier 2 testing it may be impossible to determine which chromosome harbors the SV/CNV unless additional analyses are performed.
When interrogating Tier 1 variants only, a patient may be phenotypically misclassified if they have a less common functionally relevant variant. Regarding SVs/CNVs, neither Tier 1 nor Tier 2 level testing (using one or at least two probes/targets) can discriminate which allele is duplicated when the diplotype is heterozygous, e.g., discriminate between CYP2D6*1/*4xN (IM) and *1xN/*4 (NM). Using Tier 1-based testing targeting exon 9, a CYP2D6*1/*13+*2 would most likely be called and reported as CYP2D6*1×2/*2 or *1/*2×2 (both UM), while Tier 2-based testing targeting the 5’UTR and exon 9 regions detects the presence of a CYP2D7::CYP2D6 hybrid that would be consistent with either CYP2D6*1/*13+*2 or *13+*1/*2 (both NMs). Another challenging genotype is CYP2D6*13+*2/*68+*4 which may erroneously be called as CYP2D6*2×2/*4 (NM) or *2/*4xN (IM) by Tiers 1 and 2, as all targeted regions yield CN calls of 3. CYP2D6*68+*4/*36 is an example that would be miscalled by both Tiers as CYP2D6*4/*4 (PM); however, as all gene copies are nonfunctional, this would not impact phenotype prediction. These are just a few examples illustrating how the recommended AMP Tier levels impact SV/CNV testing and diplotype assignments.
According to recent proficiency testing data, the majority of laboratories can detect the CYP2D6*5 deletion allele, while many laboratories either do not detect duplications or only utilize one position (59, 101, 102). These recommendations were published in 2021, and it may take time for laboratories to update their processes and to adopt them. Information on available laboratory tests including which alleles are tested can be found in the Genetic Testing Registry (103).
6.2. Selection of Laboratory Approach and Cost Considerations
Any of the molecular testing techniques described in this tutorial may be used by a clinical laboratory, and each has advantages and disadvantages. As such, the choice of platform is made by the laboratory director based on a variety of factors and is often driven by existing instrumentation and experience. The cost of testing, anticipated test volume, and the turn-around-time (TAT) needed to meet the clinical need are also important factors. Full runs (or “batches”) on high-throughput instrumentation are the most cost-effective in a research setting; however, if a quick TAT is required and the clinical laboratory has a modest testing volume, a lower-throughput instrument may be more appropriate. As of 2023, targeted approaches such as qPCR and dPCR tend to have lower reagent costs than sequencing and other approaches, and the cost is proportional to the number of CYP2D6 positions targeted (e.g., the cost is higher when testing both exon 1 and exon 9 for SV/CNV than testing only exon 9). As clinicians increasingly incorporate PGx into routine practice, test volumes are increasing, and more laboratories are entering the PGx testing field. For many genetic laboratories, despite the higher cost of NGS-based approaches, these may better fit into existing workflows and thus, are increasingly adopted. Although CYP2D6 SV/CNV calling algorithms have improved, techniques such as qPCR and dPCR may be required to supplement the NGS data at an added cost. Clinical tests undergo rigorous validation prior to use, which allows the laboratory to ensure that the selected technique is accurate, reproducible, and to gain familiarity with any limitations. Ultimately, the laboratory director must have expertise with both the testing platform(s) selected and the CYP2D6 gene in order to be able to understand the limitations of the approach and recommend alternative methods when challenging cases are encountered where the genotype cannot be resolved.
6.3. Reference Materials
Laboratories must demonstrate accurate and consistent CYP2D6 testing results before incorporating the test into clinical care (101). This process is facilitated, in part, by access to publicly available data and reference materials maintained by the Centers for Disease Control and Prevention’s Genetic Testing Reference Materials Program (GeT-RM) and Coriell Cell Repositories, respectively. To date, 179 Coriell samples have undergone extensive CYP2D6 characterization, and some have been found to carry rare or difficult-to-analyze CYP2D6 alleles, including SVs/CNVs (64). Per GeT-RM (64, 104), 62 of the 179 samples interrogated contain one or more SVs/CNVs. A selection of reference materials has been added to the Structural Variation document (37) for quick reference. GeT-RM identified samples for 16 (37%) of the 43 SVs/CNVs currently cataloged by PharmVar. The materials are available for laboratories to purchase as extracted genomic DNA or lymphoblastoid cell lines from the Coriell Institute for Medical Research (Camden, NJ).
7. Discussion of Selected CYP2D6 Genotypes with Structural Variants
This section discusses five samples with SVs/CNVs which have been extensively tested on various platforms as part of the CYP2D6 GeT-RM project (64) (Figure 3). Table 4 summarizes CN calls for samples with a variety of SVs/CNVs and indicates which XL-PCR reactions are expected to amplify and can be employed to ascertain the presence of SVs/CNVs. The regions amplified by XL-PCR are visualized in Figure 1.
Figure 3. Graphical overview of CYP2D6 diplotypes containing structural variants.
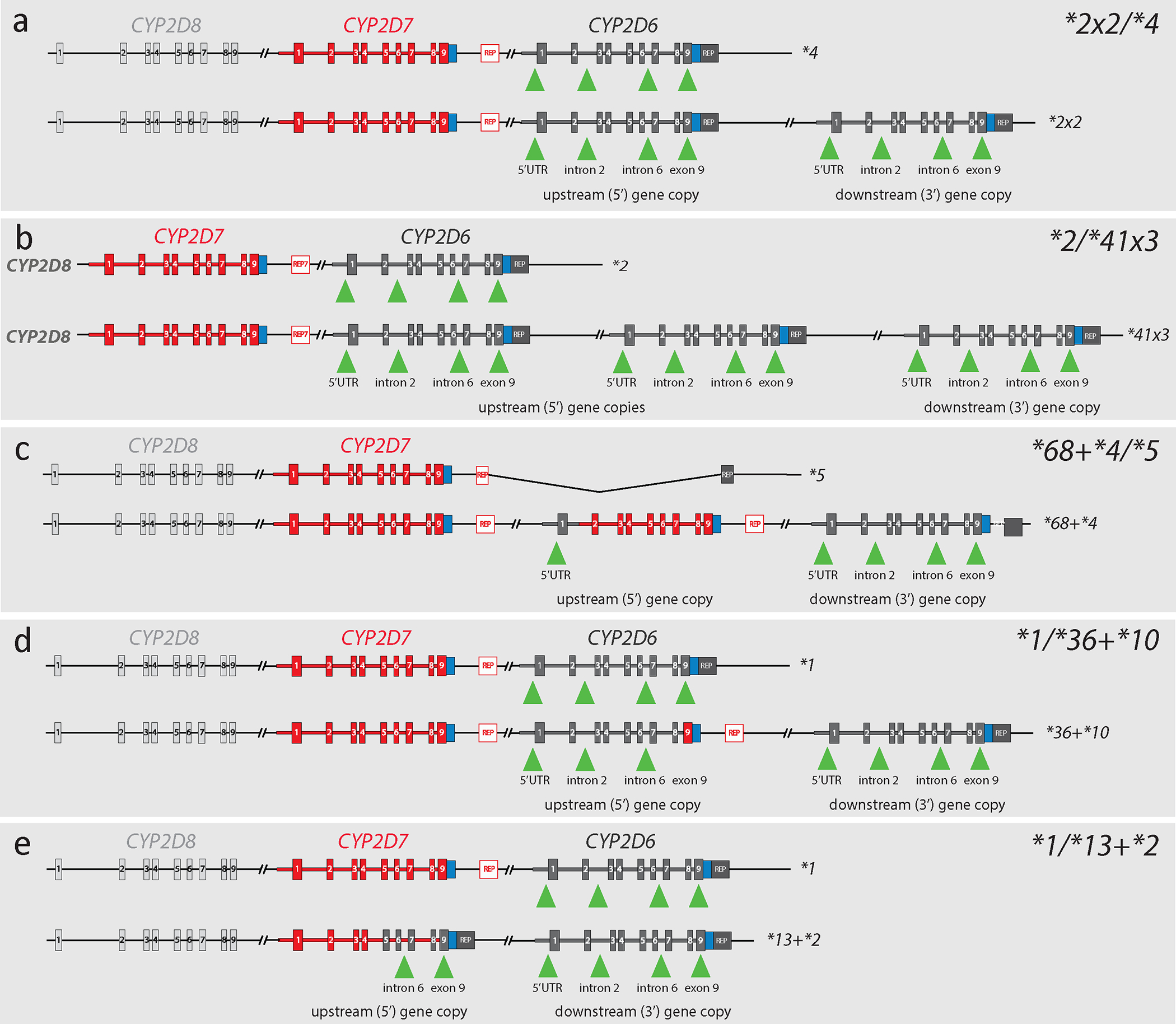
Each panel shows both alleles of samples with SVs/CNVs detailing the regions commonly targeted by quantitative copy number assays. Graphs depict CYP2D6 (dark gray), CYP2D7 (red), and CYP2D8 (light gray) with boxes representing exons. Green triangles indicate the gene regions targeted by commonly used TaqMan copy number assays. Panel a shows Coriell sample NA23296, which was genotyped as CYP2D6*2×2/*4. This sample produces copy number (CN) calls of 3 for each interrogated region. Panel b depicts Coriell sample NA24217, which has a CYP2D6*2/*41×3 diplotype. This sample produces CN calls of 4 for each of the interrogated regions. Panel c shows Coriell sample HG01190, which was genotyped as CYP2D6*68+*4/*5. Because the CYP2D6*68 hybrid switches to CYP2D7 in intron 1, this sample produces CN calls of 1 when probed with intron 2, intron 6, and exon 9 gene copy number assays; in contrast, the assay targeting the 5’UTR produces a CN call of 2. Panel d represents Coriell samples NA23093 and NA18563, which were genotyped as CYP2D6*1/*36+*10. Because CYP2D6*36 has a CYP2D7-derived exon 9, the assay targeting exon 9 produces a signal only from the *1 and *10 alleles, yielding a CN call of 2; assays probing the other regions produce CN calls of 3. Finally, panel e illustrates Coriell sample NA19790, which was genotyped as CYP2D6*1/*13+*2. The CYP2D6*13 hybrid gene in this sample has CYP2D7-derived exons 1–4 and therefore does not generate a signal when probed with assays targeting the 5’UTR or intron 2. Thus, CN calls of 2 are produced for these regions, while the other two assays produce CN calls of 3. CN calls for these and other Coriell samples with SVs/CNVs are summarized in Table 3.
7.1. Example 1: CYP2D6*2×2/*4
This example represents a diplotype with a commonly found SV/CNV (Figure 3a). The CYP2D6*2×2 duplication allele has two gene copies that are assumed to be identical (in some cases, the gene copies may represent different *2 suballeles). Both gene copies have a CYP2D6 downstream region. Quantitative CN testing is expected to return 3-copy calls regardless of which gene region is interrogated.
Neither XL-PCR nor quantitative testing alone can determine whether the duplication is on the CYP2D6*2 or *4 allele. Because CYP2D6*2, relative to *4, is the most frequently observed duplication SV/CNV, samples like this are often reported as CYP2D6*2×2/*4 by default or are reported ambiguously (e.g., CYP2D6*2/*4 (3N)) to indicate that a duplication is present (see Table 2 for recommended reporting of ambiguous genotypes). As described in section 5.3.1, real-time PCR may be used to determine whether the sample has a CYP2D6*2 or *4 duplication.
Patients with a CYP2D6*2×2/*4 diplotype (AS=2) are predicted to be NMs, while those with a *2/*4×2 diplotype (AS=1) are predicted to be IMs. To accurately predict phenotype, testing must determine which of the alleles has the duplication.
7.2. Example 2: CYP2D6*2/*41×3
This example represents a diplotype with a rare allele having three identical CYP2D6*41 gene copies, all of which have a CYP2D6 downstream region. Quantitative assays return calls of four copies for all regions interrogated.
XL-PCR amplicons are generated from both 5’ (gained) gene copies and their intergenic regions but do not discriminate between duplication and multiplication. Neither XL-PCR nor quantitative testing alone can determine whether the duplication is on the CYP2D6*2 or *41 allele. Thus, the sample’s genotype could be CYP2D6*2×3/*41, *2×2/*41×2, or *2/*41×3. Because CYP2D6*2×2 is the most common, samples like this may be reported as CYP2D6*2×3/*41 or *2/*41 (4N) to indicate that multiple gene copies are present. Real-time PCR signal ratios for 2989G>A (rs28371725) may be utilized to show that this sample has multiple CYP2D6*41 gene copies (ratio of 1:3 for 2989G/A is consistent with *2/*41×3, while ratios of 1:1 and 3:1 would be indicative of *2×2/*41×2 and *2×3/*41 diplotypes, respectively).
Patients with a CYP2D6*2/*41×3 diplotype (AS=1.75) are predicted to be NMs, while those with *2×2/*41×2 (AS=2.5) or *2×3/*41 (AS=3.25) diplotypes are predicted to be UMs.
7.3. Example 3: CYP2D6*68+*4/*5
This example represents a diplotype with SVs/CNVs on both alleles (Figure 3c). One allele has two non-identical gene copies, i.e., a CYP2D6::CYP2D7 hybrid upstream of a CYP2D6*4. The upstream CYP2D6*68 gene copy comprises CYP2D6 (exon 1) and CYP2D7 (exons 2–9 and downstream). The other allele is a CYP2D6*5 gene deletion. Both alleles are relatively common across populations. Quantitative assays return calls of two copies for the 5’UTR and one copy for intron 2, intron 6, and exon 9 (the higher CN call for the 5’UTR suggests the presence of a CYP2D6::CYP2D7 hybrid gene). This diplotype would be assigned as CYP2D6*4/*5 if only exon 9 is assayed.
The CYP2D6*5 gene deletion can readily be detected by XL-PCRs 3, 4, or 5. The CYP2D6*68 hybrid can be amplified by XL-PCR 7; due to the presence of the CYP2D7-derived spacer, the amplicon is 1.6 kb longer compared to an amplicon that is generated from a 5’ gene copy with a CYP2D6-derived downstream region such as those found in, e.g., CYP2D6*1×2, *2×2, or *4×2 duplications. Real-time PCR signal ratios are not informative since 100C>T and 1847G>A, along with other SVs/CNVs, yield homozygous results.
Because CYP2D6*68 is nonfunctional, it is unnecessary to discriminate CYP2D6*4 from *68+*4. CYP2D6*68+*4/*5 and *4/*5 diplotypes both have an AS of 0, predicting PM status.
7.4. Example 4: CYP2D6*1/*36+*10
This example represents a diplotype with one allele carrying two non-identical gene copies (Figure 3d), a CYP2D6::CYP2D7 hybrid (*36), and a CYP2D6*10. The CYP2D6*36+*10 allele is frequently found in East Asian individuals. The upstream CYP2D6*36 gene copy comprises CYP2D6 (exons 1–8) and CYP2D7 (exon 9 and downstream). Quantitative assays return calls of three copies for the 5’UTR, intron 2, and intron 6 regions and two copies for exon 9.
The CYP2D6*36 hybrid can be amplified by XL-PCR 7; amplicons are, however, longer than those generated from the 5’ gene copy in a CYP2D6*2×2 due to the presence of the CYP2D7-derived spacer. Real-time PCR signal ratios for 100C>T would be expected to be 1:2 for 100C/T since both CYP2D6*36 and *10 alleles have 100T.
Because CYP2D6*36 is nonfunctional, it is unnecessary to discriminate CYP2D6*10 from *36+*10. CYP2D6*1/*36+*10 and *1/*10 diplotypes both have an AS of 1.25, predicting NM status.
7.5. Example 5: CYP2D6*1/*13+*2
This example represents a diplotype with one allele having two non-identical gene copies, i.e., a CYP2D7::CYP2D6 hybrid upstream of a CYP2D6*2 (Figure 3e). The CYP2D6*13 hybrid in this sample switches from CYP2D7 to CYP2D6 in intron 4 and, like all other such *13 genes, has a CYP2D6-derived downstream region. Other CYP2D6*13 alleles may switch to CYP2D6 at different locations. Quantitative assays return calls of two copies for 5’UTR and intron 2, and three copies for intron 6 and exon 9.
Amplification products from XL-PCRs 6 and 8 indicate the presence of a duplication, which may be misinterpreted as a CYP2D6*2×2 in the absence of further testing. Quantitative assays performed at multiple locations within CYP2D6, however, reveal the presence of a CYP2D7::CYP2D6 hybrid (if only exon 9 is assessed, CYP2D6*13 hybrids may be misinterpreted as gene duplications). The presence of a *13 hybrid can be substantiated by amplifying the entire hybrid gene with XL-PCR 10. Sequencing of this amplicon may be considered to validate the hybrid nature of this gene copy.
Real-time PCR signal ratios may also suggest the presence of a third gene copy, but these are difficult to interpret as the CYP2D6 portions in CYP2D6*13 hybrid genes vary and may or may not harbor the variants tested.
Since all CYP2D6*13 hybrid genes are nonfunctional due to a frameshift in exon 1, individuals with the CYP2D6*1/*13+*2 diplotype (AS=2) are predicted to have an NM phenotype; if the CYP2D6*13 is not detected (e.g., only exon 9 is tested for CNV), the genotype may be reported as CYP2D6*1/*2×2 (AS=3), which falsely predicts UM status.
8. Applications beyond CYP2D6
Although this tutorial focuses on CYP2D6, the terms and recommendations for reporting can be applied to other pharmacogenes. To promote adoption, PharmVar is also updating Structural Variation documents for other genes with SVs/CNVs accordingly.
9. Conclusions
The CYP2D6 enzyme mediates the catabolism or bioactivation of over 20% of clinically used drugs and is thus an essential pharmacogene in clinical testing and research. The CYP2D6 gene, however, is extremely polymorphic, with 163 distinct defined alleles as of this writing, including numerous SVs/CNVs. In addition, the long history of CYP2D6 research without standardization of terms has added confusion for novices and experts alike. This complexity makes CYP2D6 one of the most difficult gene loci to characterize in clinical testing, while the importance of accurate genotyping and phenotype predictions for appropriate pharmacogenomics-informed prescribing cannot be overstated. With this guide, PharmVar hopes to establish a unified nomenclature, describe methods for (and limitations to) detecting CYP2D6 structural variants, and provide an educational resource for those responsible for clinical testing, interpretation, or implementation of pharmacogenetic testing, as well as those performing CYP2D6 basic and translational research.
Funding:
PharmGKB and CPIC gratefully acknowledge support from NIH (U24 HG010615 and U24HG010135).
TEK, MWC, and KS are supported by NIH/NHGRI/NICHD/NIDA (U24 HG010615). PharmGenetix GmbH acknowledges the Österreichische Forschungsförderungsgesellschaft GmbH (FFG) for support via the PGx-Next Generation Analytics Part 2 grant (FO0999891633/42175800).
Footnotes
Conflicts of interest: C.A.B. is the founder and CEO of Sequence2Script Inc. H.H. is an employee of AccessDx Laboratories and a former employee of Translational Software. B.E.R. is an employee of Let’s Get Checked and owner of Phoenix Laboratory Consulting, LLC. H.M.D. is an owner of MD Omics Consulting, LLC. C.N. and S.V. are employed by PharmGenetix GmbH, a private laboratory providing PGx testing, reporting, and interpretation services. T.E.K. is on the Scientific Advisory Board of Galatea. M.S.P. is currently employed as president of Precision Medicine Advisers, Inc. and is a former employee of Sequence Bio, Inc. and Genuity Science, Ltd. A.J.T. is supported in part by RPRD Diagnostics LLC. All other authors declared no competing interests for this work.
References
- (1).Taylor C, Crosby I, Yip V, Maguire P, Pirmohamed M & Turner RM A review of the important role of CYP2D6 in pharmacogenomics. Genes (Basel) 11:1295, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Saravanakumar A, Sadighi A, Ryu R & Akhlaghi F Physicochemical properties, biotransformation, and transport pathways of established and newly approved medications: A systematic review of the Top 200 most prescribed drugs vs. the FDA-approved drugs between 2005 and 2016. Clin Pharmacokinet 58, 1281–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Caudle KE et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 13, 116–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Whirl-Carrillo M et al. An Evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 110, 563–72 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nofziger C et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther 107, 154–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jarvis JP, Peter AP & Shaman JA An Evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Front Psychiatry 10, 432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bousman CA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A genotypes and serotonin reuptake inhibitor antidepressants. Clin Pharmacol Ther, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hicks JK et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid Therapy. Clin Pharmacol Ther 110, 888–96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Goetz MP et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther 103, 770–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Brown JT et al. Clinical Pharmacogenetics Implementation Consortium guideline for Cytochrome P450 (CYP) 2D6 genotype and atomoxetine therapy. Clin Pharmacol Ther 106, 94–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bell GC et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 102, 213–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kane M & Dean L Eliglustat therapy and CYP2D6 Genotype. In: Medical Genetics Summaries (eds. Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kattman BL and Malheiro AJ) (Bethesda (MD), 2012). [Google Scholar]
- (14).PharmGKB. Annotation of FDA Label for tetrabenazine and CYP2D6. <https://www.pharmgkb.org/labelAnnotation/PA166104829>.
- (15).PharmGKB. Annotation of FDA Label for pimozide and CYP2D6. <https://www.pharmgkb.org/labelAnnotation/PA166104870>.
- (16).Lenk HC et al. The polymorphic nuclear factor NFIB regulates hepatic CYP2D6 expression and influences risperidone metabolism in psychiatric patients. Clin Pharmacol Ther 111, 1165–74 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Boone EC et al. Long-distance phasing of a tentative “enhancer” single-nucleotide polymorphism with CYP2D6 star allele definitions. Front Pharmacol 11, 486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dinh JC et al. The impact of the CYP2D6 “enhancer” single nucleotide polymorphism on CYP2D6 activity. Clin Pharmacol Ther 111, 646–54 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Khor CC et al. Cross-ancestry genome-wide association study defines the extended CYP2D6 locus as the principal genetic determinant of endoxifen plasma concentrations. Clin Pharmacol Ther 113, 712–23 (2023). [DOI] [PubMed] [Google Scholar]
- (20).Sanchez-Spitman AB, Moes DA, Gelderblom H, Dezentje VO, Swen JJ & Guchelaar HJ The effect of rs5758550 on CYP2D6*2 phenotype and formation of endoxifen in breast cancer patients using tamoxifen. Pharmacogenomics 18, 1125–32 (2017). [DOI] [PubMed] [Google Scholar]
- (21).Wang D, Poi MJ, Sun X, Gaedigk A, Leeder JS & Sadee W Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum Mol Genet 23, 268–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Brunetti-Pierri N, Patel A, Brown CW, Rauch RA & Heptulla RA De novo terminal 22q12.3q13.3 duplication with pituitary hypoplasia. Am J Med Genet A 149A, 2554–6 (2009). [DOI] [PubMed] [Google Scholar]
- (23).Upadia J et al. A previously unrecognized 22q13.2 microdeletion syndrome that encompasses TCF20 and TNFRSF13C. Am J Med Genet A 176, 2791–7 (2018). [DOI] [PubMed] [Google Scholar]
- (24).Dyar B, Meaddough E, Sarasua SM, Rogers C, Phelan K & Boccuto L enetic findings as the potential basis of personalized pharmacotherapy in Phelan-McDermid Syndrome. Genes (Basel) 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ricciardello A, Tomaiuolo P & Persico AM Genotype-phenotype correlation in Phelan-McDermid syndrome: A comprehensive review of chromosome 22q13 deleted genes. Am J Med Genet A 185, 2211–33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).PharmVar. CYP2D6 gene page. <https://www.pharmvar.org/gene/CYP2D6>.
- (27).Kimura S, Umeno M, Skoda RC, Meyer UA & Gonzalez FJ The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet 45, 889–904 (1989). [PMC free article] [PubMed] [Google Scholar]
- (28).Gaedigk A, Blum M, Gaedigk R, Eichelbaum M & Meyer UA Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am J Hum Genet 48, 943–50 (1991). [PMC free article] [PubMed] [Google Scholar]
- (29).Gaedigk A, Bradford LD, Alander SW & Leeder JS CYP2D6*36 gene arrangements within the CYP2D6 locus: association of CYP2D6*36 with poor metabolizer status. Drug Metab Dispos 34, 563–9 (2006). [DOI] [PubMed] [Google Scholar]
- (30).Gaedigk A & Coetsee C The CYP2D6 gene locus in South African Coloureds: unique allele distributions, novel alleles and gene arrangements. Eur J Clin Pharmacol 64, 465–75 (2008). [DOI] [PubMed] [Google Scholar]
- (31).Gaedigk A, Fuhr U, Johnson C, Berard LA, Bradford D & Leeder JS CYP2D7–2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics 11, 43–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gaedigk A et al. Identification of novel CYP2D7–2D6 Hybrids: non-functional and functional variants. Front Pharmacol 1:121, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gaedigk A et al. Cytochrome P4502D6 (CYP2D6) gene locus heterogeneity: characterization of gene duplication events. Clin Pharmacol Ther 81, 242–51 (2007). [DOI] [PubMed] [Google Scholar]
- (34).Kramer WE et al. CYP2D6: novel genomic structures and alleles. Pharmacogenet Genomics 19, 813–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Steen VM, Molven A, Aarskog NK & Gulbrandsen AK Homologous unequal cross-over involving a 2.8 kb direct repeat as a mechanism for the generation of allelic variants of human cytochrome P450 CYP2D6 gene. Hum Mol Genet 4, 2251–7 (1995). [DOI] [PubMed] [Google Scholar]
- (36).Lovlie R, Daly AK, Molven A, Idle JR & Steen VM Ultrarapid metabolizers of debrisoquine: characterization and PCR-based detection of alleles with duplication of the CYP2D6 gene. FEBS Lett 392, 30–4 (1996). [DOI] [PubMed] [Google Scholar]
- (37).PharmVar. CYP2D6 Structural Variation document available at. [Google Scholar]
- (38).Chen X et al. Cyrius: accurate CYP2D6 genotyping using whole-genome sequencing data. Pharmacogenomics J 21, 251–61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Turner AJ et al. Characterization of complex structural variation in the CYP2D6-CYP2D7-CYP2D8 gene loci using single-molecule long-read sequencing. Front Pharmacol 14, 1195778 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Riffel AK et al. CYP2D7 Sequence Variation Interferes with TaqMan CYP2D6*15 and *35 genotyping. Front Pharmacol 6:312, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Del Tredici AL et al. Frequency of CYP2D6 alleles including structural variants in the United States. Front Pharmacol 9:305, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Black JL, Walker DL, O’Kane DJ & Harmandayan M Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab Dispos 40, 111–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T & Leeder JS Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 19, 69–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hosono N et al. CYP2D6 genotyping for functional-gene dosage analysis by allele copy number detection. Clinical chemistry 55, 1546–54 (2009). [DOI] [PubMed] [Google Scholar]
- (45).Chan W, Li MS, Sundaram SK, Tomlinson B, Cheung PY & Tzang CH CYP2D6 allele frequencies, copy number variants, and tandems in the population of Hong Kong. J Clin Lab Anal 33, e22634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wen YF, Gaedigk A, Boone EC, Wang WY & Straka RJ The Identification of novel CYP2D6 variants in US Hmong: results from genome sequencing and clinical genotyping. Front Pharmacol 13, 867331 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).PharmGKB. Gene-Specific Information Tables for CYP2D6. <https://www.pharmgkb.org/page/cyp2d6RefMaterials>
- (48).Collins RL et al. A structural variation reference for medical and population genetics. Nature 581, 444–51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).National Library of Medicine, National Center for Biotechnology Information (NCBI). Overview of Structural Variation. <https://www.ncbi.nlm.nih.gov/dbvar/content/overview/>.
- (50).Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ & Leeder JS The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 83, 234–42 (2008). [DOI] [PubMed] [Google Scholar]
- (51).Johansson I, Lundqvist E, Bertilsson L, Dahl ML, Sjoqvist F & Ingelman-Sundberg M Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci U S A 90, 11825–9 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Dahl ML, Johansson I, Palmertz MP, Ingelman-Sundberg M & Sjoqvist F Analysis of the CYP2D6 gene in relation to debrisoquin and desipramine hydroxylation in a Swedish population. Clin Pharmacol Ther 51, 12–7 (1992). [DOI] [PubMed] [Google Scholar]
- (53).Bertilsson L et al. Molecular basis for rational megaprescribing in ultrarapid hydroxylators of debrisoquine. Lancet 341, 63 (1993). [DOI] [PubMed] [Google Scholar]
- (54).Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F & Ingelman-Sundberg M Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther 278, 441–6 (1996). [PubMed] [Google Scholar]
- (55).McLellan RA, Oscarson M, Seidegard J, Evans DA & Ingelman-Sundberg M Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics 7, 187–91 (1997). [DOI] [PubMed] [Google Scholar]
- (56).Panserat S et al. An unequal cross-over event within the CYP2D gene cluster generates a chimeric CYP2D7/CYP2D6 gene which is associated with the poor metabolizer phenotype. Br J Clin Pharmacol 40, 361–7 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Sim SC, Daly AK & Gaedigk A CYP2D6 update: revised nomenclature for CYP2D7/2D6 hybrid genes. Pharmacogenet Genomics 22, 692–4 (2012). [DOI] [PubMed] [Google Scholar]
- (58).Bruford EA et al. HUGO Gene Nomenclature Committee (HGNC) recommendations for the designation of gene fusions. Leukemia 35, 3040–3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Pratt VM et al. Recommendations for clinical CYP2D6 genotyping allele selection: A joint consensus recommendation of the association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn 23, 1047–64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Langaee T, Hamadeh I, Chapman AB, Gums JG & Johnson JA A novel simple method for determining CYP2D6 gene copy number and identifying allele(s) with duplication/multiplication. PloS one 10, e0113808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Hosono N et al. Multiplex PCR-based real-time invader assay (mPCR-RETINA): a novel SNP-based method for detecting allelic asymmetries within copy number variation regions. Hum Mutat 29, 182–9 (2008). [DOI] [PubMed] [Google Scholar]
- (62).Gaedigk A, Twist GP & Leeder JS CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics 13, 91–111 (2012). [DOI] [PubMed] [Google Scholar]
- (63).Scantamburlo G et al. Allele drop uut conferred by a frequent CYP2D6 genetic variation for commonly used CYP2D6*3 genotyping assays. Cell Physiol Biochem 43, 2297–309 (2017). [DOI] [PubMed] [Google Scholar]
- (64).Gaedigk A et al. Characterization of reference materials for genetic testing of CYP2D6 alleles: A GeT-RM collaborative project. J Mol Diagn 21, 1034–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wang WY et al. Characterization of novel CYP2D6 alleles across Sub-Saharan African populations. J Pers Med 12, 1575 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Schmittgen TD & Livak KJ Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–8 (2008). [DOI] [PubMed] [Google Scholar]
- (67).Whale AS, Huggett JF & Tzonev S Fundamentals of multiplexing with digital PCR. Biomol Detect Quantif 10, 15–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Rae JM et al. Concordance between CYP2D6 genotypes obtained from tumor-derived and germline DNA. J Natl Cancer Inst 105, 1332–4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Hindson CM et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10, 1003–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Dingle TC, Sedlak RH, Cook L & Jerome KR Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem 59, 1670–2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Turner AJ et al. Identification of CYP2D6 haplotypes that interfere with commonly used assays for copy number variation characterization. J Mol Diagn 23, 577–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Sicko RJ et al. Rare variants in RPPH1 real-time quantitative PCR control assay binding sites result in incorrect copy number calls. J Mol Diagn 24, 33–40 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Atiq MA, Peterson SE, Langman LJ, Baudhuin LM, Black JL & Moyer AM Determination of the duplicated CYP2D6 allele using real-time PCR signal: an alternative approach. J Pers Med 13, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Soderback E, Zackrisson AL, Lindblom B & Alderborn A Determination of CYP2D6 gene copy number by pyrosequencing. Clinical chemistry 51, 522–31 (2005). [DOI] [PubMed] [Google Scholar]
- (75).Agena Bioscience. VeriDose CYP2D6 CNV panel. <https://www.agenabio.com/products/panel/veridose-cyp2d6-cnv-panel/>.
- (76).Everts RE, Lois A, Nakorchevsky A, Tao H, Jackson K & Rhodes K The VeriDose CYP2D6 CNV Panel: A one-well solution for copy number and hybrid allele detection of CYP2D6. 21 1124–5 (2019). [Google Scholar]
- (77).Qiao W et al. Integrated CYP2D6 interrogation for multiethnic copy number and tandem allele detection. Pharmacogenomics 20, 9–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Luminex Corporation, Luminex xTAG. <https://www.luminexcorp.com/>.
- (79).Thermo Fisher Scientific. PharmacoScan™ Assay Kit. <https://www.thermofisher.com/order/catalog/product/903026>.
- (80).Illumina Inc. The Infinium Global Array with Enhanced PGx. <https://www.illumina.com/products/by-type/microarray-kits/infinium-global-diversity-pgx.html>.
- (81).Illumina Inc. HiFi amplicon sequencing for pharmacogenetics: CYP2D6. <https://www.pacb.com/wp-content/uploads/Application-note-HiFi-amplicon-sequencing-for-pharmacogenetics-CYP2D6.pdf>.
- (82).Buermans HP et al. Flexible and scalable full-length CYP2D6 long amplicon PacBio sequencing. Hum Mutat 38, 310–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Charnaud S et al. PacBio long-read amplicon sequencing enables scalable high-resolution population allele typing of the complex CYP2D6 locus. Commun Biol 5, 168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Caspar SM, Schneider T, Stoll P, Meienberg J & Matyas G Potential of whole-genome sequencing-based pharmacogenetic profiling. Pharmacogenomics 22, 177–90 (2021). [DOI] [PubMed] [Google Scholar]
- (85).Liau Y, Maggo S, Miller AL, Pearson JF, Kennedy MA & Cree SL Nanopore sequencing of the pharmacogene CYP2D6 allows simultaneous haplotyping and detection of duplications. Pharmacogenomics 20, 1033–47 (2019). [DOI] [PubMed] [Google Scholar]
- (86).Hari A et al. An efficient genotyper and star-allele caller for pharmacogenomics. Genome Res 33, 61–70 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Twesigomwe D et al. StellarPGx: A Nextflow Pipeline for Calling Star Alleles in Cytochrome P450 Genes. Clin Pharmacol Ther 110, 741–9 (2021). [DOI] [PubMed] [Google Scholar]
- (88).Lee SB et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med 21, 361–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Lee SB, Shin JY, Kwon NJ, Kim C & Seo JS ClinPharmSeq: A targeted sequencing panel for clinical pharmacogenetics implementation. PloS One 17, e0272129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Twesigomwe D, Wright GEB, Drogemoller BI, da Rocha J, Lombard Z & Hazelhurst S A systematic comparison of pharmacogene star allele calling bioinformatics algorithms: a focus on CYP2D6 genotyping. NPJ Genom Med 5, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Belkadi A et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A 112, 5473–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Ng D, Hong CS, Singh LN, Johnston JJ, Mullikin JC & Biesecker LG Assessing the capability of massively parallel sequencing for opportunistic pharmacogenetic screening. Genet Med 19, 357–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Yang Y, Botton MR, Scott ER & Scott SA Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics 18, 673–85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).van der Lee M et al. Repurposing of Diagnostic Whole Exome Sequencing Data of 1,583 Individuals for Clinical Pharmacogenetics. Clin Pharmacol Ther 107, 617–27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Lanillos J, Carcajona M, Maietta P, Alvarez S & Rodriguez-Antona C Clinical pharmacogenetic analysis in 5,001 individuals with diagnostic Exome Sequencing data. NPJ Genom Med 7, 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Tafazoli A et al. Development of an extensive workflow for comprehensive clinical pharmacogenomic profiling: lessons from a pilot study on 100 whole exome sequencing data. Pharmacogenomics J 22, 276–83 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Wang L et al. Implementation of preemptive DNA sequence-based pharmacogenomics testing across a large academic medical center: The Mayo-Baylor RIGHT 10K Study. Genet Med 24, 1062–72 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Lopes JL et al. Targeted Genotyping in Clinical Pharmacogenomics: What Is Missing? J Mol Diagn 24, 253–61 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Rubben K, Tilleman L, Deserranno K, Tytgat O, Deforce D & Van Nieuwerburgh F Cas9 targeted nanopore sequencing with enhanced variant calling improves CYP2D6-CYP2D7 hybrid allele genotyping. PLoS Genet 18, e1010176 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Gaedigk A, Riffel AK, Berrocal BG, Solaesa VG, Davila I & Isidoro-Garcia M Characterization of a complex CYP2D6 genotype that caused an AmpliChip CYP450 Test no-call in the clinical setting. Clin Chem Lab Med 52, 799–807 (2014). [DOI] [PubMed] [Google Scholar]
- (101).Moyer AM et al. Genotype and Phenotype Concordance for Pharmacogenetic Tests Through Proficiency Survey Testing. Arch Pathol Lab Med 144, 1057–66 (2020). [DOI] [PubMed] [Google Scholar]
- (102).Bousman CA, Jaksa P & Pantelis C Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenet Genomics 27, 387–93 (2017). [DOI] [PubMed] [Google Scholar]
- (103).National Center for Biotechnology Information (NCBI). GTR:Genetic Testing Registry. <https://www.ncbi.nlm.nih.gov/gtr/>
- (104).Centers for Disease Control and Prevention. Genetic Testing Reference Materials (GeT-RM) Coordination Program. Reference Materials for Pharmacogenetics. <https://www.cdc.gov/labquality/get-rm/inherited-genetic-diseases-pharmacogenetics/pharmacogenetics.html>.


