The level of expression achieved by a gene is often simply associated with how efficiently it is transcribed. However, other factors can also have dramatic effects on gene expression levels. A mutant version of the human prothrombin gene that shows over-expression of its protein product through activation of its poly(A) signal has recently been studied by Gehring et al. (2001). This apparently perturbs the delicate balance of the clotting cascade and may cause serious problems of thrombosis in affected individuals.
Producing a mature, translatable messenger (m)RNA from a mammalian gene requires an amazingly complex series of molecular tricks. First, the gene must be exposed from its hidden state in chromatin (Workman and Kingston, 1998). Then, RNA polymerase II (Pol II) must be seduced by multiple activation domains of transcription factors bound and lined up on the gene’s promoter or enhancers to land on the transcription start site (Blackwood and Kadonaga, 1998). This of course occurs in conjunction with numerous other proteins (general transcription factors), so that the final initiation complex is in the mDa size range (Orphanides et al., 1996). The effectiveness of any one of these transcription initiation steps can directly determine not only the specificity, but also the level of gene expression. Complex rearrangements then occur to allow the polymerase to escape from the promoter and begin elongating through the gene (McKnight, 1996). However, with potential gene sizes of over a million nucleotides, the elongation process can be a marathon experience both in time (some 16 h) and in terms of RNA processing. Extensive intron splicing, both constitutive and alternative, occurs co-transcriptionally (Lopez, 1998; Hirose and Manley, 2000), playing an essential role in generating protein diversity (Graveley, 2001) as well as assuring that the message produced is the exact sequence required to make the correct protein product (Hentze and Kulozik, 1999). Although splicing efficiency does not appear to have a major quantitative effect on gene expression, some splicing needs to take place to allow efficient nuclear export of mRNA (Zenklusen and Stutz, 2001). Transcriptional termination eventually stops the polymerase juggernaut through a process that is triggered by recognition of poly(A) signals in the nascent transcript (Proudfoot, 2000). However, poly(A) signals have another critical role: defining the site at which the nascent RNA is cleaved prior to the addition of the poly(A) tail. Only polyadenylated mRNA is exported from the nucleus to the cytoplasm, so that failure to add the poly(A) tail means that less mRNA will escape the highly active nuclear degradation processes (Bousquet-Antonelli et al., 2000). Therefore, the efficiency of polyadenylation can have significant quantitative effects on gene expression. Furthermore, polyadenylation also affects the efficiency of translation in the cytoplasm. The poly(A) binding protein (PABPI), which coats the poly(A) tail, helps to recruit ribosomes to the mRNA cap site (at the other end of the mRNA). This process occurs through mRNA circularization mediated by PABPI interaction with cap binding proteins (Sachs et al., 1997).
Poly(A) signals have a quite precise arrangement (Zhao et al., 1999). The sequence AAUAAA is the most obvious and essential element. It is placed ∼20 nt upstream of the actual site of cleavage, which is usually an A residue. A GU- or U-rich sequence then immediately follows on from the cleavage site, and its extent is often the main determinant of how efficiently this RNA processing signal will work. As shown in Figure 1, specific proteins bind to these two elements; cleavage polyadenylation specificity factor (CPSF) interacts with AAUAAA through its largest (160 kDa) subunit, and cleavage stimulation factor (CstF) interacts with the GU/U element through its 64 kDa subunit. CPSF also interacts with poly(A) polymerase (PAP), thereby recruiting it to the complex in preparation for polyadenylation once cleavage has taken place. Interactions between CPSF and CstF enhance their binding to the RNA signals and also somehow position two further cleavage factors (CFI and CFII) over the actual site of cleavage (Colgan and Manley, 1997; Wahle and Rüegsegger, 1999). This site must be precisely placed within the now very large protein complex to allow cleavage at the poly(A) site, by an as yet unidentified endonuclease. A further twist to the mechanism of factor assembly on the poly(A) signal and subsequent cleavage at the poly(A) site is that this whole process occurs in association with the Pol II elongation complex. In particular, the C-terminal domain (CTD) of the Pol II large subunit, a seven amino acid sequence repeated 52 times, strongly activates cleavage and polyadenylation (Hirose and Manley, 1998). This effect of the Pol II CTD effectively couples polyadenylation to transcription as discussed in recent reviews (Hirose and Manley, 2000; Proudfoot, 2000). Once cleavage has occurred, poly(A) polymerase, already recruited to the protein complex, adds the poly(A) tail and is aided by PABPII, which stimulates poly(A) synthesis of up to 200 nt by successive binding to the growing poly(A) tail (Wahle, 1991). Other sequence features around the poly(A) signal have a modest effect on how well this final cleavage–polyadenylation complex binds to the poly(A) signal and in this way also influence how much polyadenylation actually occurs. In particular, the nucleotide to which the poly(A) tail is added following endonucleolytic cleavage (normally an A) has a small but significant effect on efficiency (Chen et al., 1995).
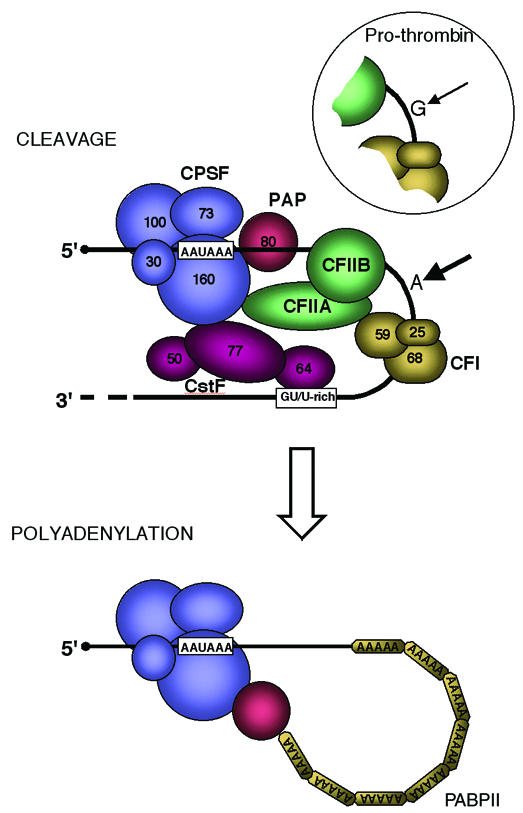
Fig. 1. Molecular arrangement of poly(A) signals on the pre-mRNA together with their associated cleavage and polyadenylation factors. First cleavage occurs (thick arrow) normally at an A nucleotide. A variant G is found at this position in the prothrombin gene, resulting in weaker cleavage efficiency (thin arrow). The mutant form of this gene reverts G to the normal and more efficient A nucleotide (Gehring et al., 2001). CPSF denotes tetrameric cleavage polyadenylation stimulatory factor and CstF denotes trimeric cleavage stimulation factor (Colgan and Manley, 1997). CFI is a trimeric cleavage factor (Wahle and Ruegsegger, 1999) and CFII is a recently characterized second cleavage factor that separates into two multi-subunit proteins, CFIIA and CFIIB (de Vries et al., 2000). Following the cleavage reaction, polyadenylation occurs, requiring CPSF, PAP and PABPII. PAP denotes poly(A) polymerase and PABPII denotes poly(A) binding protein II (Wahle, 1991).
It is this exact nucleotide to which the poly(A) tail is added that has been highlighted quite unexpectedly in a commonly occurring mutation of the prothrombin gene in a recent publication by Gehring et al. (2001). This mutation (present in 1–2% of the human population) consists of a G→A substitution that makes the normally less than perfect poly(A) signal slightly more efficient (Figure 1). Furthermore, the presence of this nucleotide substitution correlates with an increased risk of thrombosis (Lane and Grant, 2000). Patients with this mutation appear to have elevated levels of prothrombin in their plasma, and this is likely to be the cause of their increased risk of thrombosis. To prove the point, Gehring et al. (2001) describe the isolation of the prothrombin poly(A) signal and its immediate sequence environment for both the wild type (G version) and mutant (A version). These two poly(A) signals were then used to replace the 3′ untranslated sequence of the human β-globin gene and the hybrid genes so designed were transfected into tissue culture cells to measure gene expression. Sure enough, almost twice as much message is made by the A version as by the G version construct, which in turn results in twice as much protein expression. Further work on the A and G hybrid globin/prothrombin gene constructs revealed that cleavage at the poly(A) site is twice as efficient but that once cleavage has occurred, polyadenylation is apparently unchanged. What these data graphically illustrate is that the strength of a poly(A) signal can directly influence the level of gene expression. This fact had already been documented (e.g. Gil and Proudfoot, 1987), but the occurrence of a common genetic disorder associated with poly(A) signal efficiency underlines and adds a new dimension to this observation.

Footnotes
Tel: +44 1865 275566; Fax: +44 1865 275556; E-mail: nicholas.proudfoot@path.ox.ac.uk
REFERENCES
- Blackwood E.M. and Kadonaga, J.T. (1998) Going the distance: a current view of enhancer action. Science, 281, 61–63. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti, C. and Tollervey, D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Chen F., MacDonald, C.C. and Wilusz, J. (1995) Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res., 23, 2614–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D. and Manley, J. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- de Vries H., Rüegsegger, U., Hübner, W., Friedlein, A., Langen, H. and Keller, W. (2000) Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J., 19, 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Frede, U., Neu-Yilik, G., Hundsdoerfer, P., Vetter, B., Hentze, M.W. and Kulozik, A.E. (2001) Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nature Genet., 28, 389–392. [DOI] [PubMed] [Google Scholar]
- Gil A. and Proudfoot, N.J. (1987) Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA 3′ end formation. Cell, 49, 399–406. [DOI] [PubMed] [Google Scholar]
- Gravely B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik, A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley, J.L. (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley, J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Lane D.A. and Grant, P.J. (2000) Role of hemostatic gene polymorphisms in venous and arterial thrombotic disease. Blood, 95, 1517–1532. [PubMed] [Google Scholar]
- Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange, T. and Reinberg, D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- McKnight S.L. (1996) Transcription revisited: a commentary on the 1995 Cold Spring Harbor Laboratory meeting, ‘Mechanisms of Eukaryotic Transcription’. Genes Dev., 10, 367–381. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. (2000) Connecting transcription to RNA processing. Trends Biochem. Sci., 25, 105–110. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow, P. and Hentze, M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell, 66, 759–768. [DOI] [PubMed] [Google Scholar]
- Wahle E. and Rüegsegger, U. (1999) 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- Workman J.L. and Kingston, R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- Zenklusen D. and Stutz, F. (2001) Nuclear export of mRNA. FEBS Lett., 498, 150–156. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman, L. and Moore, C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]


