Abstract
Studies on the control of eukaryotic translation initiation by a cap-independent recruitment of the 40S ribosomal subunit to internal messenger RNA sequences called internal ribosome entry sites (IRESs) have shown that these sequence elements are present in a growing list of viral and cellular RNAs. Here we discuss their prevalence, mechanisms whereby they may function and their uses in regulating gene expression.
Introduction
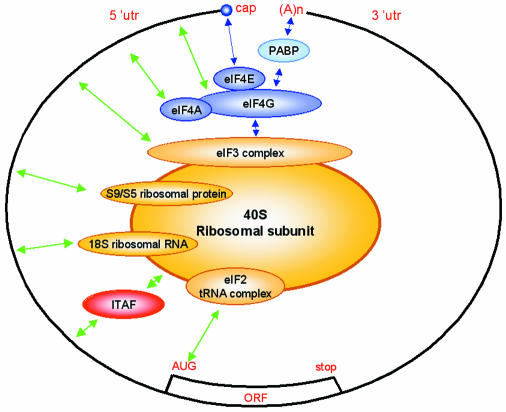
In prokaryotes, the process of ribosome recruitment to the mRNA results from direct base-pairing between the 18S ribosomal RNA and the mRNA Shine–Dalgarno sequence. This RNA–RNA interaction brings the prokaryotic 30S subunit to an internal position on the mRNA, in the vicinity of an AUG initiation codon. In eukaryotes, the situation has proved to be more complicated. The conventional mode of attachment of the 40S ribosomal subunit to the mRNA requires the eIF4F protein adaptor complex, which bridges the 7methyl guanosine capped structure located at the 5′ end of the mRNA and the 40S subunit carrying the eIF3 and eIF2-GTP–Met-tRNA complexes (see Figure 1). In addition, studies on the translation of viral and eukaryotic mRNAs bearing uncommon features in their 5′ untranslated regions (UTRs) have shed light on an alternative mode of 40S recruitment to the mRNA. This process is called internal entry of ribosomes and is mediated by internal sequences called internal ribosome entry sites (IRESs). (An IRES database website is available at http://www.rangueil.inserm.fr/IRESdatabase.)
Fig. 1. Mechanisms of ribosome recruitment to mRNAs. In the cap- and poly(A) tail-mediated mechanism, the eIF4F complex (composed of eIF4E, eIF4G and eIF4A) targets the eIF3-associated 40S ribosomal subunit to the mRNA 5′ end. The cap and the poly(A) tail act synergistically through the interaction between eIF4G and PABP. In the IRES-mediated mechanism, the 40S ribosomal subunit can interact with mRNAs, even in the absence of canonical translation initiation factors. Green arrows indicate direct or indirect [through ITAFs(IRES trans-acting factors)] interactions between IRES elements and the translation initiation machinery identified thus far.
How widespread is internal ribosome entry?
Studies on viral gene translation were essential for the initial discovery of internal entry of ribosomes. Unlike their cellular counterparts, picornaviral mRNAs are naturally uncapped at their 5′ end. Their 5′ UTRs also have complex features predicted to impair ribosome recruitment and linear scanning: (i) a long leader sequence; (ii) stable secondary structures; and (iii) potential upstream initiation codons. Nevertheless, these 5′ UTRs confer efficient 40S joining. The poliovirus and encephalomyocarditis virus (EMCV) 5′ UTRs were the first to be described to ‘break the rule’ of translation initiation (Jackson, 1988; Jang et al., 1988; Pelletier and Sonenberg, 1988). Bicistronic RNAs with two non-overlapping open reading frames (ORFs) were shown to be good models to test cap-independent translation initiation. This was first shown for poliovirus, where inserting a segment of the 5′ UTR of a poliovirus genome between the two ORFs allows translation of the downstream cistron, independent of the cap-mediated translation of the first cistron. This strategy can be considered the ‘gold standard’ for characterizing IRESs (Sachs, 2000) if one considers the presence of cryptic RNA processing signals or promoter sequences in the intercistronic space as having been ruled out (Kozak, 2001). Using this assay as the basis for defining IRESs, these elements have been found in all picornavirus genera. Their presence in viruses as diverse as flaviviruses, retroviruses and even DNA viruses such as the Kaposi’s sarcoma-associated herpesvirus reveals the widespread nature of these RNA elements (see Supplementary data, available at EMBO reports Online).
As is the case for many viral mRNAs, a number of cellular mRNAs possess structural features in their 5′ UTRs that make them unlikely to be translated by a 5′ cap-dependent ribosome-scanning mechanism. Moreover, a few cellular mRNAs are translated preferentially when cap-dependent initiation of translation is impaired. These discoveries argued for an alternative mechanism such as the internal entry of ribosomes. Indeed, the first cellular IRES was identified in the 220-nucleotide-long 5′ UTR of the immunoglobulin heavy chain-binding protein (BiP) mRNA, whose translation is maintained in poliovirus-infected cells at a time when cap-dependent translation is severely inhibited (Macejak and Sarnow, 1991). Since then, and particularly over the last three years, IRES activities have been detected in a restricted but increasing number of cellular mRNAs from yeast, Drosophila, birds and mammals, showing that the internal ribosome entry process is far more extensive than previously thought (see Supplementary data).
Insights into the molecular events underlying internal entry of ribosomes
A chimeric protein formed from a domain of the translation initiation factor eIF4G and the iron responsive element binding protein-1 (IRP-1) is sufficient to promote translation of the downstream cistron of a bicistronic mRNA bearing the iron responsive element (IRE) RNA sequence in the intercistronic space (De Gregorio et al., 1999). This is due to the ability of eIF4G to connect to the 40S subunit via the eIF3 translation initiation factor (Figure 1). This artificial recruitment of eIF4G is reminiscent of that used in 5′ end cap- and poly(A)-dependent translation, where eIF4G is recruited to the mRNA through its interaction with the cap-binding protein eIF4E and the poly(A) tail-binding protein (PABP). A direct ribosome recruitment process is therefore possible for IRES-based translation. However, natural IRESs have developed complex interaction networks as alternatives to the cap/poly(A)–eIF4E/PABP connection.
Various attempts to define cis-elements required for IRES activity revealed that the three-dimensional RNA fold, rather than its primary sequence, is the major determinant of IRES function. To operate as an IRES, an RNA should form a structural scaffold in which precisely positioned RNA tertiary structures contact the 40S ribosomal subunit through a number of specific intermolecular interactions. In a reconstituted translation initiation system, purified 40S ribosomal subunits are able to form a binary complex with the hepatitis C virus (HCV) IRES, even in the absence of the canonical translation initiation factors (Pestova et al., 1998). One site that had previously been defined as a contact point between the 40S subunit and the HCV IRES is the ribosomal protein S9. However, mutations that reduce S9 binding do not affect binary complex formation (Pestova et al., 1998), suggesting that multiple contact points act together to stabilize the complex. On the other hand, a more recent study showed the ribosomal protein S5, but not S9, to interact with the HCV IRES (Fukushi et al., 2001). Thus, the precise role of both proteins should be re-evaluated. Strikingly, a cryo-electron microscopy map of the HCV IRES bound to the 40S subunit revealed that the RNA induces a conformational change that closes the protein’s mRNA-binding cleft (Spahn et al., 2001), but it is currently unclear whether this change is specific to translation initiation that does not require canonical initiation factors. In other IRESs, such as those of cricket paralysis-like viruses, translation is initiated at non-AUG codons without the help of any proteins and even without initiator Met-tRNA (Sasaki and Nakashima, 2000; Wilson et al., 2000), suggesting a strong dependence on RNA structure. Indeed, phylogenetic and mutational analyses have identified a pseudoknot structure to be essential for IRES function (Wilson et al., 2000; Kanamori and Nakashima, 2001).
One might expect that when mRNA is not correctly folded to establish contacts with ribosomal proteins or RNA, some non-ribosomal cofactors are required either to create additional interactions with the 40S subunit or to act as RNA chaperones controlling the functional configuration of the IRES. Studies on picornaviral IRESs have revealed unexpected mRNA-binding properties for various canonical translation initiation factors including eIF3 and eIF4G. Furthermore, a reconstituted 40S ribosome-binding assay has demonstrated the functional involvement of eIF4G and eIF4A in picornaviral IRES function. Non-canonical translation initiation factors with known functions in other processes were shown to interact with various IRESs. The functional roles of these additional IRES trans-acting factors (ITAFs) are generally assessed by in vitro translation assays of IRES-containing reporter constructs supplemented with recombinant proteins. Such assays have led to the assignment of heterogeneous nuclear ribonucleoprotein (hnRNP) I/PTB (Kaminski and Jackson, 1998; a polypyrimidine-tract-binding protein known for its role as a splicing regulator), hnRNP E2/PCBP2 (Hunt et al., 1999; Walter et al., 1999), La (Meerovitch et al., 1993; Holcik and Korneluk, 2000; an autoantigen with diverse RNA metabolism activities), unr (Hunt et al., 1999; upstream of N-ras), ITAF45/Mpp1 (Pilipenko et al., 2000; a protein whose expression is up-regulated in response to mitogen stimulation and is not detectable in differentiated cells), DAP5/NAT1/p97 (Henis-Korenblit et al., 2000; an eIF4G homolog) and nucleolin (Izumi et al., 2001) as ITAFs. These ITAFs are not active on all IRESs and they act either alone or in combination to mediate IRES-dependent translation (Hunt et al., 1999; Pilipenko et al., 2000; Mitchell et al., 2001). However, in vivo assays are required to confirm their ITAF function. Indeed, disruption of the DAP5 gene in mouse embryonic stem cells does not affect the IRES activities of various bicistronic transfected genes (Yamanaka et al., 2000).
Interestingly, the predominant nuclear localization of several ITAFs led to the hypothesis that either their binding to IRES-containing mRNAs is a nuclear process or they relocalize to the cytoplasm to bind their target mRNAs. The observation that several cellular, but not viral, IRES-containing mRNAs are translated only when expressed within the nucleus suggest that there is a requirement for a ‘nuclear history’ in the functionality of certain cellular IRESs (Stoneley et al., 2000a; B. Galy and S. Vagner, unpublished). However, the ability of some ITAFs, e.g. hnRNPs, to shuttle between the nucleus and the cytoplasm could also reflect their putative role in translation initiation. Discoveries of new ITAFs and the definition of the complexes involved in IRES-dependent translation will help in the precise understanding of the initiation process. Whereas the biochemical purification of a complex on such a long and incompletely defined RNA is not an easy task, the recent discovery of IRESs in Saccharomyces cerevisiae (Zhou et al., 2001) makes a genetic approach possible and this will certainly speed up the discovery process.
Cap-mediated translation is stimulated not only by 5′ UTR-interacting proteins, but also by the poly (A) binding protein (PABP), which links the poly(A) tail to the translation machinery via a direct association with eIF4G (Figure 1). Two mammalian in vitro systems that support cap-poly(A) tail synergistic stimulation of translation were recently developed and used to show that translation mediated by some picornaviral IRESs is also stimulated by the poly(A) tail (Bergamini et al., 2000; Michel et al., 2000) and requires the integrity of the eIF4G–PABP interaction (Michel et al., 2001). Interestingly, instead of using a poly(A) tail to enhance translation, the HCV RNA harbors a 98-nucleotide-long region at its 3′ end for this purpose (Ito et al., 1998; Michel et al., 2001). It is not firmly established whether this translation activation is specific to the HCV IRES but it binds various proteins that could be important for IRES function. Among them, the L22 ribosomal protein seems to modulate IRES activity (Wood et al., 2001).
Finally, because of the fact that direct binding of the small ribosomal subunit to the mRNA in prokaryotes is largely governed by base pairing between the 16S rRNA and the short Shine–Dalgarno sequence (Shine et al., 1974), it has been hypothesized that, instead of RNA–protein interactions, mRNA–18S rRNA contacts would help in 40S subunit recruitment in eukaryotes. Indeed, the Gtx homeodomain protein 5′ UTR contains a 9-nucleotide segment that can function as an IRES (Chappell et al., 2000a) and crosslink to the 18S rRNA (Hu et al., 1999). The contribution of this interaction in IRES activity can now be evaluated precisely.
Although IRESs may be viewed simply as functional equivalents of the prokaryotic Shine–Dalgarno sequence, the possible and as yet incompletely understood molecular mechanisms leading to internal entry of ribosomes on eukaryotic or viral mRNAs are far more complicated than those used by prokaryotes. The advantage of this may be that various mechanisms can form multiple targets for the fine-tuning of gene expression at the translational level.
IRES control of gene expression?
The mechanisms that viruses have evolved to promote internal entry of ribosomes are some of the most effective for hijacking the translational apparatus of the host cell to favor the expression of foreign transcripts. For example, upon infection with certain picornaviruses, a virally encoded protease proteolytically cleaves eIF4G, thus dissociating the cap-binding activity of the eIF4F complex adaptor from its 40S subunit-binding activity. This not only blocks cap-dependent translation of host transcripts, but also enhances IRES-mediated translation of viral mRNAs. Some picornaviruses can also block cap-dependent translation through the manipulation of the signaling pathways that normally control eIF4F complex asssembly. For instance, EMCV and poliovirus inhibit the phosphorylation of 4E-BP1, whose hypophosphorylated forms prevent eIF4F assembly and repress cap-dependent translation, but have no effect on IRES-mediated translation of viral mRNAs.
Most cellular IRESs have been shown to function preferentially when cap-dependent translation is physiologically impaired, either by dephosphorylation of 4E-BP1 (during mitosis, quiescence, differentiation or stress) or by proteolytic cleavage of eIF4G (during apoptosis). The molecular mechanisms that redirect the ribosome from the cap structure to IRES sequences under such conditions are unknown. One attractive possibility is that the inhibition of cap-dependent translation frees initiation factors for IRES-mediated processes. However, this would apply indiscriminately to all IRES-containing mRNAs, and does not explain how, depending on cell type or cell status, only a subset of IRES-containing mRNAs is translated. It is thus likely that, in addition to canonical translation initiation factors, specific trans-acting proteins capable of selectively targeting the ribosome to a particular set of cellular mRNAs are involved. There is some evidence to support this.
For instance, cap-dependent translation is impaired when cells undergo various forms of stress. Consistent with this, IRES elements were found to be active during γ-irradiation (XIAP; Holcik et al., 1999), hypoxia (VEGF; Stein et al., 1998) or amino acid starvation (Fernandez et al., 2001). This led to the current hypothesis that IRES-mediated translation of certain mRNAs represents a regulatory mechanism that helps the cell to cope with transient stress. Variation in trans-acting factors binding to IRESs could mediate this regulation. Indeed, the profile of proteins bound to the FGF2 5′ UTR, which bears an IRES, varies upon heat shock treatment (Vagner et al., 1996).
Some mRNAs encoding pro-apoptotic proteins, including Apaf-1 (Coldwell et al., 2000) and DAP5 (Henis-Korenblit et al., 2000), are also translated via an IRES element. This suggests that pro- and anti-apoptotic (or survival) factors utilize a similar IRES-mediated translational mechanism. The outcome for the cell could then depend on the balance between the relative strengths of the IRESs under a particular condition. Accordingly, when the cell undergoes apoptosis, c-myc and DAP5 translation persists even following caspase cleavage of eIF4G (Henis-Korenblit et al., 2000; Stoneley et al., 2000b). Among the ITAFs that would control translation during apoptosis, the La autoantigen seems to be a good candidate (Holcik and Korneluk, 2000). Although it is unknown whether the Apaf-1 IRES actually functions during apoptosis, it binds unr and PTB (Mitchell et al., 2001), two proteins already known to bind other IRESs.
An IRES-dependent mechanism is also used by mRNAs that encode proteins whose biological function is restricted to differentiated cells. For instance, while gap junctions are not present in stem cells, they serve a vital function in the maintenance of the differentiated state of certain epithelial cells at a time when cap-dependent translation is impaired. It is thus not too surprising that IRESs in the 5′ UTRs of several Connexin (gap junction proteins) encoding mRNAs have been identified and are suspected to operate preferentially in differentiated cells (Werner, 2000). In neurons, de novo protein synthesis within dendrites is required for lasting changes in synaptic activity. Nevertheless, the components of the cap-binding complex are present at low concentration in dendrites as compared with the cell body. Interestingly, five different neuronal mRNAs can be translated in dendrites through an IRES-mediated mechanism (Pinkstaff et al., 2001). These findings suggest that IRES-mediated processes are utilized by differentiated cells to control the translation of mRNAs that are localized to specific regions within the cell. In oligodendrocytes, one IRES-containing mRNA whose protein product accumulates during differentiation despite the reduction in global translation rates is that encoding p27kip1, the inhibitor of G1 cyclins (Miskimins et al., 2001). The putative ITAFs HuR and hnRNP C1/C2 have been shown to interact specifically with a U-rich element in the p27kip1 5′ UTR (Millard et al., 2000). Since the presence of U-rich elements in other cellular mRNAs, including ornithine decarboxylase (ODC) and XIAP mRNAs, is critical for IRES activity, it is likely that U-rich binding proteins play a role in IRES-mediated translation initiation. That hnRNP C1/C2 interact with the c-sis IRES during differentiation further supports this hypothesis (Sella et al., 1999).
IRES-dependent initiation is sometimes utilized during mitosis, when cap-dependent translation is impeded. Translation of poliovirus and HCV is unaffected, or even enhanced, during mitosis (Bonneau and Sonenberg, 1987; Honda et al., 2000). Similarly, two cellular mRNAs, encoding ODC (Pyronnet et al., 2000) and the cdk-like p58PITSLRE (Cornelis et al., 2000), possess IRESs whose activities are restricted to mitosis. No ITAFs with a specific activity at that time of the cell cycle have yet been described.
Perspectives
IRESs may continue to be identified in the near future. Certainly, the numerous mRNAs whose 5′ UTR structures likely interfere with the 5′ cap-dependent ribosome are good candidates for the presence of an IRES. However, the prediction of an IRES from just looking at the 5′ UTR could be strengthened by a better understanding of the structural elements that comprise these elements. The maintenance or the activation of translation of some mRNAs under conditions of reduced cap-dependent translation can also be used as a predictive tool. Finally, an experimental genome-wide approach such as a cDNA microarray identification of polysome-associated mRNAs could be applied to the various conditions in which the cap-mediated initiation is affected (e.g. Johannes et al., 1999).
Cellular IRES-containing mRNAs often encode regulatory proteins whose expression is tightly regulated. Therefore, any disturbance in IRES function (e.g. due to IRES mutation and/or inadequate ITAF activity) could have dramatic physiological and pathological consequences. For instance, a C→T transition in the Connexin-32 5′ UTR detected in some patients with the Charcot–Marie–Tooth disease, a neurodegenerative disorder, has been shown to severely impair IRES activity (Hudder and Werner, 2000). Conversely, a C→T transition in the c-myc IRES oncogene has been proposed to increase IRES activity and the consequent translation of c-Myc in multiple myeloma (Chappell et al., 2000b). Thus, strategies designed to control IRES activity could prove to be of therapeutic interest.
In addition to being potential therapeutic targets, IRESs can serve as biotechnological tools, particularly for gene therapy. A direct application is the synthesis of several proteins of interest from a single, multicistronic mRNA. Furthermore, the property of certain IRESs to be active in certain tissues (Creancier et al., 2000, 2001) and to be regulated under defined conditions could represent an advantage for the targeting of transgene expression.
Admittedly, translation is a key step in the regulation of gene expression. In this respect, the unexpectedly broad distribution of IRESs suggests a major role in rapidly setting up the cellular equipment during mitosis and under stress. No doubt future descriptions of the IRES structures will help to unravel the intricate mechanisms whereby they operate, particularly in terms of the trans-acting factors that participate in the internal ribosome entry process. How individual ITAFs interact with particular IRESs to modulate their activities and how IRES function can be controlled artificially are irresistible problems to tackle in the future.
Supplementary data. Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
Acknowledgements
Work performed in the authors’ laboratories was supported by INSERM (Institut National de la Santé et de la Recherche Médicale) (S.V. and S.P.), ARC (Association pour la Recherche sur le Cancer) (S.V.), La Ligue Nationale Contre le Cancer (S.P.) and EMBL (European Molecular Biology Laboratory) (B.G.). B.G. is the recipient of a long-term EMBO fellowship (ALTF 312-1999).
References
- Bergamini G., Preiss, T. and Hentze, M.W. (2000) Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA, 6, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau A.M. and Sonenberg, N. (1987) Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem., 262, 11134–11139. [PubMed] [Google Scholar]
- Chappell S.A., Edelman, G.M. and Mauro, V.P. (2000a) A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA, 97, 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell S.A., LeQuesne, J.P., Paulin, F.E., deSchoolmeester, M.L., Stoneley, M., Soutar, R.L., Ralston, S.H., Helfrich, M.H. and Willis, A.E. (2000b) A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene, 19, 4437–4440. [DOI] [PubMed] [Google Scholar]
- Coldwell M.J., Mitchell, S.A., Stoneley, M., MacFarlane, M. and Willis, A.E. (2000) Initiation of Apaf-1 translation by internal ribosome entry. Oncogene, 19, 899–905. [DOI] [PubMed] [Google Scholar]
- Cornelis S., Bruynooghe, Y., Denecker, G., Van Huffel, S., Tinton, S. and Beyaert, R. (2000) Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell, 5, 597–605. [DOI] [PubMed] [Google Scholar]
- Creancier L., Morello, D., Mercier, P. and Prats, A.C. (2000) Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol., 150, 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creancier L., Mercier, P., Prats, A.C. and Morello, D. (2001) c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol. Cell. Biol., 21, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Preiss, T. and Hentze, M.W. (1999) Translation driven by an eIF4G core domain in vivo. EMBO J., 18, 4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J., Yaman, I., Mishra, R., Merrick, W.C., Snider, M.D., Lamers, W.H. and Hatzoglou, M. (2001) Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem., 276, 12285–12291. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Okada, M., Stahl, J., Kageyama, T., Hoshino, F.B. and Katayama, K. (2001) Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem., 276, 20824–20826. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S., Strumpf, N.L., Goldstaub, D. and Kimchi, A. (2000) A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol., 20, 496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M. and Korneluk, R.G. (2000) Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol., 20, 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Lefebvre, C., Yeh, C., Chow, T. and Korneluk, R.G. (1999) A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nature Cell Biol., 1, 190–192. [DOI] [PubMed] [Google Scholar]
- Honda M., Kaneko, S., Matsushita, E., Kobayashi, K., Abell, G.A. and Lemon, S.M. (2000) Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology, 118, 152–162. [DOI] [PubMed] [Google Scholar]
- Hu M.C., Tranque, P., Edelman, G.M. and Mauro, V.P. (1999) rRNA-complementarity in the 5′ untranslated region of mRNA specifying the Gtx homeodomain protein: evidence that base-pairing to 18S rRNA affects translational efficiency. Proc. Natl Acad. Sci. USA, 96, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudder A. and Werner, R. (2000) Analysis of a Charcot–Marie–Tooth disease mutation reveals an essential internal ribosome entry site element in the connexin-32 gene. J. Biol. Chem., 275, 34586–34591. [DOI] [PubMed] [Google Scholar]
- Hunt S.L., Hsuan, J.J., Totty, N. and Jackson, R.J. (1999) unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev., 13, 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Tahara, S.M. and Lai, M.M. (1998) The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol., 72, 8789–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi R.E., Valdez, B., Banerjee, R., Srivastava, M. and Dasgupta, A. (2001) Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res., 76, 17–29. [DOI] [PubMed] [Google Scholar]
- Jackson R.J. (1988) RNA translation. Picornaviruses break the rules. Nature, 334, 292–293. [DOI] [PubMed] [Google Scholar]
- Jang S.K., Krausslich, H.G., Nicklin, M.J., Duke, G.M., Palmenberg, A.C. and Wimmer, E. (1988) A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol., 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G., Carter, M.S., Eisen, M.B., Brown, P.O. and Sarnow, P. (1999) Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA, 96, 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A. and Jackson, R.J. (1998) The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA, 4, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y. and Nakashima, N. (2001) A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA, 7, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2001) New ways of initiating translation in eukaryotes? Mol. Cell. Biol., 21, 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak D.G. and Sarnow, P. (1991) Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature, 353, 90–94. [DOI] [PubMed] [Google Scholar]
- Meerovitch K., Svitkin, Y.V., Lee, H.S., Lejbkowicz, F., Kenan, D.J., Chan, E.K., Agol, V.I., Keene, J.D. and Sonenberg, N. (1993) La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol., 67, 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel Y.M., Poncet, D., Piron, M., Kean, K.M. and Borman, A.M. (2000) Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem., 275, 32268–32276. [DOI] [PubMed] [Google Scholar]
- Michel Y.M., Borman, A.M., Paulous, S. and Kean, K.M. (2001) Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis c virus translation. Mol. Cell. Biol., 21, 4097–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard S.S., Vidal, A., Markus, M. and Koff, A. (2000) A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol., 20, 5947–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskimins K., Wang, G., Hawkinson, M. and Miskimins, R. (2001) Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol., 21, 4960–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.A., Brown, E.C., Coldwell, M.J., Jackson, R.J. and Willis, A.E. (2001) Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol., 21, 3364–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J. and Sonenberg, N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J. and Hellen, C.U. (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev., 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Pestova, T.V., Kolupaeva, V.G., Khitrina, E.V., Poperechnaya, A.N., Agol, V.I. and Hellen, C.U. (2000) A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev., 14, 2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Pinkstaff J.K., Chappell, S.A., Mauro, V.P., Edelman, G.M. and Krushel, L.A. (2001) Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl Acad. Sci. USA, 98, 2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S., Pradayrol, L. and Sonenberg, N. (2000) A cell cycle-dependent internal ribosome entry site. Mol. Cell, 5, 607–616. [DOI] [PubMed] [Google Scholar]
- Sachs A.B. (2000) Cell cycle-dependent translation initiation: IRES elements prevail. Cell, 101, 243–245. [DOI] [PubMed] [Google Scholar]
- Sasaki J. and Nakashima, N. (2000) From the cover: methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl Acad. Sci. USA, 97, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella O., Gerlitz, G., Le, S.Y. and Elroy-Stein, O. (1999) Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell. Biol., 19, 5429–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno, L. and Hunt, J.A. (1974) Fingerprinting of eukaryotic ribosomal RNA labelled with tritiated nucleosides. Anal. Biochem., 59, 360–365. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A. and Frank, J. (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science, 291, 1959–1962. [DOI] [PubMed] [Google Scholar]
- Stein I., Itin, A., Einat, P., Skaliter, R., Grossman, Z. and Keshet, E. (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol., 18, 3112–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M., Subkhankulova, T., Le Quesne, J.P., Coldwell, M.J., Jopling, C.L., Belsham, G.J. and Willis, A.E. (2000a) Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res., 28, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M., Chappell, S.A., Jopling, C.L., Dickens, M., MacFarlane, M. and Willis, A.E. (2000b) c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol., 20, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S., Touriol, C., Galy, B., Audigier, S., Gensac, M.C., Amalric, F., Bayard, F., Prats, H. and Prats, A.C. (1996) Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J. Cell Biol., 135, 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter B.L., Nguyen, J.H., Ehrenfeld, E. and Semler, B.L. (1999) Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA, 5, 1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner R. (2000) IRES elements in connexin genes: a hypothesis explaining the need for connexins to be regulated at the translational level. IUBMB Life, 50, 173–176. [DOI] [PubMed] [Google Scholar]
- Wilson J.E., Pestova, T.V., Hellen, C.U. and Sarnow, P. (2000) Initiation of protein synthesis from the A site of the ribosome. Cell, 102, 511–520. [DOI] [PubMed] [Google Scholar]
- Wood J., Frederickson, R.M., Fields, S. and Patel, A.H. (2001) Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol., 75, 1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Zhang, X.Y., Maeda, M., Miura, K., Wang, S., Farese, R.V., Jr, Iwao, H. and Innerarity, T.L. (2000) Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J., 19, 5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Edelman, G.M. and Mauro, V.P. (2001) Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl Acad. Sci. USA, 98, 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.