Abstract
Modification of histones can have a dramatic impact on chromatin structure and function. Acetylation of lysines within the N-terminal tail of the histone octamer marks transcriptionally active regions of the genome whereas deacetylation seems to play a role in transcriptional silencing. Recently, the methylation of the histone tails has also been shown to be important for transcriptional regulation and chromosome structure. Here we show by immunoaffinity purification that two activities important for chromatin-mediated gene silencing, the histone methyltransferase SU(VAR)3-9 and the histone deacetylase HDAC1, associate in vivo. The two activities cooperate to methylate pre-acetylated histones. Both enzymes are modifiers of position effect variegation and interact genetically in flies. We suggest a model in which the concerted histone deacetylation and methylation by a SU(VAR)3-9/HDAC1-containing complex leads to a permanent silencing of transcription in particular areas of the genome.
INTRODUCTION
The packaging of DNA into chromatin plays a very important role during establishment and maintenance of stable gene expression patterns. Post-translational modifications of the nucleosome have a major regulatory function during this process by generating an epigenetic code (Grant et al., 1998b; Strahl and Allis, 2000; Imhof and Becker, 2001). Acetylation of lysine residues within the N-terminal histone tails is associated with transcriptionally active regions within the genome and plays a causal role during the activation process (Wade et al., 1997; Kuo and Allis, 1998; Mizzen and Allis, 1998). Conversely the removal of acetyl groups from the histones has a negative effect on gene expression in multiple model systems (Zhang et al., 1997, 1999; Brehm et al., 1998; Jones et al., 1998; Wade et al., 1999). The recent identification of the first lysine-specific histone methyltransferase (HIM) as the heterochromatin-associated protein SUV39H1 (Rea et al., 2000) led to the hypothesis that histone methylation might also play a role during gene silencing. SUV39H1 selectively methylates lysine 9 within the H3 N-terminus, which can then serve as a docking site for another well characterized heterochromatin-associated protein, HP1 (Bannister et al., 2001; Lachner et al., 2001), connecting lysine 9 methylation with transcriptional repression.
A way of studying transcriptional silencing in vivo is the analysis of position effect variegation (PEV) in flies (Spofford, 1967; Henikoff, 1990; Reuter and Spierer, 1992). In extensive screens for modifiers of this phenomenon mutant alleles of the methyltransferase Su(var)3-9 as well as the deacetylase HDAC1 have been isolated as potent suppressors of PEV (Tschiersch et al., 1994; Mottus et al., 2000). This suggests a functional role for these histone-modifying enzymes during the establishment and/or maintenance of functional chromatin domains.
Here we report the isolation of SU(VAR)3-9 from Drosophila embryo extracts. We show that SU(VAR)3-9 isolated from embryos is associated with HIM as well as histone deacetylase (HDAC) activity. We show that this deacetylase activity is due to the association of SU(VAR)3-9 with HDAC1. The deacetylase activity is essential for methylation of a pre-acetylated peptide. This functional synergism between HDAC1 and SU(VAR)3-9 is also observed in vivo as a dominant-negative HDAC1 mutant effectively represses a triplo-enhancer effect of Su(var)3-9 on PEV. Recently, a genetic interaction between the methyltransferase clr4 and the deacetylase clr3 in Schizosaccharomyces pombe has been reported (Nakayama et al., 2001). It thus seems likely that a comparable interaction between a HIM and a HDAC also exists in S. pombe and that an evolutionarily conserved deacetylation/methylation reaction serves to establish a specific epigenetic mark for heterochromatin formation.
RESULTS
Recombinant SUV39H1 has intrinsic HIM activity (Rea et al., 2000). However, little is known about SU(VAR)3-9 in vivo and whether it acts as a single molecule or within a macromolecular assembly analogous to the acetyltransferase complexes SAGA and NuA4 (Grant et al., 1998a; Allard et al., 1999). In order to study this we fractionated nuclear extracts from Drosophila embryos (0–12 h after egg laying) (Figure 1A) prepared from a fly strain expressing a myc-tagged version of SU(VAR)3-9 (Aagaard et al., 1999). In this strain the Su(var)3-9 gene is expressed under the control of a heat shock promoter and can be switched on by a simple heat shock pulse (Aagaard et al., 1999). Myc-tagged SU(VAR)3-9 binds to a resource Q column and can be eluted at a salt concentration between 150 and 250 mM KCl (Figure 1A and B). The anti-myc antibody immunoprecipitates HIM activity from a nuclear extract made from the tagged fly strain but not from wild-type flies (Figure 1C) and from active column fractions containing myc-SU(VAR)3-9 (Figure 1C).
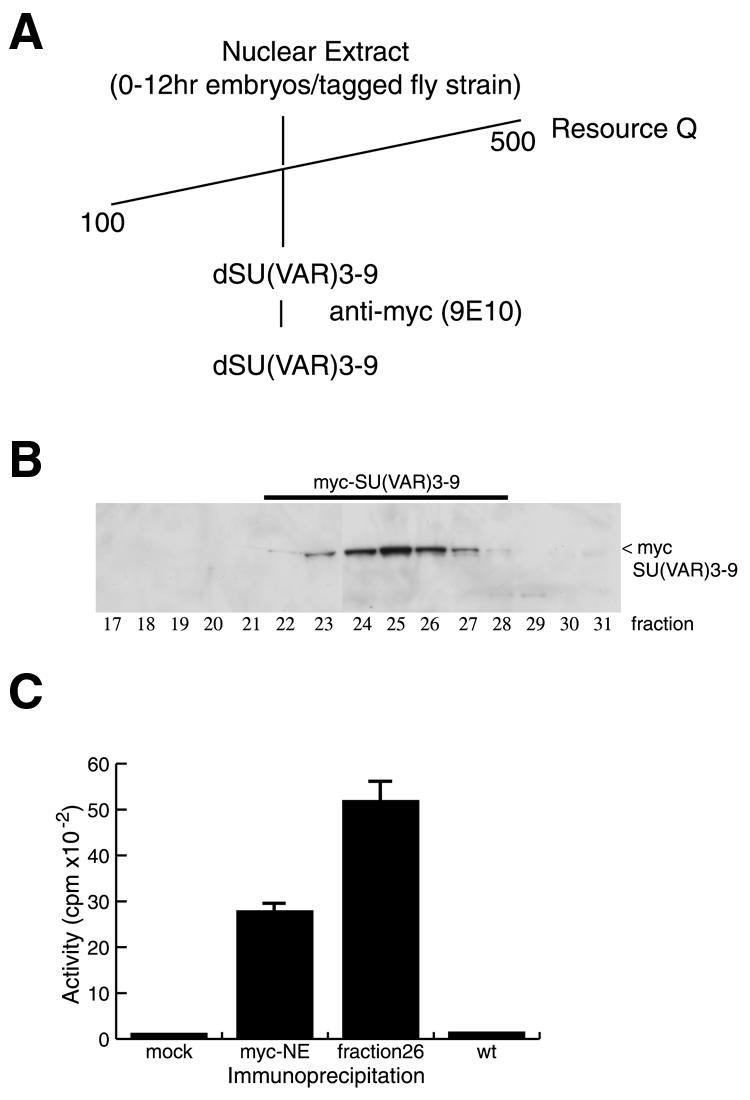
Fig. 1. Purification of a HIM from Drosophila embryo extracts. (A) Purification scheme for the purification of myc-tagged SU(VAR)3-9 from Drosophila embryonal extracts. (B) Purification of myc-SU(VAR)3-9 from nuclear extracts prepared from flies expressing a myc-tagged SU(VAR)3-9 (Aagaard, 1999) on a resource Q column. Western blot of fractions eluted from the resource Q column probed with an anti-c-myc antibody (9E10). (C) HIM activity assay with immunoprecipitates of SU(VAR)3-9 from an extract (myc-NE) or from partially purified myc-SU(VAR)3-9 fractions (fraction 26) prepared from flies expressing triple-myc-tagged SU(VAR)3-9 using an anti-myc antibody (9E10). As controls immunoprecipitations were also performed without the addition of antibody (mock) or from an extract prepared from wild-type embryos (wt).
To analyse the lysine specificity of immunoprecipitated SU(VAR)3-9 we used peptide substrates resembling the N-terminus of histone H3 that were either unmodified or premodified at specific amino acid residues (Figure 2A). SU(VAR)3-9 isolated from embryonic extracts could methylate a peptide premethylated at lysine 4 but was unable to methylate a peptide methylated at lysine 9, which matches the lysine specificity observed with recombinant SUV39H1 (Rea et al., 2000) (Figure 2A). However, a peptide acetylated on lysine 9 could be methylated by immunoprecipitated SU(VAR)3-9, but not by recombinant SUV39H1 (Figure 2A). In order to find out whether this effect was due to a deacetylase activity associated with SU(VAR)3-9 we repeated the assay in the presence of a specific HDAC inhibitor, Trichostatin A (TSA). These experiments showed that TSA significantly reduces the observed HIM activity of SU(VAR)3-9 on a peptide acetylated at lysine 9 (Figure 2B). This inhibition by TSA suggests that the deacetylation of lysine 9 is indeed a prerequisite for a subsequent methylation of lysine 9 by SU(VAR)3-9.
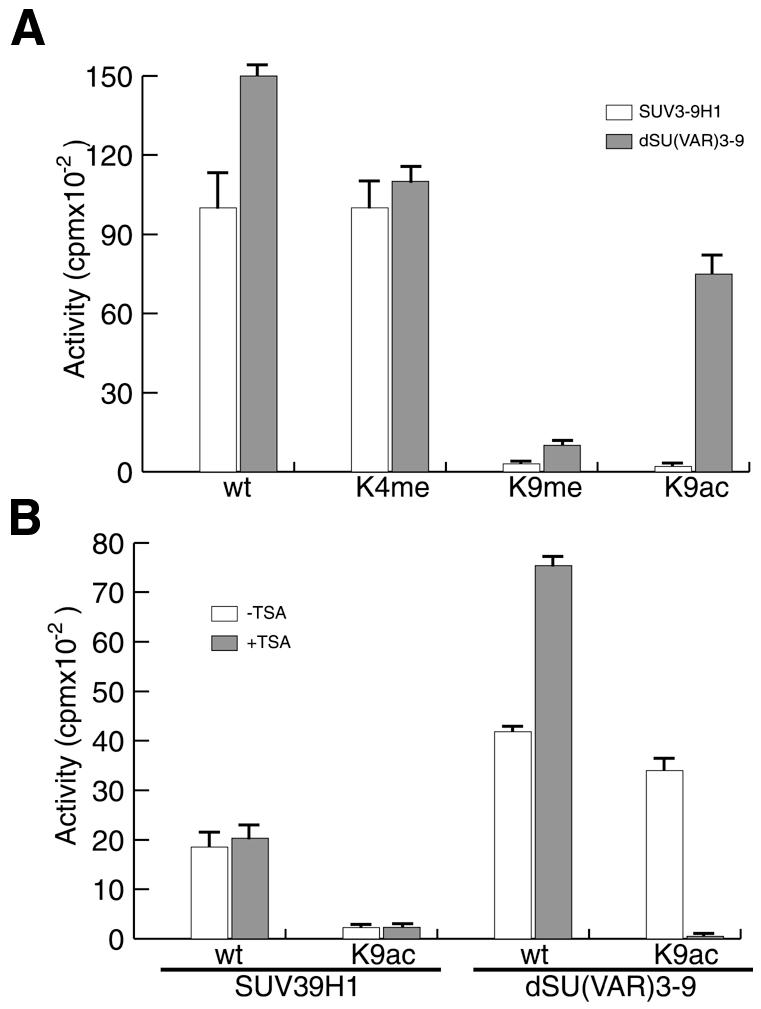
Fig. 2. Lysine specificity of purified SU(VAR)3-9. (A) Specificity assay of recombinant SUV3-9H1 (white bars) and SU(VAR)3-9 prepared as described in Figure 1A (dark bars) on various modified peptides. Modifications used were unmodified (wt), methylated on lysine 4 (K4me), methylated on lysine 9 (K9me) and acetylated on lysine 9 (K9ac). (B) Reactions were performed as in (A) with the exception that reactions contained either 50 nM Trichostatin A (+TSA) dissolved in ethanol or only ethanol (–TSA).
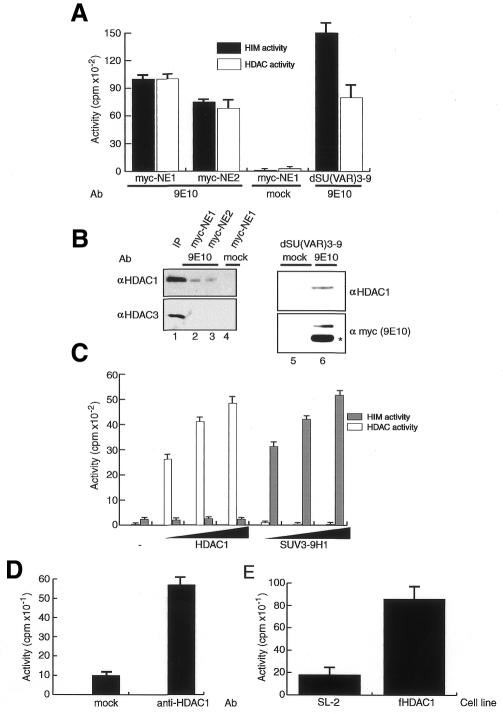
Based on these observations we analysed the immunoprecipitated SU(VAR)3-9 for HDAC activity (Figure 3A). With the anti-myc antibody, we could indeed recover HDAC activity from extracts and partially purified SU(VAR)3-9-containing fractions prepared from the tagged fly strain but not from wild-type flies (Figure 3A). This HDAC activity is probably due to the presence of HDAC1 in these fractions as we could detect HDAC1 in the immunopurified material but not HDAC3 (Figure 3B). We cannot eliminate, however, the possibility that other HDACs different from HDAC1 and 3 contribute to the observed HDAC activity, but as we have identified HDAC1 as being associated with SU(VAR)3-9 we decided to further investigate this association. Neither recombinant HDAC1 nor SUV39H1 has a dual activity in our standard methyltransferase and deacetylase assays (Figure 3C), therefore we could exclude the possibility that a single polypeptide is responsible for the observed effects.
Fig. 3. Association of SU(VAR)3-9 with HDAC1. (A) HDAC activity assay using immunoprecipitated material from two independently prepared nuclear extracts (myc-NE1 and myc-NE2) or a partially purified fraction (fraction 26) from the tagged fly strain precipitated with an anti-c-myc antibody (9E10) or no antibody (mock). (B) Western blot of immunoprecipitated material from (A) probed either with anti-HDAC1 (top panel), anti-HDAC3 antibodies (bottom panel, lanes 1–3) or anti-c-myc antibody (9E10) (bottom panel, lanes 5–6); the asterisk indicates the light chain of the antibody used for immunoprecipitation. The input lane (lane 1) contains ∼30% of the material used for immunoprecipitations. (C) HIM and HDAC activity assays of recombinantly expressed HDAC1 (left panel) and SUV3-91 (right panel). (D) HIM activity with immunoprecipitates using an anti-HDAC1 antibody from a partially purified fraction (Figure 1B, fraction 26). (E) HIM activity assay using material immunoprecipitated with an anti-flag antibody. Immunoprecipitates were purified from Drosophila control cells (SL-2) or cells stably expressing flag-tagged HDAC1 (fHDAC1).
To further confirm the interaction between HDAC1 and SU(VAR)3-9, we used an anti-HDAC1 antibody to immunoprecipitate HIM activity from the partially purified SU(VAR)3-9 fractions mentioned above (Figure 3D). Independent evidence for an interaction of HDAC and HIM activities was obtained by using a cell line that was stably transfected with flag-HDAC1 and expresses it to high levels. Immunoprecipitation with anti-FLAG antibodies precipitated HDAC1 and, in addition to a strong HDAC activity, a pronounced HIM activity (Figure 3E). Although these experiments suggest a physical interaction between SU(VAR)3-9 and HDAC1 we were unable to show a direct interaction using GST pull-down assays (data not shown). Therefore, we suggest the presence of at least one additional factor bridging between SU(VAR)3-9 and HDAC1. Interestingly we have partially purified a very similar activity from extracts prepared from wild-type flies, which further confirms the observed interaction and argues against the possibility that the association we see is artificial due to an overexpression of SU(VAR)3-9 (B. Czermin et al., manuscript in preparation).
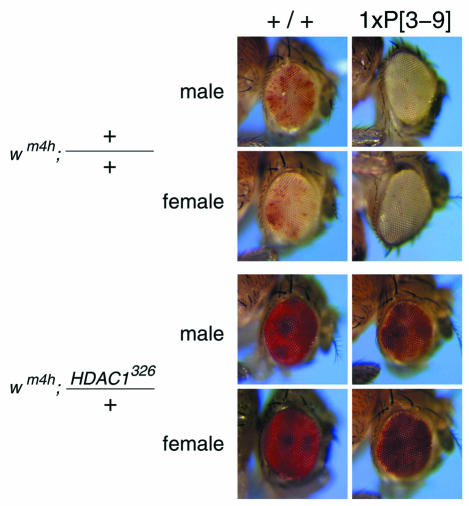
To study the functional interaction of HDAC1 and SU(VAR)3-9 in vivo we made use of the well characterized PEV tester strain In(1)wm4h (Reuter et al., 1986), which contains an inversion placing the white marker gene adjacent to pericentric X heterochromatin. In this experimental setting extra copies of Su(var)3-9 significantly enhance silencing of the white gene (Tschiersch et al., 1994) (Figure 4, top panel). This so-called triplo-enhancer effect of Su(var)3-9 displays a dominant effect over most other known Su(var)s. However, when the strain expressing an extra copy of Su(var)3-9 is crossed with a strain carrying the HDAC1326 mutation (Mottus et al., 2000) we still observe a strong suppression of PEV leading to a red eye phenotype (Figure 4, bottom panel). Because Drosophila has four different HDACs with sequence similarities to HDAC1 (Adams et al., 2000) we used a point mutation of HDAC1, which is still expressed (Mottus et al., 2000) and will get incorporated into multiprotein complexes where it exerts dominant-negative functions. We concluded from these experiments that HDAC1 lies upstream of Su(var)3-9 in the regulatory cascade leading to the formation of pericentric heterochromatin.
Fig. 4. The HDAC326 mutation dominates the PEV enhancer effect of an additional genomic copy of Su(var)3-9. The P[3–9] carries one additional genomic copy of Su(var)3-9 introduced by P[(ry+) 11kb SU(VAR)3-9] at 100E.
DISCUSSION
Here we report the association of the HIM SU(VAR)3-9 and the deacetylase HDAC1 within Drosophila embryo extracts. We show that this interaction plays an essential role in SU(VAR)3-9’s ability to methylate acetylated histone tails, which is probably important during the ‘invasion’ of euchromatic regions of the genome by heterochromatin that is observed when a gene is placed closed to heterochromatin (Spofford, 1967).
In addition to their biochemical interaction, we also see a strong genetic interaction between Su(var)3-9 and HDAC1. A point mutation within the HDAC1 gene, which has a strong Su(var) phenotype, very efficiently dominates the ‘triplo-enhancer effect’ usually seen in flies carrying additional copies of Su(var)3-9. The effect of different HDAC1 mutations on PEV has been described as enhancing, suppressing or neutral (De Rubertis et al., 1996; Mannervik and Levine, 1999; Mottus et al., 2000) depending on the experimental setup. These controversial results are probably due to the different nature of the mutants used in the different laboratories. A functional redundancy of HDAC1 with other known HDACs could, for example, obscure the contribution of HDAC1 in a hypomorphic strain. We postulate that in the HDAC1326 strain a mutant protein is made that is able to interact with SU(VAR)3-9 but fails to deacetylate the histone substrate and therefore acts as a dominant-negative suppressor.
Based on these findings we propose a model in which deacetylation precedes methylation of lysine 9 in the N-terminus of histone H3. Methylated H3 would then serve as a docking site for HP1 (Bannister et al., 2001; Lachner et al., 2001), which in turn could help in assembling a specialized higher order chromatin structure. As the turnover of methylated histones is slow in comparison to acetylation (Waterborg, 1993), we suggest that histone methylation serves as a permanent epigenetic mark that freezes a particular chromatin conformation.
The concerted action of deacetylation and methylation of lysine 9 in histone H3 could allow the generation of a permanently repressed chromatin structure within otherwise more accessible, acetylated chromatin of the early embryo. The interaction between SU(VAR)3-9 and HDAC1 is probably especially important at boundaries between eu- and heterochromatin and under circumstances when heterochromatin has been shown to ‘invade’ euchromatic regions like the white gene in the In(1)wm4h strain (Reuter et al., 1986).
The existence of a complex containing a HDAC and a HIM provides a molecular mechanism for the close connection between histone deacetylation and histone methylation. This link between deacetylation and methylation is also evident in S. pombe, where inhibition of deacetylases by TSA as well as mutations of the Su(var)3-9 orthologue clr4 leads to defects in centromere function (Ekwall et al., 1997; Ivanova et al., 1998). It will be very interesting to see whether the coupling of histone deacetylation and histone methylation is a mechanism commonly used to stably repress gene expression not only around the centromere but also in euchromatic regions of the genome.
METHODS
Histone methylation. Peptide HIM assays were carried out in 25 µl of methyltransferase buffer [50 mM Tris–HCl pH 8.0, 0.5 mM DTT] containing 1 µg of peptide (ARTKQTARKSTGGKAPRKQL, synthesised by Peptide Speciality Laboratories, Heidelberg) as substrate and 500 nCi S-adenosyl-[methyl-3H]-l-methionine (25 µCi/ml) (Amersham) as methyl donor. Reactions were stopped by spotting 20 µl on P81 filter paper. Filter papers were then washed three times for 10 min in 50 mM carbonate buffer pH 9.2, dried and 3H-incorporation was measured by scintillation counting. All methylation assays were repeated at least three times. Error bars on the graphs represent the SEM values derived from at least three experiments.
Histone deacetylation. For HDAC activity 20 µl aliquots of column fractions were incubated at 30°C for 90 min with 1 µg of 3H-labelled core histones (25 000 c.p.m./µg). Then 230 µl of IPH buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 0.5% NP-40, 0.2 mM PMSF) and 65 µl of 1 M HCl/0.16 M acetic acid were added and the released acetate was extracted with 700 µl of ethylacetate. A 500 µl aliquot of the upper ethylacetate phase was used for scintillation counting. All deacetylation assays were repeated at least three times. Error bars on the graphs represent the SEM values derived from at least three experiments.
Immunoprecipitation. To immunoprecipitate SU(VAR)3-9-associated proteins, 200 µl of nuclear extract (NE) from myc-SU(VAR)3-9 flies were incubated with 10 µl of 9E10 antibody crosslinked to protein G–agarose beads. After incubation for 3 h at 4°C, the beads were washed three times in 1 ml of IPH 300 buffer and resuspended in 45 µl of buffer A (16 mM NaCl, 20 mM Tris–HCl pH 8.0, 1 mM DTT). For activity assays 20 µl of this material were used.
Fly culture. Drosophila cultures and stocks were reared on standard medium at 25°C. Except where noted chromosomes and mutations are described in FLYBASE. Heterochromatin-induced gene silencing has been studied in flies carrying the white variegating rearrangement ln(1)wm4h (Wade et al., 1997). The HDAC326 mutation was isolated as a dominant-negative suppressor of PEV (Nakayama et al., 2001). The mutation is caused by a P204S amino acid exchange. Extra genomic copies of Su(var)3-9 were introduced by P[(ry+) 11kb Su(var)3-9] (Mottus et al., 2000). A cross of wm4h; P[(ry+) 11kb Su(var)3-9] (100E)/TM3, Sb Ser females to wm4h/Y; HDAC1326/TM3, Sb males produced wm4h; P[(ry+) 11kb Su(var)3-9] (100E)/HDAC1326 flies with one additional genomic copy of Su(var)3-9. In the reciprocal crosses identical results were obtained.
Acknowledgments
ACKNOWLEDGEMENTS
This manuscript is dedicated to the memory of Dr Alan P. Wolffe. We thank Brian Turner and Jürg Müller for providing the anti-HDAC3 and the anti-HDAC1 antibody, Albert Courey for the baculovirus expressing dHDAC1, Tony Kouzarides for the bacterial expression vector for SUV3-9H1, Tom Grigliatti for the HDAC1326 strain and various members of the Becker laboratory for critical reading of the manuscript. B.C. is supported by the German research foundation (IM 23/3-1).
REFERENCES
- Aagaard L. et al. (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Allard S., Utley, R.T., Savard, J., Clarke, A., Grant, P., Brandl, C.J., Pillus, L., Workman, J.L. and Cote, J. (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J., 18, 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C. and Kouzarides, T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J. and Kouzarides, T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- De Rubertis F., Kadosh, D., Henchoz, S., Pauli, D., Reuter, G., Struhl, K. and Spierer, P. (1996) The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature, 384, 589–591. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Olsson, T., Turner, B.M., Cranston, G. and Allshire, R.C. (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell, 91, 1021–1032. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz, D., Pray-Grant, M.G., Steger, D.J., Reese, J.C., Yates, J.R., III and Workman, J.L. (1998a) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Sterner, D.E., Duggan, L.J., Workman, J.L. and Berger, S.L. (1998b) The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol., 8, 193–197. [DOI] [PubMed] [Google Scholar]
- Henikoff S. (1990) Position-effect variegation after 60 years. Trends Genet., 6, 422–426. [DOI] [PubMed] [Google Scholar]
- Imhof A. and Becker, P.B. (2001) Modifications of the histone N-terminal domains. Evidence for an ‘epigenetic code’? Mol. Biotechnol., 17, 1–13. [DOI] [PubMed] [Google Scholar]
- Ivanova A.V., Bonaduce, M.J., Ivanov, S.V. and Klar, A.J. (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nature Genet., 19, 192–195. [DOI] [PubMed] [Google Scholar]
- Jones P.L., Veenstra, G.J., Wade, P.A., Vermaak, D., Kass, S.U., Landsberger, N., Strouboulis, J. and Wolffe, A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kuo M.H. and Allis, C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll, D., Rea, S., Mechtler, K. and Jenuwein, T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Mannervik M. and Levine, M. (1999) The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 6797–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C.A. and Allis, C.D. (1998) Linking histone acetylation to transcriptional regulation. Cell. Mol. Life Sci., 54, 6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottus R., Sobel, R.E. and Grigliatti, T.A. (2000) Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics, 154, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice, J.C., Strahl, B.D., Allis, C.D. and Grewal, S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Reuter G. and Spierer, P. (1992) Position effect variegation and chromatin proteins. BioEssays, 14, 605–612. [DOI] [PubMed] [Google Scholar]
- Reuter G. et al. (1986) Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet., 202, 481–487. [Google Scholar]
- Spofford J.B. (1967) Single-locus modification of position-effect variegation in Drosophila melanogaster. I. White variegation. Genetics, 57, 751–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann, A., Krauss, V., Dorn, R., Korge, G. and Reuter, G. (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J., 13, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade P.A., Pruss, D. and Wolffe, A.P. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci., 22, 128–132. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne, A., Jones, P.L., Ballestar, E., Aubry, F. and Wolffe, A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Waterborg J.H. (1993) Dynamic methylation of alfalfa histone H3. J. Biol. Chem., 268, 4918–4921. [PubMed] [Google Scholar]
- Zhang Y., Iratni, R., Erdjument-Bromage, H., Tempst, P. and Reinberg, D. (1997) Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell, 89, 357–364. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng, H.H., Erdjument-Bromage, H., Tempst, P., Bird, A. and Reinberg, D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]