COMMENTARY
In this issue of JID Najidh et al. expanded our understanding of PD1 regulation in Sezary syndrome (SS) (Najidh et al., 2023) demonstrating an inverse correlation between DNA methylation status of PDCD1 gene promotor and PDCD1 gene and PD1 protein expression that strongly indicates a role for DNA methylation in regulating PDCD1 gene expression.
PD1 IS EXPRESSED BY TUMOR CELLS FROM SÉZARY SYNDROME
SS is the erythrodermic and leukemic variant within the cutaneous T cell lymphoma (CTCL) spectrum characterized by specific phenotypic and molecular aberrations, affecting tumor growth and dissemination. PD1 is commonly overexpressed by tumor cells of SS that contributed to improved diagnosis and detection by immunohistochemistry or flow cytometry (Cetinozman et al., 2012, Lewis et al., 2022). Moreover, PD1 inhibits T cell activation in the CTCL microenvironment and by binding to PD-L1 modulates disease growth and progression by downregulating anti-tumor responses (Han et al., 2023 ). Hence, checkpoint blockade has emerged as a highly promising treatment strategy via modulation of tumor cell-immune cell interactions with remarkable tumor responses, albeit only in a subset of patients (Khodadoust et al., 2020). The underlying mechanisms for treatment response and resistance are not completely elucidated. Overall, advanced stages of mycosis fungoides (MF)/SS have a poor prognosis with an estimated 5-year survival of approximately 20 - 60% (Mourad and Gniadecki, 2020).
DYSREGULATION OF THE EPIGENOME IS A HALLMARK OF CTCL
Epigenetic changes such as DNA methylation are known to alter gene expression of oncogenes and tumor suppressors contributing to malignant transformation and tumor phenotype (Baylin and Jones, 2016). In addition, epigenetic changes of genes particularly involved in DNA repair, and cell cycle checkpoint genes have been shown to influence cancer chemotherapy responses via signaling pathway alterations. Various molecular mechanisms including transcriptomic and epigenomic regulation have been recognized to drive PD1 expression during acute and chronic antigenic stimuli, and anti-tumors responses. Both high aberrant DNA hyper- and hypomethylation signatures have also been observed in CTCL that are indicative of the specific role of epigenetics in the CTCL pathogenesis (Choi et al., 2015). CTCL cells demonstrated widespread hypermethylation of CpG islands in promotor regions of tumor suppressor gene CDKN2A and recurrent loss of function mutations in epigenetic regulators such as DNMT3A (Kiel et al., 2015, van Doorn et al., 2016).
EPIGENETIC REGULATION OF PD1 IN SÉZARY SYNDROME
To this end, Najidh et al. performed combined analyses of DNA methylation, gene, and protein expression in a large cohort of SS patients (n=38), patients with benign erythroderma (BE; n=10) and healthy controls (HC; n=4) (Najidh et al., 2023). Qualitative assessment of the DNA methylation status of the gene promoter locus PDCD1 in CD4+ T cells from peripheral blood samples revealed a markedly hypomethylated promotor in SS samples compared to methylated promotor in BE/HC samples. In addition, the authors found a consistent inverse correlation between PDCD1 gene promoter methylation, PDCD1 gene expression, and PD1 membranous protein expression. PDCD1 promoter mRNA and protein levels among SS samples with hypomethylated gene promoters demonstrated a significantly higher expression when compared to methylated gene promoters from SS or BE samples. Furthermore, the authors could show the reversible character of PDCD1 methylation with low PD1 protein expression on circulating immunophenotypically aberrant CD4+ T cells in 2 follow-up peripheral blood samples.
PD1 AS IMPORTANT THERAPEUTIC TARGET IN SÉZARY SYNDROME
Immune checkpoints are important targets for immunotherapies. However, knowledge on the epigenetic modification of immune checkpoint genes is sparse. Pembrolizumab demonstrated efficacy in inducing anti-tumor responses in a subset of patients with advanced MF/SS (Khodadoust et al., 2020). Notably, a transient worsening of erythroderma in SS patients correlated with high PD1 expression on circulating Sezary cells that did not associate with subsequent clinical responses. Najidh et al. emphasized the need to utilize PD1 checkpoint blockade in combination with therapeutic strategies that involve direct targeting of epigenetic alterations to augment anti-tumor responses (Najidh et al., 2023). Considerations are given to the histone deacetylase inhibitors (HDACi) romidepsin and vorinostat that were approved by the FDA and are currently utilized for CTCL (Lopez et al., 2018). Their efficacies and epigenetic drug effects on DNA methylation and histone deacetylation are well-studied, albeit its effects on the PD1/PD-L1 axis is not known.
Although Najidh et al. have identified epigenetic alterations of PD1 in SS and convincingly demonstrated that these alterations correlated with high PDCD1 mRNA and PD1 protein expression (Najidh et al., 2023), PD1 regulation is likely to be much more complex, because PD1 is not only expressed on CTCL cells, but on other immune cells in the CTCL tumor microenvironment (TME) and targeting PD1 can have immunomodulatory effects. In addition to providing an understanding of which active immune TME components may induce and be affected by PD1 function, knowledge of other molecular mechanisms such as transcription factors or other epigenetic parameters in regulating PD1 may help tailor future immunotherapies. miRNAs have been shown to regulate immune checkpoint expression in MF that may be driven by the TME (Han et al., 2022) but is not known for SS. Nevertheless, epigenetic abnormalities in SS may have diverse implications, because they may be heritable and do not only affect gene transcription but regulating cell reprogramming. Larger data sets are needed to validate the predictive value of DNA methylation status of PD1.
Figure 1:
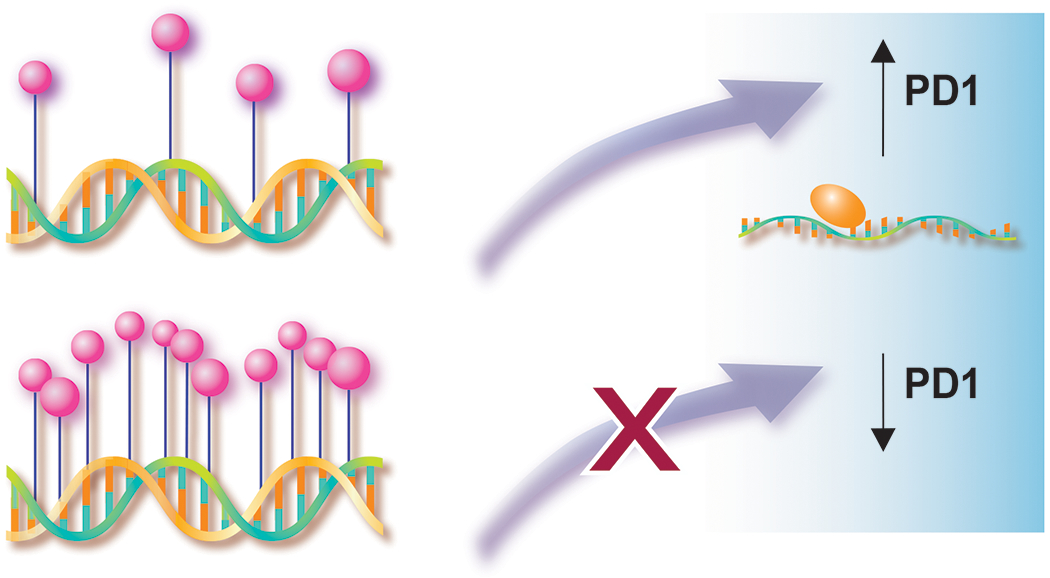
Inverse correlation of DNA methylation status of PDCD1 gene promotor region and PD1 mRNA and protein expression
Illustration assistance provided by Jan Ruvido Stebbins, Ruvido Medical Illustration, Dexter, MI.
Clinical Implications:
PD1 is expressed by tumor cells from Sézary syndrome distinct from benign erythrodermas.
The inverse correlation between DNA methylation and PD1 expression indicates a role for epigenetic control of PDCD1 gene expression.
The DNA methylation status of PD1 may serve as biomarker for response and resistance.
CONFLICT OF INTEREST
Advisory board/steering committee: Kyowa Kirin, Helsinn, Citius Pharm; investigator: Kyowa Kirin, Bristol Myers Squibb, Sorrento, Helsinn; research grants: Helsinn, Celgene
Biography

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 2016;8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetinozman F, Jansen PM, Vermeer MH, Willemze R. Differential expression of programmed death-1 (PD-1) in Sezary syndrome and mycosis fungoides. Arch Dermatol 2012;148(12):1379–85. [DOI] [PubMed] [Google Scholar]
- Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet 2015;47(9):1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Estephan RJ, Wu X, Su C, Yuan YC, Qin H, et al. MicroRNA Regulation of T-Cell Exhaustion in Cutaneous T Cell Lymphoma. J Invest Dermatol 2022;142(3 Pt A):603–12 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Wu X, Qin H, Yuan YC, Schmolze D, Su C, et al. Reprogramming of PD1+ M2-like tumor-associated macrophages with anti-PD-L1 and lenalidomide in cutaneous T cell lymphoma. JCI Insight (in press) 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadoust MS, Rook AH, Porcu P, Foss F, Moskowitz AJ, Shustov A, et al. Pembrolizumab in Relapsed and Refractory Mycosis Fungoides and Sezary Syndrome: A Multicenter Phase II Study. J Clin Oncol 2020;38(1):20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Sahasrabuddhe AA, Rolland DCM, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun 2015;6:8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NE, Gao Q, Petrova-Drus K, Pulitzer M, Sigler A, Baik J, et al. PD-1 improves accurate detection of Sezary cells by flow cytometry in peripheral blood in mycosis fungoides/Sezary syndrome. Cytometry B Clin Cytom 2022;102(3):189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AT, Bates S, Geskin L. Current Status of HDAC Inhibitors in Cutaneous T-cell Lymphoma. Am J Clin Dermatol 2018;19(6):805–19. [DOI] [PubMed] [Google Scholar]
- Mourad A, Gniadecki R. Overall Survival in Mycosis Fungoides: A Systematic Review and Meta-Analysis. J Invest Dermatol 2020;140(2):495–7 e5. [DOI] [PubMed] [Google Scholar]
- Najidh S, Zoutman WH, Schrader AMR, Willemze R, Tensen CP, Vermeer MH. PD-1 overexpression in Sezary syndrome is epigenetically regulated. J Invest Dermatol 2023. [DOI] [PubMed] [Google Scholar]
- van Doorn R, Slieker RC, Boonk SE, Zoutman WH, Goeman JJ, Bagot M, et al. Epigenomic Analysis of Sezary Syndrome Defines Patterns of Aberrant DNA Methylation and Identifies Diagnostic Markers. J Invest Dermatol 2016;136(9):1876–84. [DOI] [PubMed] [Google Scholar]


