Abstract
SHP (short heterodimer partner, NROB2) is an atypical orphan member of the mammalian nuclear receptor family that consists only of a putative ligand-binding domain and thus cannot bind DNA. Instead, SHP acts as a transcriptional coregulator by inhibiting the activity of various nuclear receptors (downstream targets) via occupation of the coactivator-binding surface and active repression. However, repression mechanisms have remained elusive and may involve coinhibitory factors (upstream targets) distinct from known nuclear receptor corepressors. Here, we describe the isolation of mouse E1A-like inhibitor of differentiation 1 (EID1) as a candidate coinhibitor for SHP. We characterize the interactions between SHP and EID1 and identify two repression-defective SHP mutations that have lost the ability to bind EID1. We suggest histone acetyltransferases and histones as targets for EID1 action and propose that SHP inhibition of transcription involves EID1 antagonism of CBP/p300-dependent coactivator functions.
INTRODUCTION
Nuclear receptors comprise a large family of signal-regulated transcription factors with ∼50 individual members that have critical roles in mammalian development and adult physiology (Mangelsdorf and Evans, 1995). Typical nuclear receptors consist of a DNA-binding domain and a multi-functional ligand-binding and transcription regulation domain (LBD), which plays a distinctive role in signal transformation and represents the primary docking site for coregulatory proteins (for a review, see Wurtz et al., 1996). Prototypic transcriptional coregulators, defined as non-DNA-binding proteins that connect activators or repressors with chromatin modifying enzymatic activities and the basal transcription machinery, are crucial components of any nuclear receptor-signalling pathway (for a review, see Glass and Rosenfeld, 2000).
Two-hybrid interaction screenings aimed at identifying novel coregulators for nuclear receptors led to the isolation of the atypical orphan nuclear receptor SHP (short heterodimer partner, NROB2), which consists of a putative LBD but lacks a DNA-binding domain (Seol et al., 1996; Johansson et al., 1999). Previous research has focused on different aspects of SHP structure and function to elucidate the basis for its inhibitory role on the signalling of various active nuclear receptors. SHP has a physiological function as a negative regulator of the conversion of cholesterol to bile acids in the liver (Goodwin et al., 2000; Lu et al., 2000) and may play roles in the reproductive system by inhibiting oestrogen signalling (Seol et al., 1998; Johansson et al., 1999). These findings emphasize the physiological importance of the inhibitory capacity of SHP. Mechanistically, SHP acts as a coregulator by direct binding to the ligand-inducible activation domain AF-2 (characteristic of coactivators) and by active repression mechanisms (characteristic of corepressors) (Johansson et al., 2000; Lee et al., 2000). However, the basis for active repression by SHP and the cofactors involved remained unknown. Here, we describe the isolation and characterization of mouse E1A-like inhibitor of differentiation 1 (EID1) as a candidate coinhibitory factor for SHP and propose a mechanism for transcriptional inhibition that is exceptional for a member of the nuclear receptor family.
RESULTS AND DISCUSSION
Cloning of EID1
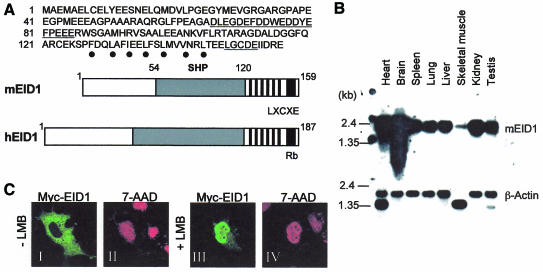
The yeast two-hybrid approach was applied to screen for SHP-interacting proteins. Among various positive clones, including the known SHP-interacting orphan receptor HNF4 (Lee et al., 2000), the most frequently isolated cDNA contained a 477 bp open reading frame encoding a novel mouse protein of 159 amino acids. During the progress of our study, a putative human orthologue EID1 was independently cloned as an Rb-interacting protein (MacLellan et al., 2000; Miyake et al., 2000), so we designated our protein mouse EID1. The sequence features and domain structure of EID1 are shown in Figure 1A. Highly conserved regions include the Rb-binding motif and a putative heptad repeat domain in the C-terminal region, a central SHP-interaction domain including an acidic region, and the first 30 residues in the N-terminus. The ∼1.8 kb mouse EID1 mRNA appeared ubiquitously expressed, albeit at different levels, in all adult tissues examined and during mouse embryogenesis (Figure 1B; data not shown), resembling the expression and size of the human orthologue (Miyake et al., 2000; Wen and Ao, 2001). At the intracellular level, we found Myc-tagged EID1 to be localized nearly exclusively in the cytoplasm of transfected Cos-7 cells (Figure 1C). However, in the presence of the nuclear export inhibitor leptomycin B (Figure 1C, +LMB), EID1 was nuclear in ∼50% of the cells, which is consistent with the presence of putative leucine-rich/hydrophobic nuclear export signals in EID1. This suggests that EID1 presumably represents a nuclear protein, consistent with its postulated functions within the nucleus (see below), which is actively exported out of the nucleus in a CRM1/exportin-dependent manner.
Fig. 1. EID1 primary structure and expression analysis. (A) Upper part: delineated EID1 protein sequence. Underlined are the acidic stretch and the Rb-binding motif. Dots indicate a putative heptad repeat (α-helix positions 1 and 4). Lower part: domain structure of mouse and human EID1. Highlighted are conserved C-terminal motifs including the Rb-binding motif and heptad repeat (black) and a conserved central part involved in SHP binding (grey). (B) Tissue distribution of EID1 mRNA. A mouse multiple tissue blot containing 2 µg poly(A) mRNA (Clontech) was probed with radiolabelled EID1 or β-actin cDNA. (C) Intracellular localization of EID1. Myc-tagged EID1 was expressed in Cos-7 cells and analysed by indirect immunofluorescence using Myc antibody (green) in the absence (I) or presence (III) of leptomycin B (LMB, 5 nM for 5 h). Nuclei were stained with 7-aminoactinomycin D (7-AAD) (red, II and IV). Fluorescence images are representative for 99% (–LMB) and 50% (+LMB); >50 positive cells were studied and the experiment was independently reproduced at least three times.
Characterization of EID1 interactions with SHP
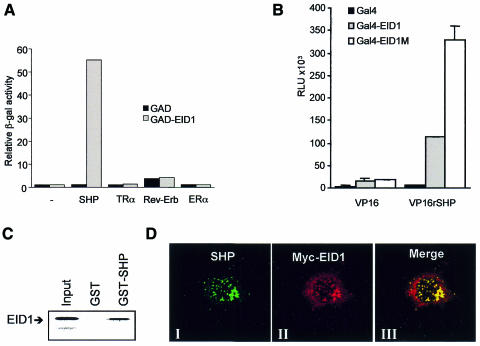
The following experiments were performed to characterize interactions between EID1 and SHP. First, yeast two-hybrid analysis revealed that EID1 only interacted strongly with SHP and not with other repressing nuclear receptors, including the unliganded thyroid hormone receptor TRα, the orphan receptor Rev-Erb and antagonist-bound oestrogen receptor ERα (Figure 2A). Secondly, mammalian two-hybrid assays demonstrated interactions between SHP and EID1 and were used to delineate the SHP-interaction domain of EID1 (amino acids 54–120, Figure 2B). Apparently enhanced interaction of the central domain compared with wild-type EID1 suggests that N- or C-terminal EID1 domains act in an inhibitory manner in this system. Indeed, the C-terminus has been implicated in the recruitment of cellular cofactors such as Rb and CBP/p300 (Figure 1; MacLellan et al., 2000; Miyake et al., 2000). As in yeast, no interaction was observed with various other nuclear receptors tested, including PPARα, TRα, ERα and HNF4 (data not shown). This suggests that EID1, unlike many other LBD-associated proteins, does not promiscuously interact with multiple nuclear receptors and thus displays receptor selectivity. Thirdly, in vitro-translated EID1 readily bound to GST–SHP in a pull-down assay (Figure 2C). Fourthly, we performed co-immunoprecipitations from mammalian cell extracts expressing Myc-tagged EID1 and SHP alone or in combination using an SHP-specific antiserum. As judged from western analysis using a Myc-specific antiserum, Myc-EID1 could be precipitated only in the presence of coexpressed SHP (see Supplementary figure 1 available at EMBO reports Online). Fifthly, we studied the influence of SHP on the intracellular localization of EID1 by confocal microscopy. We observed that both proteins were colocalized in the nucleus in 20% of coexpressing cells (Figure 2D). Interestingly, EID1 adopted a dot-like pattern typical for that seen with SHP alone but distinct from that observed with EID1 alone (Figure 1C), suggesting that SHP, at least in the case of overexpression, can relocalize EID1 to distinct areas within the nucleus. Together, these results indicate a specific and direct interaction of EID1 with SHP in vitro and in vivo.
Fig. 2. Analysis of EID1 interactions with SHP. (A) Yeast two-hybrid analysis of EID1 interactions with repressing nuclear receptors. Gal4–receptor fusions were tested for interaction with activation-domain tagged (GAD) EID1. Gal4–ERα was analysed in the presence of 1 µM 4-hydroxytamoxifen. (B) Mammalian two-hybrid analysis in Cos-7 cells. Gal4 expression plasmids (200 ng) encoding Gal4–EID1 (amino acids 1–159), Gal4–EID1M (amino acids 54–120) or Gal4 alone (for control) were cotransfected with VP16–SHP (amino acids 1–260) or VP16 expression plasmids (500 ng) and the UAS–tk–luc reporter plasmid (500 ng). Relative luciferase units (RLUs) represent the mean ± SD of duplicate transfections and were reproduced in at least three independent experiments. (C) EID1 interacts with SHP in vitro. Binding of [35S]methionine-labelled EID1 (25 kDa) to GST–SHP or GST alone was analysed in a pull-down assay. The input represents 20%. (D) Nuclear colocalization of EID1 with SHP. Myc-tagged EID1 and SHP were coexpressed and analysed by indirect immunofluorescence using antibodies for SHP and Myc. Representative fluorescence images for SHP (green) are shown in panel I, for EID1 (red) in panel II and merged images (yellow) following superimposition in panel III.
EID1 binds to acetyltransferases and histones in vitro
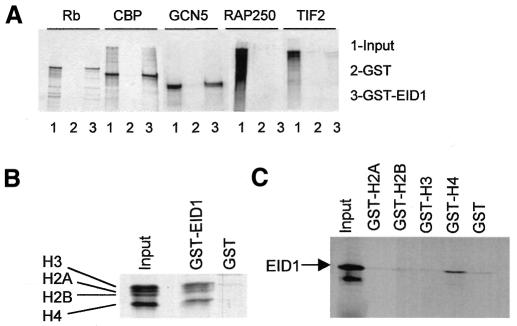
Transcriptional inhibition by SHP is believed to involve antagonism of coactivator function via cofactors that are distinct from previously characterized coactivators or corepressors (Seol et al., 1997; Johansson et al., 1999, 2000). Therefore, we considered whether EID1 directly binds to coactivators relevant for nuclear receptor function and performed GST pull-down assays. Figure 3A shows binding of the histone acetyltransferase (HAT) CBP and of Rb (for control) to GST–EID1, which is consistent with the previously reported binding of these two proteins to human EID1 (MacLellan et al., 2000; Miyake et al., 2000). Furthermore, we observed binding of human GCN5, the orthologue of the HAT coactivator PCAF (Yamauchi et al., 2000), while the coactivators RAP250 (Caira et al., 2000) and TIF2 (Leers et al., 1998) did not bind, indicating EID1 selectivity towards HATs. Moreover, we noted that EID1 and several small CBP/p300 cofactors are highly acidic and thus considered histones as additional EID1 targets. First, GST–EID1 easily absorbed purified histones H2A, H2B, H3 and H4 (Figure 3B). Secondly, GST–H4 tails (Georgel et al., 1997) retained EID1 (Figure 3C), which is intriguing because H4 acetylation by CBP/p300 and GCN5/PCAF is associated with nuclear receptor activation (Chen et al., 1999).
Fig. 3. EID1 interacts with acetyltransferases and histones in vitro. (A) Indicated radiolabelled proteins were analysed in pull-down assays for binding to GST–EID1 or GST alone. (B) Binding of pure histones (500 ng) was analysed in pull-down assays as in (A), except bound proteins were stained with Coomassie Blue. (C) Binding of radiolabelled EID1 to GST–histone tail fusions. The input represents 20% of the amount of protein used in each pull-down.
EID1 inhibits the function of CBP/p300-dependent coactivators in vivo
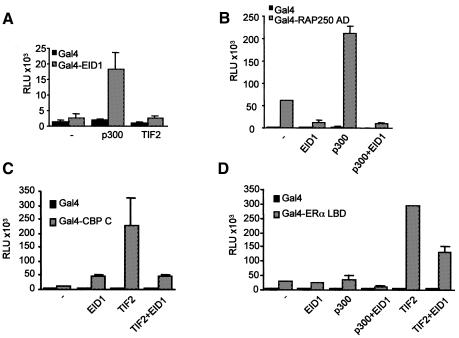
To evaluate functional implications of the above EID1 interactions for the in vivo situation, we analysed the influence of EID1 on coactivator function in transient transfection studies (Figure 4). First, we found that p300, but not TIF2, potentiated Gal4–EID1 activity by more than 7-fold (Figure 4A), which is consistent with the above binding studies. Secondly, we analysed the effect of EID1 on the activity of RAP250, which did not bind to EID1 but was demonstrated to cooperate with CBP/p300 in a HAT-independent manner (Lee et al., 2000). We observed that EID1 inhibited a GAL–RAP250 activation domain fusion, whereas p300 potentiated it ∼4-fold (Figure 4B). This induction was abolished after addition of EID1, suggesting antagonism of p300-dependent RAP250 activation by EID1. Thirdly, we analysed EID1 effects on the functional cooperation of CBP with TIF2, a p160 coactivator known to associate with p300/CBP (Voegel et al., 1998; Ma et al., 1999). We found that EID1 could inhibit TIF2-mediated transcriptional activity of the CBP C-terminus lacking the HAT domain (Figure 4C). Finally, we investigated EID1 effects on transcriptional activity of ERα (Figure 4D). We found that TIF2 stimulated the ligand-dependent AF-2 activity of Gal4–ERα LBD up to 6-fold, which was reduced by up to 50% in the presence of EID1, in concordance with previous data that TIF2 utilizes both CBP/p300-dependent and -independent pathways (Voegel et al., 1998). Similarly, p300-mediated coactivation of ERα was inhibited by EID1, which is in concordance with related data on the inhibition of glucocorticoid receptor activity by human EID1 (Miyake et al., 2000). Under our conditions, TIF2 appeared more effective in enhancing ERα AF-2 than p300, which is consistent with the model where binding of p160 coactivators to AF-2 is a prerequisite for the subsequent recruitment of CBP/p300 to the complex (as discussed in Voegel et al., 1998). Collectively, these data imply that EID1 antagonizes nuclear receptor activation by inhibiting CBP/p300-dependent transcription activation functions of nuclear receptor-associated coactivators such as TIF2 and RAP250.
Fig. 4. EID1 inhibits coactivator function in vivo. Transient transfections in Cos-7 cells were performed using the UAS–tk–luc reporter (500 ng) and expression plasmids as indicated. (A) p300, but not TIF2, potentiates Gal4–EID1 activity. Amounts of the expression plasmids: Gal4–EID1, 100 ng; coactivators, 500 ng. (B) EID1 inhibits p300-dependent RAP250 activity. The effect of p300 (500 ng plasmid) and EID1 (500 ng plasmid), alone or in combination, on the activity of a Gal4–RAP250 activation domain fusion (AD, amino acids 577–855, 5 ng plasmid) was analysed. (C) EID1 interferes with TIF2–CBP cooperation. The effect of TIF2 (500 ng plasmid) and EID1 (500 ng plasmid), alone or in combination, on the activity of a Gal4–CBP C-terminus (amino acids 1678–2441, 100 ng plasmid) was analysed. (D) EID1 antagonizes AF-2 activity of ERα. The effect of TIF2 or p300 (500 ng plasmid) and EID1 (500 ng plasmid), alone or in combination, on the activity of a Gal4–ERα LBD fusion (amino acids 349–595, 100 ng plasmid) was analysed in the presence of 10 nM 17β-oestradiol. For control, in all experiments Gal4 fusions were analysed in comparison with the Gal4 DNA-binding domain (amino acids 1–147) alone. RLUs represent the mean ± SD of duplicate transfections and were reproduced in at least three independent experiments.
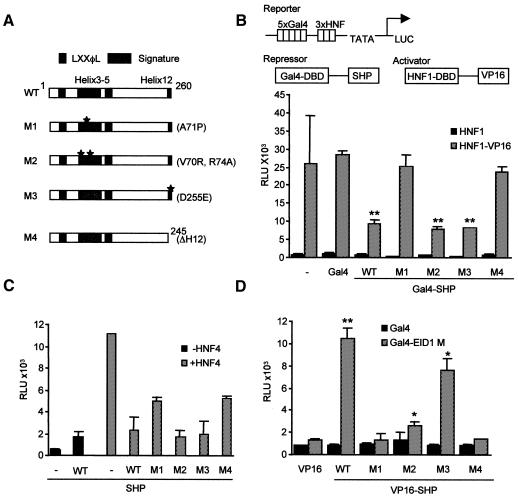
Repressor analysis of mutations within the putative LBD of SHP
To gain first insight into structural requirements for SHP repression and interactions with EID1, various mutations within the putative LBD were generated based on homology considerations (Figure 5A) and then analysed in the different experimental settings. First, the intrinsic repression potential of SHP variants was analysed in a modified assay that measures repression of the strong DNA-bound VP16 activator (Xue et al., 1998). As shown in Figure 5B, SHP WT reproducibly resulted in up to 60% repression, which was unaffected by the deacetylase inhibitor trichostatin A (data not shown; Seol et al., 1997). Importantly, mutations M1 and M4, but not M2 and M3, had lost the intrinsic repressor function, which has implications as discussed below. Next, we wanted to analyse SHP inhibition of nuclear receptor activity in a physiologically relevant setting and utilize an HNF4-responsive reporter system. The orphan receptor HNF4 is one of the most intriguing SHP targets in liver and pancreas (Lee et al., 2000), and natural SHP mutations have been identified that possess impaired inhibition of HNF4 activity (Nishigori et al., 2001). As shown in Figure 5C, inhibition of HNF4-dependent activity by the different SHP variants correlated well with the results of the repressor assay (Figure 1B), although the receptor-based system does not necessarily distinguish between coactivator competition and active repression mechanisms (Johansson et al., 1999; Lee et al., 2000). Finally, we investigated binding of EID1 to the SHP variants in mammalian two-hybrid assays (Figure 5D) and found that M1 and M4, but not M2 and M3, had completely lost the ability to interact with EID1. While all interactions were significant and could be confirmed using yeast two-hybrid assays (data not shown), M2 apparently displayed reduced interaction with EID1 in the mammalian system, possibly suggesting that additional factors are involved in repression by SHP M2. On the basis of the results presented in Figure 5, we suggest that the inability of SHP M1 and M4 to repress and inhibit nuclear receptor activity could be linked with the inability to interact with EID1.
Fig. 5. Mutational analysis of the SHP repressor domain. (A) Schematic representation of SHP structure and mutations. Highlighted are the signature motif (helices 3–5 and 12), LXXφL motifs, and mutations introducing amino acid substitutions (M1–3) and a deletion (M4) as indicated. (B) Repressor assay. Gal4–SHP fusions (1 µg plasmid) were analysed for repression of the VP16 activator (500 ng). Expression of all Gal4 fusion proteins was confirmed by western analysis (data not shown). ** P < 0.01 was obtained by comparing the Gal4–SHP variants with Gal4 and indicates significant difference. (C) Analysis of nuclear receptor inhibition. The effect of SHP variants (300 ng plasmid) on the HNF4 (200 ng plasmid)-dependent activity of an apolipoprotein CIII–luc reporter (500 ng plasmid) was analysed. Note that SHP inhibition of reporter activity was dependent on HNF4 coexpression and not observed in the absence of HNF4. (D) Analysis of EID1 interactions with SHP mutants. Interactions of VP16-tagged SHP variants with Gal4–EID1 M (amino acids 54–120) were analysed in a mammalian two-hybrid assay essentially as described in Figure 1. * P < 0.05 and ** P < 0.01 were obtained by comparing Gal4 and Gal4–EID1 M values and indicate significant differences.
Conclusions
In summary, we conclude the following. (i) EID1 represents the first SHP-associated upstream target protein that can be directly linked to transcription inhibitory mechanisms and is different from conventional corepressors. (ii) EID1 significantly inhibited CBP/p300-dependent functions, which may partly be HAT-independent (Perissi et al., 1999), of coactivators relevant for nuclear receptor activation. Inhibitory mechanisms may include the disruption of CBP/p300 coactivator interactions, the direct inhibition of HAT activity (MacLellan et al., 2000; Miyake et al., 2000) and possibly histone binding (Seo et al., 2001). (iii) Mutational analysis of SHP repression and EID1 binding highlights the divergence of repression mechanisms between SHP and repressing receptors that depend on N-CoR/SMRT corepressors (as discussed in Seol et al., 1997; Glass and Rosenfeld, 2000) and identifies two mutations that abolished both repression and EID1 binding. These results indicate that SHP has a coregulator-binding surface encompassing putative helices 3 and 12. It is distinct from those of other nuclear receptors (for a review, see Glass and Rosenfeld, 2000), presumably because critical residues (yet to be identified) account for the selectivity of SHP towards EID1. (iv) The homology of SHP M1 to the naturally occurring DAX-1 mutation R267P, as well as the requirement of helix 12 (M4) for repression (Lalli et al., 1997), highlights the close functional relationship of these two atypical nuclear receptors. (v) Finally, naturally occurring SHP mutations (Nishigori et al., 2001) possibly manifest in physiological disorders because they affect the inhibitory capacity and interactions with associated cofactors such as EID1.
METHODS
Plasmids.
All plasmids were generated using standard cloning procedures and verified by DNA sequencing. Mutations were generated using PCR mutagenesis. Details are available upon request.
Yeast two-hybrid screening and interaction assay.
Screenings using Gal4–rat SHP (amino acids 1–260) and a mouse embryo cDNA library (Clontech) were performed essentially as described previously (Johansson et al., 1999; Caira et al., 2000). Three different clones encoding EID1 were isolated 11 times. The largest cDNA of 1.7 kb corresponds well to the approximate size of the mRNA in northern blots, and an in-frame stop codon precedes the start methionine, indicating the cDNA to be full length. Yeast interaction assays were performed in liquid culture and measured as relative β-galactosidase activity.
Northern blot analysis.
A mouse multiple tissue northern blot [2 µg poly(A)+ RNA per lane] was sequentially hybridized with radioactively labelled cDNA for mouse EID1 and β-actin (for control) according to the manufacturer’s protocol (Clontech).
GST pull-down assays.
GST fusion proteins were partially purified from Escherichia coli BL21 (pLys), and pull-down assays (except Figure 3B) were as described previously (Johansson et al., 1999, 2000). Briefly, 1 µg GST fusion proteins were incubated with 35S-labelled proteins for 2 h at 4°C in a buffer containing 200 mM NaCl. After extensive washing, bound proteins were analysed by SDS–PAGE and visualized by autoradiography.
Cell cultures and transient transfections.
Cos-7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 100 µg/ml penicillin and 100 µg/ml streptomycin. For transient transfection assays, cells were plated onto 6-well plates (Falcon) 24 h prior to transfection. Cells were transfected using Lipofectin as instructed by the manufacturer (Life Technologies) and cultured for 24 h. Cell extracts were analysed for luciferase activity as described previously (Johansson et al., 1999). Data are presented as relative luciferase units (RLUs), the values represent the mean ± SD of duplicate transfections, and the results are representative for at least three independent experiments. Statistical analysis was performed using the Student’s t-test.
Confocal microscopy.
Cos-7 cells were seeded on glass cover slips in six-well plates transfected as above. Cells were fixed in 3% paraformaldehyde in 5% sucrose/phosphate-buffered saline (PBS), permeabilized with PBS/Tween 20 (0.1%) and blocked with 5% goat serum (Jackson ImmunoResearch). Mouse monoclonal anti-Myc (Myc 1-9E10.2, ATCC CRL-1729) or rabbit polyclonal anti-SHP were detected with appropriate secondary antibodies conjugated to fluorescein (FITC), tetramethyl rhodamin isothiocyanate (TRITC) or lissamine rhodamin (LRSC) (Jackson ImmunoResearch). Nuclei were stained using 2 µM 7-aminoactinomycin D (7-AAD) (Molecular Probes). To block nuclear export, 5 nM leptomycin B (Sigma) was added 5 h before fixation. Subcellular images were determined using a TCS SP multiband confocal imaging system (Leica).
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Steffan Ho, Weidong Wang, Carl Wu, Tony Kouzarides, Shelley Berger, David Livingston, Victoria Green and John Ladias for providing plasmids. We thank group members, Dr Anders Ström in particular, for supplying materials. This work was supported by grants from the Swedish Cancer Society and from the KaroBio AB.
REFERENCES
- Caira F., Antonson, P., Pelto-Huikko, M., Treuter, E. and Gustafsson, J.A. (2000) Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem., 275, 5308–5317. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin, R.J., Xie, W., Wilpitz, D. and Evans, R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Georgel P.T., Tsukiyama, T. and Wu, C. (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J., 16, 4717–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld, M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Goodwin B. et al. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1 and LRH-1 represses bile acid biosynthesis. Mol. Cell, 6, 517–526. [DOI] [PubMed] [Google Scholar]
- Johansson L., Thomsen, J.S., Damdimopoulos, A.E., Spyrou, G., Gustafsson, J.A. and Treuter, E. (1999) The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERα and ERβ. J. Biol. Chem., 274, 345–353. [DOI] [PubMed] [Google Scholar]
- Johansson L., Bavner, A., Thomsen, J.S., Farnegardh, M., Gustafsson, J.A. and Treuter, E. (2000) The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol., 20, 1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli E., Bardoni, B., Zazopoulos, E., Wurtz, J.M., Strom, T.M., Moras, D. and Sassone-Corsi, P. (1997) A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Mol. Endocrinol., 11, 1950–1960. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Jung, S.Y., Kim, Y.S., Na, S.Y., Lee, Y.C. and Lee, J.W. (2001) Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol. Endocrinol., 15, 241–254. [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Dell, H., Dowhan, D.H., Hadzopoulou-Cladaras, M. and Moore, D.D. (2000) The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell. Biol., 20, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leers J., Treuter, E. and Gustafsson, J.Å. (1998) Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. Mol. Cell. Biol., 18, 6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.T., Makishima, M., Repa, J.J., Schoonjans, K., Kerr, T.A., Auwerx, J. and Mangelsdorf, D.J. (2000) Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell, 6, 507–515. [DOI] [PubMed] [Google Scholar]
- Ma H., Hong, H., Huang, S.M., Irvine, R.A., Webb, P., Kushner, P.J., Coetzee, G.A. and Stallcup, M.R. (1999) Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol., 19, 6164–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan W.R., Xiao, G., Abdellatif, M. and Schneider, M.D. (2000) A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol., 20, 8903–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J. and Evans, R.M. (1995) The RXR heterodimers and orphan receptors. Cell, 83, 841–850. [DOI] [PubMed] [Google Scholar]
- Miyake S. et al. (2000) Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol., 20, 8889–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishigori H. et al. (2001) Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc. Natl Acad. Sci. USA, 98, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Dasen, J.S., Kurokawa, R., Wang, Z., Korzus, E., Rose, D.W., Glass, C.K. and Rosenfeld, M.G. (1999) Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl Acad. Sci. USA, 96, 3652–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.B., McNamara, P., Heo, S., Turner, A., Lane, W.S. and Chakravarti, D. (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell, 104, 119–130. [DOI] [PubMed] [Google Scholar]
- Seol W., Choi, H.S. and Moore, D.D. (1996) An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science, 272, 1336–1339. [DOI] [PubMed] [Google Scholar]
- Seol W., Chung, M. and Moore, D.D. (1997) Novel receptor interaction and repression domains in the orphan receptor SHP. Mol. Cell. Biol., 17, 7126–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol W., Hanstein, B., Brown, M. and Moore, D.D. (1998) Inhibition of estrogen receptor action by the orphan receptor SHP. Mol. Endocrinol., 12, 1551–1557. [DOI] [PubMed] [Google Scholar]
- Voegel J.J., Heine, M.J., Tini, M., Vivat, V., Chambon, P. and Gronemeyer, H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J., 17, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H. and Ao, S. (2001) Identification and characterization of a novel human cDNA encoding a 21 kDa pRb-associated protein. Gene, 263, 85–92. [DOI] [PubMed] [Google Scholar]
- Wurtz J.M., Bourguet, W., Renaud, J.P., Vivat, V., Chambon, P., Moras, D. and Gronemeyer, H. (1996) A canonical structure for the ligand-binding domain of nuclear receptors. Nature Struct. Biol., 3, 87–94. [DOI] [PubMed] [Google Scholar]
- Xue Y., Wong, J., Moreno, G.T., Young, M.K., Cote, J. and Wang, W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Yamauchi, J., Kuwata, T., Tamura, T., Yamashita, T., Bae, N., Westphal, H., Ozato, K. and Nakatani, Y. (2000) Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl Acad. Sci. USA, 97, 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.