Abstract
Yra1p is an essential and conserved mRNA export factor in yeast. Strikingly, removal of the intron from YRA1 causes a dominant-negative growth phenotype and a concomitant inhibition of mRNA export. However, both defects are neutralized by replacement of the intron of YRA1 by a different intron. Significantly, Yra1p is overproduced in yeast when expressed from its intronless gene, but Yra1p levels are the same as the wild type when expressed from an intron-containing YRA1 gene. Thus, an intron in YRA1 controls Yra1p expression and mRNA export.
INTRODUCTION
In eukaryotes, the presence of the nuclear envelope necessitates transport between the nucleus and the cytoplasm. Specifically, mRNAs synthesized in the nucleus need to be exported to the cytoplasm before being translated. This mRNA export process is mediated by an evolutionarily conserved mRNA export machinery. The mRNA exporter Mex67p/Mtr2p in yeast and its metazoan homologue, TAP/p15, interact directly with their transport cargo mRNA and with FG repeat nucleoporins (Segref et al., 1997; Bear et al., 1999; Braun et al., 1999; Kang and Cullen, 1999; Kang et al., 1999; Katahira et al., 1999; Bachi et al., 2000; Sträßer et al., 2000), which line the nuclear pore channel. Mex67p/Mtr2p also binds directly to Yra1p, an RNA-binding protein that is mainly located inside the nucleus (Sträßer and Hurt, 2000; Stutz et al., 2000; Zenklusen et al., 2001). Thus, it is thought that Yra1p serves as a coupling protein that mediates recruitment of Mex67p/Mtr2p to the mRNP. Interestingly, the lethality caused by deletion of YRA1 in yeast can be rescued by heterologous expression of its murine homologue, Aly (Sträßer and Hurt, 2000). Aly was first identified to be a transcriptional coactivator (Bruhn et al., 1997) and a protein chaperone (Virbasius et al., 1999). More recently, Aly has been shown to be involved in splicing-coupled mRNA export in metazoans (Le Hir et al., 2000, 2001; Zhou et al., 2000). Furthermore, Yra1p and Aly belong to a conserved protein family called RNA export factor binding (REF) proteins (Sträßer and Hurt, 2000; Stutz et al., 2000). All REF proteins share a common domain organization. They possess a central RNA recognition motif (RRM), which is linked to a highly conserved N-terminal domain and a highly conserved C-terminal domain by variable linker regions. Recently, it has been shown that the recruitment of Yra1p/Aly to the mRNA is mediated by its direct interaction with Sub2p/UAP56 (Luo et al., 2001; Sträßer and Hurt, 2001), which was first identified as a splicing factor (Fleckner et al., 1997; Kistler and Guthrie, 2001; Libri et al., 2001; Zhang and Green, 2001). Surprisingly, this conserved splicing factor Sub2p/UAP56 is needed for export of intron-containing, as well as intronless, transcripts (Gatfield et al., 2001; Jensen et al., 2001; Sträßer and Hurt, 2001). How Sub2p is recruited to mRNAs lacking introns is not yet known. Interestingly, Sub2p and Mex67p/Mtr2p compete for binding to Yra1p (Sträßer and Hurt, 2001). Thus, Sub2p might first recruit Yra1p to the mRNP and could then be displaced from the mRNA, when Mex67p/Mtr2p binds to Yra1p. All of this suggests that Yra1p is a key player in mRNA export.
Here, we show that deletion of the intron from YRA1 causes a dominant-negative phenotype and an mRNA export defect. Both defects can be rescued by insertion of an heterologous intron in place of the YRA1 intron. Yra1p is expressed at wild-type levels from YRA1 genes containing the RPL25 or UBC8 intron, whereas Yra1p is overexpressed from the intronless YRA1 gene. Thus, the ‘toxicity’ of intron removal is most likely due to the resulting overexpression of Yra1p. Taken together, an intron in YRA1 is crucial for Yra1p expression, which in turn controls mRNA export.
RESULTS AND DISCUSSION
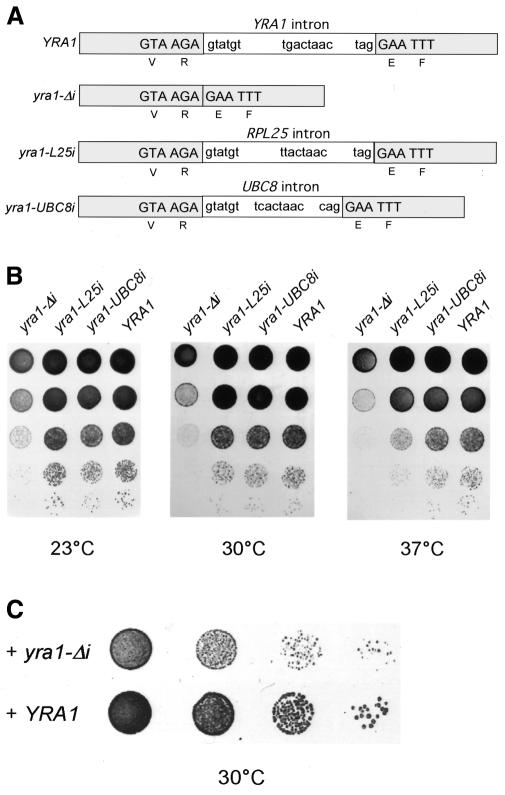
YRA1 belongs to the small group of yeast genes that contain an intron. The intron of YRA1 is unusual, as it is the second largest intron of Saccharomyces cerevisiae (766 nucleotides). In addition, it is located far downstream of the start codon (in the sequence coding for the RRM domain; for domain organization of Yra1, see Sträßer and Hurt, 2000), and its branchpoint sequence does not correspond to the consensus motif (Figure 1A). In the course of studying the function of Yra1p, we noticed that the intron in YRA1 is important for yeast cell growth (Figure 1B). Specifically, an intronless version of YRA1 (yra1-Δi; Figure 1A) complements the lethal yra1 null mutant, but cells grow significantly slower than cells expressing the corresponding intron-containing version of YRA1 (Figure 1B). To test whether yra1-Δi causes a dominant-negative phenotype, the YRA1 shuffle strain was transformed with a plasmid-borne copy of either yra1-Δi or YRA1. This revealed that an extra copy of yra1-Δi, but not of YRA1, inhibits cell growth in yeast (Figure 1C). Thus, removal of the intron from YRA1 causes a dominant-negative growth defect.
Fig. 1. Removal of the intron of YRA1 results in a dominant-negative growth defect. (A) A schematic showing the YRA1 gene, yra1-Δi, yra1-L25i and yra1-UBC8i (exons and an intron). The two exons are indicated in grey and the intron in white. Furthermore, the DNA sequence at the 5′and 3′ splice site and the branchpoint sequence are shown. The deduced amino acid sequence is indicated below the boxes. (B) Cells expressing YRA1 from the wild-type gene (YRA1), the intronless version (yra1-Δi), the YRA1 gene containing RPL25 (yra1-L25i) or the UBC8 (yra1-UBC8i) intron instead of the YRA1 intron were spotted in 1:10 dilutions on YPD plates, and growth was assessed after 2 days incubation at 23, 30 and 37°C. (C) The YRA1 shuffle strain (Dyra1 + pRS316–YRA1) was transformed with ARS/CEN plasmids containing the LEU2 marker and a copy of either YRA1 or yra1-Δi. Transformants were spotted on SDC (-ura/-leu) plates to select for both plasmids, and growth was assessed after 3 days incubation at 30°C.
Since Yra1p is essential for mRNA export, we tested whether removal of the YRA1 intron, which impairs cell growth, also causes an mRNA export defect. Hence, localization of poly(A)+ RNA was detected in the YRA1 and yra1-Δi strains by in situ hybridization. As shown in Figure 2, poly(A)+ RNA accumulates in the nucleus of cells expressing YRA1 from the intronless gene but not in the nucleus of cells expressing Yra1p from the intron-containing YRA1 gene. Thus, the intron of YRA1 is necessary for efficient mRNA export.

Fig. 2. An intron in YRA1 is crucial for mRNA export. YRA1, yra1-Δi and yra1-L25i cells were grown at 30°C, and localization of poly(A)+ RNA was assessed by in situ hybridization with a fluorescence-labelled oligo(dT) probe. The DNA was stained with 4′-6-diamidine-2-phenylindole (DAPI).
Since the dominant-negative phenotype and the mRNA export defect are caused solely by intron removal from YRA1 (note that the deduced amino acid sequence of yra1-Δi is not altered; see Figure 1A), we next sought to test whether a foreign intron can substitute for the YRA1 intron. We therefore replaced exactly the intron of YRA1 with an intron derived from the RPL25 gene, which encodes the ribosomal protein Rpl25p. The RPL25 intron is also comparibly large (414 nucleotides) but exhibits a branchpoint sequence that matches the consensus (Figure 1A). Strikingly, insertion of the RPL25 intron into yra1-Δi (Figure 1A) restored cell growth (Figure 1B; yra1-L25 i) and the mRNA export (Figure 2; yra1-L25 i). However, the intron of RPL25 did not fully substitute for the intron of YRA1, as cells expressing yra1-L25i grew slightly more slowly at elevated temperatures (e.g. 37°C; Figure 1B). In addition, the mRNA export defect was not fully abolished, as a small number of cells (10%) accumulate poly(A)+ RNA in the nucleus (Figure 2; yra1-L25 i). The introns of YRA1 and RPL25 are both large. To test whether the growth defect can also be abolished by the presence of a small intron in YRA1, the intron of UBC8 was inserted in the coding region of YRA1 (Figure 1A). The UBC8 intron is very small (123 nucleotides) and also contains a non-canonical branchpoint. As observed for insertion of the RPL25 intron, insertion of the UBC8 intron also restores cell growth to wild-type levels (Figure 1B; yra1-UBC8 i). Therefore, the presence of a foreign intron within the YRA1 coding region restores the function of YRA1 to a large extent. We conclude that an intron within the YRA1 gene can regulate the function of YRA1.
To find out how the presence of an intron in YRA1 can affect cell growth and mRNA export, we sought to test whether Yra1p levels are controlled by the YRA1 intron. It has been reported previously that overexpression of YRA1 from a cDNA construct (i.e. lacking the intron) is toxic for the cells (Espinet et al., 1995). In addition, it has been noted that expression from an intronless YRA1 construct results in overproduction of Yra1p (Zenklusen et al., 2001). We therefore tested whether an intron in YRA1 can modulate Yra1p expression by western blot analysis using anti-Yra1p antibodies. Strikingly, the Yra1p is overexpressed (∼3-fold) in yeast when expressed from the yra1-Δi construct, but it is normal when expressed from either the intact YRA1 gene or the yra1-L25i and yra1-UBC8i constructs (Figure 3A). We conclude that the authentic intron or a foreign intron inserted into the YRA1 gene is responsible for downregulation of Yra1p expression.
Fig. 3. An intron within the YRA1 gene downregulates expression of Yra1p. (A) Equivalent amounts of whole-cell lysates of yra1-Δi (lane 1), yra1-L25i (lane 2), yra1-UBC8i (lane 3) and YRA1 (lane 4) cells were analysed by SDS–PAGE and western blotting using Yra1p and Arc1p antibodies. (B) Growth curve of wild-type yeast cells (RS453) transformed with plasmids encoding GAL1::YRA1, GAL1::yra1-Δi, GAL1::yra1-L25i or GAL1. Transformants were grown in galactose-containing medium and the optical density was determined at 600 nm. (C) Whole-cell lysates of GAL1::yra1-Δi (lane 1), GAL1::yra1-L25i (lane 2) and GAL1::YRA1 (lane 3) cells were analysed by SDS–PAGE and western blotting using Yra1p and Arc1p antibodies. (D) Northern blot analysis of YRA1 mRNA in yra1-Δi (lane 1), yra1-L25i (lane 2) and YRA1 cells (lane 4). Two bands corresponding to the spliced (YRA1 s) and unspliced (YRA1 u) mRNA were detected with a YRA1 probe. After quantification, an increase of 15% of spliced YRA1 mRNA for the yra1-Δi was detected. The value was obtained after normalization with ACT1 mRNA levels as a loading control.
These findings prompted us to test whether the intron-dependent downregulation of Yra1p expression depends on the promoter that drives YRA1 transcription. Therefore, we placed the YRA1, yra1-Δi and yra1-L25i constructs under control of the strong GAL1 promoter, which is switched on by galactose and switched off by glucose (Guarente, 1984). Unexpectedly, cells expressing Yra1p from the GAL1::YRA1 or GAL1::yra1-L25i constructs grew normally in galactose-containing medium (Figure 3B). However, GAL1::yra1-Δi cells grew significantly slower in galactose-containing medium (Figure 3B). In contrast, all three constructs, GAL1::YRA1, GAL1::yra1-L25i and GAL1::yra1-Δi, did not affect cell growth in glucose-containing medium (data not shown). To find out whether growth inhibition in galactose-containing medium observed for the GAL1::yra-Δi construct results from Yra1p overproduction, western blot analysis was performed using anti-Yra1p antibodies. As seen in Figure 3C, the amount of Yra1p is significantly higher in GAL1::yra1-Δi cells than in GAL1::YRA1 and GAL1::yra1-L25i cells when grown in galactose-containing medium. Thus, the presence of an intron in YRA1 can downregulate Yra1p expression to physiological, i.e. non-toxic, levels even when its expression is driven from the strong GAL1 promoter.
This last result also suggests that Yra1p production is not transcriptionally regulated. Therefore, Yra1p expression appears to be regulated post-transcriptionally. To assess whether the amount of YRA1 mRNA is altered in YRA1, yra1-Δi and yra1-L25i cells, we performed northern blot analysis of poly(A)+ RNA isolated from these three different strains. The levels of mature YRA1 mRNA were not strikingly different in these strains, although yra1-Δi had the tendency to exhibit slightly enhanced amounts of YRA1 mRNA (Figure 3D). Moreover, unspliced YRA1 pre-mRNA could be detected in the YRA1 and yra1-L25i strains. Taken together, northern and western blotting data revealed that a post-transcriptional regulatory mechanism that depends on splicing downregulates Yra1p expression.
In this study, we have shown that Yra1p expression in yeast is regulated by the intron in YRA1. Strikingly, removal of the intron from the YRA1 gene causes overexpression of Yra1p, which results in a dominant-negative phenotype and an mRNA export defect. Thus, Yra1p expression must be tightly regulated in cells, otherwise excess Yra1p might titrate a factor(s) involved in mRNA export. Candidate proteins that may require a balanced concentration of Yra1p are the essential mRNA export factors Mex67p/Mtr2p and Sub2p, which form separate complexes with Yra1p (Sträßer and Hurt, 2000, 2001; Stutz et al., 2000). However, overexpression of Sub2p or Mex67p from a high copy plasmid in the yra1-Δi strain did not suppress the dominant-negative phenotype (data not shown).
The finding that downregulation of Yra1p expression is not transcriptionally regulated and does not correlate with the amount of mRNA could point to a translationally controlled process. In higher eukaryotes, it is known that the nuclear history of a pre-mRNA can determine the translational activity of cytoplasmic mRNAs (Matsumoto et al., 1998). Accordingly, the position of an intron within a pre-mRNA determines the translational efficiency of the mature mRNA in the cytoplasm. When the intron is close to the 5′ end of the transcript, expression is stimulated. On the other hand, when the intron is close to the 3′ end of the transcript, translation is repressed. In analogy, the intron in YRA1, which is located far downstream of the start codon (286 nucleotides), could repress translation. In contrast to YRA1, most yeast pre-mRNAs have the intron close to the 5′ end of the open reading frame, which may stimulate translation (Fink, 1987; Woolford, 1989; Rymond and Rosbash, 1992). However, there could also be other ways to control Yra1p expression in vivo at the level of translation, e.g. by autoregulation of the splicing-derived YRA1 transcript upon Yra1p binding. Further work is required to unravel the exact mechanism that controls Yra1p expression and therefore mRNA export in yeast.
METHODS
Yeast strains and plasmids. Saccharomyces cerevisiae strains were grown in YPD (1% yeast extract, 2% peptone and 2% glucose) or minimal SC medium [0.67% yeast nitrogen base without amino acids (Difco) supplemented with the appropriate ‘drop-out mix’ (Difco)] containing raffinose or galactose as a carbon source. Growth curves of strains expressing YRA1 from the GAL1 promoter were obtained after transformation of strain RS453 with plasmids YCplac22G–YRA1, YCplac22G–yra1-Δi or YCplac22G–yra1-L25i. Cells were grown in selective raffinose-containing medium, washed and resuspended in SC medium containing galactose as a carbon source.
Wild-type RS453 and YRA1 shuffle strains were described previously (Sträßer and Hurt, 2000). DNA recombinant work such as restriction analysis, ligation, PCR amplification and Escherichia coli transformation were performed according to Maniatis et al. (1982).
The plasmid pUN100–YRA1 has been described previously (Sträßer and Hurt, 2000). pRS315–yra1-Δi, pRS315–yra1-L25i and pRS315–yra1-UBC8i were constructed by fusion PCR resulting in an exact deletion of the intron or its exact replacement by the intron of RPL25 or UBC8, respectively. GAL1::YRA1, GAL1::yra1-Δi and GAL1::yra1-L25i were cloned by amplifying the YRA1 coding region from pUN100–YRA1, pRS315–yra1-Δi or pRS315–yra1-L25i, respectively, creating BamHI and Pst I sites and cloning into the same sites of plasmid YCplac22G.
Northern blot, western blot and fluorescence microscopy. Isolation of poly(A)+ mRNA was performed following the recommendations of the provider (PolyATract System 1000, Promega). Electrophoresis, blotting, radioactive probe labelling and signal detection were performed as described in Maniatis et al. (1982).
SDS–PAGE and western blot analysis were performed according to Siniossoglou et al. (1996). Poly(A)+ RNA export was analysed by in situ hybridization (Santos-Rosa et al., 1998).
Acknowledgments
ACKNOWLEDGEMENTS
E.H. is a recipient of grants from the Deutsche Forschungsgemeinschaft, and S.R.-N. is a holder of a Marie Curie fellowship.
REFERENCES
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J., Tan, W., Zolotukhin, A.S., Tabernero, C., Hudson, E.A. and Felber, B.K. (1999) Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol., 19, 6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach, E., Schmitt, C. and Izaurralde, E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L., Munnerlyn, A. and Grosschedl, R. (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRa enhancer function. Genes Dev., 11, 640–653. [DOI] [PubMed] [Google Scholar]
- Espinet C., de la Torre, M.A., Aldea, M. and Herrero, E. (1995) An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast, 11, 25–32. [DOI] [PubMed] [Google Scholar]
- Fink G.R., (1987) Pseudogenes in yeast? Cell, 49, 5–6. [DOI] [PubMed] [Google Scholar]
- Fleckner J., Zhang, M., Valcarcel, J. and Green, M.R. (1997) U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev., 11, 1864–1872. [DOI] [PubMed] [Google Scholar]
- Gatfield D., Le Hir, H., Schmitt, C., Braun, I.C., Kocher, T., Wilm, M. and Izaurralde, E. (2001) The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol., 11, 1716–1721. [DOI] [PubMed] [Google Scholar]
- Guarente L. (1984) Yeast promoters: positive and negative elements. Cell, 36, 799–800. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Boulay, J., Rosbash, M. and Libri, D. (2001) The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol., 11, 1711–1715. [DOI] [PubMed] [Google Scholar]
- Kang Y.B. and Cullen, B.R. (1999) The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev., 13, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.B., Bogerd, H.P., Yang, J. and Cullen, B.R. (1999) Analysis of the RNA binding specificity of the human Tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology, 262, 200–209. [DOI] [PubMed] [Google Scholar]
- Katahira J., Sträβer, K., Podtelejnikov, A., Mann, M., Jung, J.J. and Hurt, E.C. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A.L. and Guthrie, C. (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev., 15, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde, E., Maquat, L.E. and Moore, M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield, D., Izaurralde, E. and Moore, M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Graziani, N., Saguez, C. and Boulay, J. (2001) Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev., 15, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.-J., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M. and Reed, R. (2001) Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature, 413, 644–647. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch, E.T. and Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Matsumoto K., Montzka Wassarman, K. and Wolffe, A.P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B. and Rosbash, M. (1992) Yeast pre-mRNA splicing. In Broach, J.R., Pringle, J. and Jones, E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae (Vol. 2). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 143–191.
- Santos-Rosa H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Panté, N. and Hurt, E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R. and Hurt, E.C. (1997) Mex67p which is an essential factor for nuclear mRNA export binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer, C., Rieger, M., Doye, V., Tekotte, H., Weise, C., Emig, S., Segref, A. and Hurt, E.C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell, 84, 265–275. [DOI] [PubMed] [Google Scholar]
- Sträßer K. and Hurt, E.C. (2000) Yra1p, a conserved nuclear RNA binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. and Hurt, E.C. (2001) Splicing factor Sub2p is required for nuclear export through its interaction with Yra1p. Nature, 413, 648–652. [DOI] [PubMed] [Google Scholar]
- Sträßer K., Baßler, J. and Hurt, E.C. (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol., 150, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi, A., Doerks, T., Braun, I.C., Séraphin, B., Wilm, M., Bork, P. and Izaurralde, E. (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius C.M., Wagner, S. and Green, M.R. (1999) A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell, 4, 219–228. [DOI] [PubMed] [Google Scholar]
- Woolford J.L. (1989) Nuclear pre-mRNA splicing in yeast. Yeast, 5, 439–457. [DOI] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra, P., Strahm, Y. and Stutz, F. (2001) The Yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol., 21, 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. and Green, M.R. (2001) Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev., 15, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.L., Luo, M., Straesser, K., Katahira, J., Hurt, E. and Reed, R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]