Abstract
During replication, mitochondrial DNA (mtDNA) takes on a triple-stranded structure called a D-loop. Although their physiological roles are not understood, D-loops are implicated in replication and transcription of mtDNA. Little is known about the turnover of D-loops. We investigated the effects of mitochondrial transcription factor A (TFAM) and single-stranded DNA-binding protein (mtSSB) on D-loops. In human HeLa cells, TFAM and mtSSB are, respectively, 1700- and 3000-fold more abundant than mtDNA. This level of TFAM is two orders of magnitude higher than reported previously and is sufficient to wrap human mtDNA entirely. TFAM resolves D-loops in vitro if added in similar stoichiometries. mtSSB inhibits the resolution of mtDNA by TFAM but enhances resolution by RecG, a junction-specific helicase from Escherichia coli. Hence, mtSSB functions in both stabilization and resolution. We propose that TFAM and mtSSB are cooperatively involved in stabilizing D-loops and in the maintenance of mtDNA.
INTRODUCTION
Most vertebrate cells contain 103–104 copies of mitochondrial DNA (mtDNA), depending on the cellular energy demand (Shadel and Clayton, 1997). To meet energy demands, maintenance of the mtDNA copy number is important. However, it has not been elucidated fully how copy number is maintained. Clearly, replication of mtDNA could play a central role in the maintenance of copy number. Replication of mtDNA is initiated by synthesis of the heavy strand and proceeds to displace the parental heavy strand. Synthesis frequently terminates ∼700 bp downstream, giving rise to 7S DNA (D-loop strand). This premature termination produces a characteristic triple-stranded structure, called the D-loop (Kasamatsu et al., 1971). Although the precise function of mitochondrial D-loops is not known, D-loops are assumed to be involved in regulating mtDNA replication. A Parkinsonism-causing toxin, 1-methyl-4-phenylpyridium ion (MPP+), accumulates at levels of up to 20 mM in mitochondria in vivo (Ramsay and Singer, 1986) and inhibits mtDNA replication (Miyako et al., 1997). This inhibition results from the destabilization of D-loops (Umeda et al., 2000), which suggests that D-loop stability is involved in the regulation of replication.
Mitochondrial transcription factor A (TFAM), a member of the high mobility group (HMG) of proteins, contains two HMG-box domains. Many HMG family proteins, including TFAM, are able to bind, wrap, bend and unwind DNA with a low degree of sequence specificity. Disruption of the mouse TFAM gene is an embryonic lethal mutation due to depletion of mtDNA (Larsson et al., 1998). This indicates that TFAM is essential for the maintenance of mtDNA. ABF2, a TFAM homolog of Saccharomyces cerevisiae, is abundant in mitochondria. Unlike TFAM in mammals, ABF2 is not required for transcription initiation of mtDNA (Diffley and Stillman, 1992). When ABF2 is deleted, S. cerevisiae loses mtDNA in the presence of fermentable carbon sources (Diffley and Stillman, 1991). This mtDNA depletion can be rescued by a bacterial histone-like protein, HU (Megraw and Chae, 1993), which suggests that ABF2 maintains mtDNA by wrapping mtDNA. Because ABF2 can also be replaced functionally by human TFAM (Parisi et al., 1993), human TFAM shares common properties with ABF2 and HU. Disruption of a mitochondrial single-stranded DNA-binding protein (mtSSB) gene causes mtDNAs to be lost in yeast (van Dyck et al., 1992) and Drosophila (Maier et al., 2001), thereby suggesting that mtSSB is essential for the maintenance of mtDNA. mtSSB enhances processivity by DNA polymerase γ of Xenopus laevis (Mignotte et al., 1988) and Drosophila (Farr et al., 1999).
Here, we show that TFAM and mtSSB exist at levels, respectively, 1700- and 3000-fold higher than mtDNA in human HeLa cells. At these levels, D-loops were destabilized by application of purified recombinant TFAM. mtSSB inhibited the resolution of D-loops by TFAM or MPP+ but enhanced the resolution of D-loops by RecG, a junction-structure-specific helicase of Escherichia coli. This raises the possibility that mtSSB not only stabilizes D-loops, but may also participate in the resolution of D-loops. These results suggest that TFAM and mtSSB are cooperatively involved in the turnover of D-loops.
RESULTS
Quantification of TFAM, mtSSB and mtDNA
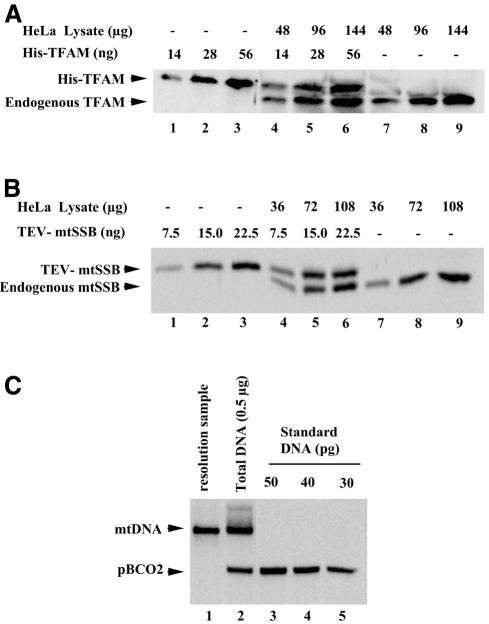
We quantified TFAM, mtSSB and mtDNA in HeLa MRV11 cells (Figure 1A–C). On average, individual HeLa cells contained 1.7 × 106, 2.8 × 106 and 1.0 × 103 molecules of TFAM, mtSSB and mtDNA, respectively (Table I). The amount of mtDNA was confirmed by quantitative PCR analysis (data not shown). The ratio of TFAM to mtDNA was 1700:1 (Table I), which means that mitochondria of HeLa cells contain one molecule of TFAM for every 10 bp of mtDNA.
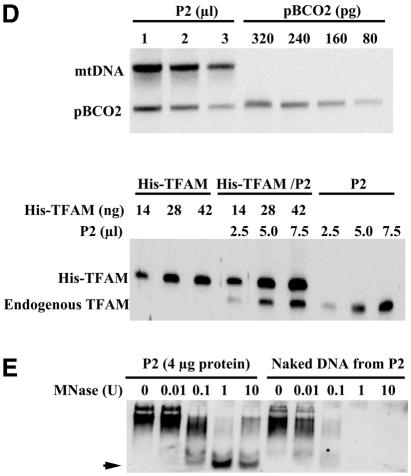
Fig. 1. Quantification of TFAM, mtSSB and mtDNA. (A) The purified recombinant His-TFAM and HeLa cell lysates were run in the same gel. The amount of His-TFAM was adjusted to yield a signal intensity similar to that of endogenous TFAM in western blots. One microgram protein of the lysate corresponds to 1.7 × 103 cells. (B) Western blots for quantification of mtSSB. Procedures are the same as in (A). The N-terminal His tag of His-mtSSB was removed with TEV protease (TEV-mtSSB). (C) Southern blots for quantification of mtDNA. Signals of the standard DNA pBCO2 are shown (lanes 3–5). In lane 2, endogenous mtDNA and an internal standard are shown. mtDNA used for the resolution assay is in lane 1. (D) Total DNA extracted from the indicated volume of P2 (1 mg protein/ml) was analyzed as in (C) (upper). His-TFAM, P2 mixed with His-TFAM and P2 alone were analyzed in western blots (lower). (E) P2 or DNA extracted from P2 was incubated with various concentrations of micrococcal nuclease (MNase) and then mtDNA was detected in Southern blots using a probe covering nucleotide position (np) 7441–8286. One unit is an activity producing one OD260 of acid-soluble materials from calf thymus DNA at 37°C for 30 min.
Table I. Amounts of TFAM, mtSSB and mtDNA in HeLa cellsx.
| Molecules per cell | Molecules per mtDNAa | |
|---|---|---|
| TFAM | 1.7 ± 1.3 × 106 | 1.7 ± 1.3 × 103 |
| mtSSB | 2.8 ± 1.0 × 106 | 3.0 ± 1.0 × 103 |
| mtDNA | 1.0 ± 0.08 × 103 | – |
Each value is expressed as the mean ± SD of three independent experiments.
aThe ratio of protein to mtDNA was calculated in each experiment and then averaged.
To examine TFAM–mtDNA complexes, we prepared an NP-40-insoluble fraction (P2) from human placental mitochondria. In this fraction, the iron sulfur protein of complex II was removed completely, but almost all TFAM and mtDNA was recovered (data not shown). The TFAM:mtDNA ratio was 940:1 in this fraction (Figure 1D). Consistent with the idea that mtDNA has a nucleoid structure, mtDNA in the P2 fraction was 10-fold more resistant to nuclease digestion than was naked mtDNA (Figure 1E). In addition, we observed a band of ∼500 bp in the P2 fraction (arrow in Figure 1E) that persisted even in the presence of 10 U nuclease. Essentially the same pattern was obtained using a different probe corresponding to np 2573–4431 (data not shown), raising the possibility that ∼500 bp mtDNA reside in a nucleosome-like unit.
TFAM resolves the D-loop structure
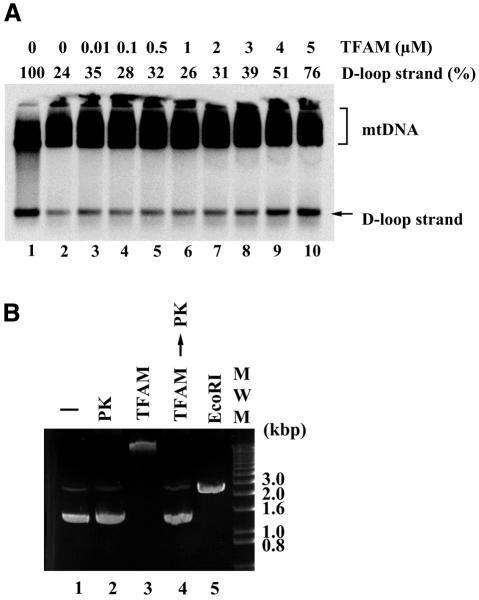
Because HMG proteins alter DNA structure, we examined the effects of TFAM on D-loops. Treatment with purified recombinant His-TFAM released nascent strands ∼700 bases in length (D-loop strands) in a dose-dependent manner. TFAM maximally resolved D-loops at 5 µM (Figure 2A) and higher (data not shown) in a 10 µl reaction mixture containing 1 µg of DNA. Five micromolar TFAM is equivalent to one molecule of TFAM per 30 bp DNA in this assay system, thus indicating that TFAM can resolve D-loops at physiologically relevant concentrations. Although mtDNA accounted for only 10% of the DNA used in the reactions (based on Southern blot analysis; data not shown), this is a reasonable conclusion, considering the ratio of TFAM to base-pair number rather than the molar ratio to mtDNA, because TFAM can bind to DNA non-specifically (Fisher and Clayton, 1988; see also Figure 2B, lane 3). Because contamination of the purified TFAM with nucleases could cause strand breaks leading to D-loop resolution, we assayed our preparations for nuclease activity. Purified TFAM at concentrations as high as 8 µM never introduced strand breaks in supercoiled plasmids (Figure 2B, lane 4), thus ruling out nuclease contamination. In these experiments, TFAM retarded markedly the mobility of the supercoiled plasmids, probably due to binding to DNA (Figure 2B, lane 3).
Fig. 2. Effect of TFAM on D-loop structure. (A) mtDNA was incubated at 37°C for 30 min with various concentrations of TFAM (lanes 2–10). In lane 1, mtDNA was incubated at 90°C for 10 min. Each sample was analyzed in Southern blots. (B) Ethidium bromide staining of supercoiled plasmid pBR322. The plasmids were incubated with (lanes 3 and 4) or without (lanes 1 and 2) 8 µM TFAM at 37°C for 30 min, followed by incubation without (lanes 1 and 3) or with (lanes 2 and 4) proteinase K (PK) at 37°C for 15 min. In lane 5, plasmids were linearized with EcoRI.
Effects of mtSSB on D-loop resolution by TFAM, MPP+ and RecG
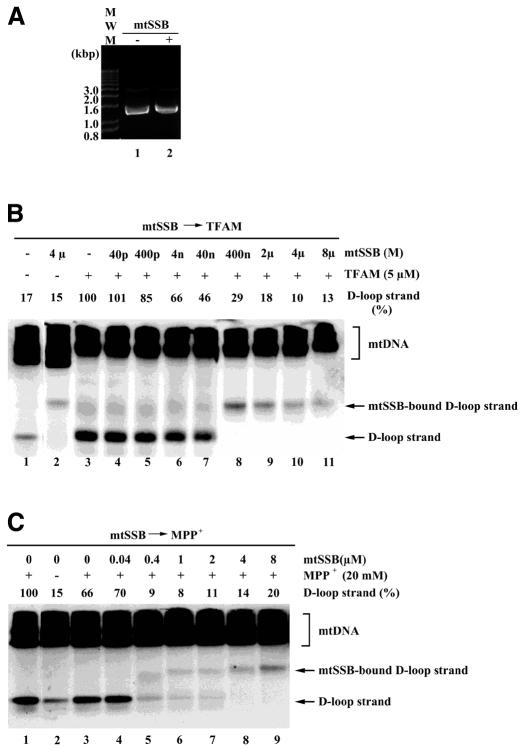
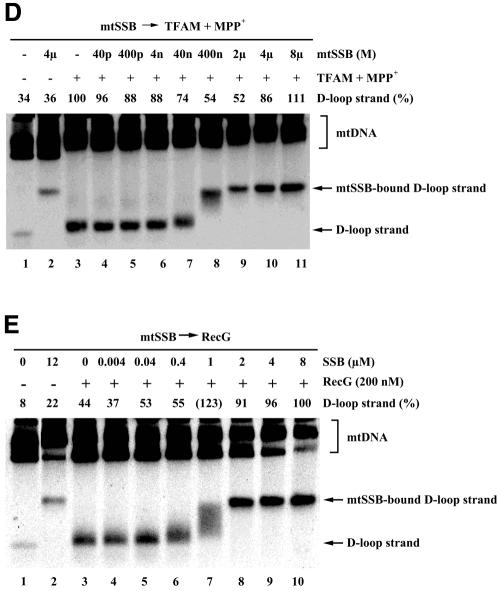
mtSSB may stabilize D-loops by binding to their single-stranded portions (Pavco and van Tuyle, 1985). In our experiments, treatment with recombinant His-mtSSB did not cause any mobility shift of supercoiled pBR322 plasmids (Figure 3A), which excludes the possibility that this mtSSB sample contained nucleases or double-stranded DNA-binding proteins. Pretreatment with mtSSB inhibited the TFAM-induced resolution of D-loops (Figure 3B, lanes 4–11). Because the released D-loop strand is single-stranded DNA, mtSSB bound to the released D-loop strand and slowed its mobility. In the presence of 2 µM and higher mtSSB, TFAM-induced resolution of D-loops was reduced to control levels (Figure 3B, lanes 1 and 9–11). Similarly, resolution by MPP+ was inhibited by preincubation with mtSSB (Figure 3C). D-loops were resolved by the combination of 5 µM TFAM and 10 mM MPP+ (Figure 3D, lane 3). Pretreatment with mtSSB did not notably inhibit resolution by the combination of TFAM and MPP+ (Figure 3D, lanes 4–11), unlike its effects on TFAM and MPP+ alone. In other words, MPP+, with the assistance of TFAM, can resolve D-loops even in the presence of mtSSB. This is consistent with the fact that MPP+ resolves D-loops in organello (Umeda et al., 2000).
Fig. 3. Effects of mtSSB on D-loop resolution. (A) pBR322 plasmids were incubated with 8 µM mtSSB (lane 2) and a vehicle (lane 1), electrophoresed on an agarose gel and then stained with ethidium bromide. mtDNA was preincubated with the indicated concentration of mtSSB, followed by incubation with 5 µM TFAM (B), 20 mM MPP+ (C), 5 µM TFAM and 10 mM MPP+ (D) and 200 nM of RecG (E).
RecG (200 nM), a junction-specific helicase of E. coli, also resolved D-loops (Figure 3E, lanes 1 and 3). Stimulation of helicase activity by single-stranded DNA-binding proteins has been observed frequently in various systems. Indeed, mtSSB at concentrations of 2 µM and higher enhanced the resolution of D-loops by RecG (Figure 3E, lanes 3–10).
DISCUSSION
The TFAM:mtDNA ratio has been reported to be between 15:1 and 30:1 in human cells (Fisher et al., 1991). However, this ratio was calculated based on an indirect estimate. The TFAM:mtDNA ratio increases up to 2000:1 in Xenopus oocytes (Shen and Bogenhagen, 2001), and one copy of the TFAM homolog ABF2 exists for every 15 bp mtDNA in yeast (Diffley and Stillman, 1991). Our estimate of the ratio of TFAM to mtDNA in HeLa cells is rather close to these values. Given that TFAM occupies ∼25 bp DNA (Fisher and Clayton, 1988), the amount of TFAM in HeLa cells is sufficient to cover the entire region of mtDNA. Oocytes are transcriptionally silent, but yeast is not. Therefore, high levels of mitochondrial HMG-family proteins are not necessarily related to transcriptional state. TFAM may be essential for initiation of transcription. We hypothesize, however, that there are more TFAM molecules in human mitochondria than are actually required for initiation. Excess TFAM, in part, explains recent findings that TFAM levels can be substantially reduced without significant inhibition of mtDNA transcription (Goto et al., 2001). Human TFAM may actually constitute a main component of the mtDNA nucleoid (Spelbrink et al., 2001).
In this study, we analyzed short-type D-loops, which we refer to simply as D-loops. Extended D-loops, in which nascent strands exceed the premature termination site, lead to complete mtDNA replication. Extended D-loops would naturally be resolved by TFAM and stabilized by mtSSB. The majority of short-type D-loops turn over rapidly and are ‘abortive’ for replication (Bogenhagen and Clayton, 1978), which means that short-type D-loops are resolved for a restart of DNA synthesis. If mtSSB continued to stabilize short-type D-loops, ‘true’ replication would decline. We propose that the turnover of short-type D-loops is also involved in the regulation of replication. It is interesting that RecG, a junction-specific helicase, resolved D-loops and that this effect was enhanced by the addition of mtSSB. A junction-specific helicase seems like a reasonable candidate for an endogenous factor that resolves D-loops. Assuming that such a helicase is involved in the resolution of D-loops, mtSSB may play a key role in regulating the turnover of D-loops because it could participate in both stabilization and resolution.
METHODS
Expression and purification of mtSSB and TFAM. To express N-terminal His6-tagged human mtSSB (His-mtSSB), DNA encoding mature human mtSSB (17–148 amino acid residues) was inserted between the BamHI and EcoRI sites of pProEXHTb (Gibco-BRL); the vector was designated pHis-mtSSB. The recombinant His-mtSSB was expressed in BL21 cells. Aggregated His-mtSSB was denatured, then renatured, and His-mtSSB was purified using Ni2+-bound chelating Sepharose resin (Amersham Pharmacia Biotech). Recombinant TFAM with N-terminal glutathione S-transferase (GST–TFAM) and N-terminal His (His-TFAM) were purified as described previously (Ohno et al., 2000). After separation by SDS–PAGE, protein concentrations were determined by Coomassie Brilliant Blue staining using bovine serum albumin as a standard and an LAS-1000 CCD camera and Image Gauge image analysis software (Fuji Photo Film). Protein concentrations were also confirmed both by SYPRO orange (Molecular Probes) staining and by amino acid analysis.
Resolution of D-loops. D-loop resolution was performed as described previously (Umeda et al., 2000). Briefly, a crude mitochondrial fraction from HeLa MRV11 cells was prepared by differential centrifugation. Total DNA was extracted from the mitochondrial fraction, treated with RNase A, precipitated with ethanol and solubilized in buffer A containing 20 mM HEPES–KOH pH 7.5 and 25 mM KCl. For the reaction with mtSSB, 1 µg of DNA was incubated in 10 µl of buffer A in the presence of the indicated concentration of His-mtSSB at 37°C for 20 min. For the reaction with TFAM or MPP+, 1 µg of DNA was incubated in 10 µl of buffer A in the presence of the indicated concentration of His-TFAM or MPP+ at 37°C for 30 min. Final salt concentrations in reactions were adjusted with NaCl, when indicated. The reaction with RecG was performed as described previously (Ohsato et al., 1999). Released D-loop strands were detected on Southern blots. DNA from the D-loop region (np 16 036–394) was used as a probe.
Quantification of TFAM, mtSSB and mtDNA in cultured cells. HeLa MRV11 cells (108; Kang et al., 1995) were lysed in 1 ml of buffer containing 50 mM Tris–HCl pH 8.0, 20 mM NaCl, 1 mM EDTA and 1% SDS. Aliquots from the same lysate were used for quantification of TFAM, mtSSB and mtDNA in each round of experiments. TFAM and mtSSB were quantified by western blotting. Antibodies against TFAM and mtSSB were produced by immunizing rabbits with GST–TFAM and His-mtSSB, respectively. The N-terminal histidine tag of His-mtSSB was removed with TEV protease (Gibco-BRL) to avoid immunoreaction with the His tag on western blots. The mtSSB without the His tag was used as an mtSSB standard in protein assays.
To prepare an internal standard that would allow us to accurately quantify mtDNA, an mtDNA region (np 7441–8286) was cloned into pBlueScript KS (Stratagene); this clone was designated pBCO2. pBCO2 was linearized with XhoI, separated by electrophoresis and purified. The purified DNA was quantified by measuring A260. Total DNA in the HeLa cell lysate was extracted with a DNAzol kit (Gibco-BRL). To estimate the recovery of DNA, 6.4 ng of pBCO2 was added to 40 µl (4.0 × 106 cells) of the lysate prior to DNA extraction. On average, ∼30 µg of total DNA was extracted from 4 × 106 cells. The extracted DNA was digested with XhoI, treated with phenol/chloroform, precipitated with ethanol and solubilized in distilled water. The DNA was quantified by measuring A260. The XhoI-digested DNA was analyzed by Southern blotting using DNA from np 7441–8286 as a probe. The calculation of the amount of mtDNA per cell was corrected for losses that occurred during extraction by examining the amount of pBCO2 lost during the procedure. The amount of mtDNA was also measured by quantitative PCR analysis using a LightCycler system (Roche).
NP-40-insoluble fraction. Human placental mitochondria were prepared by a combination of differential and Percoll gradient centrifugation (Gasnier et al., 1993). A non-ionic detergent (NP-40) was used to prepare an insoluble fraction essentially according to the method of Miyakawa et al. (1987). Briefly, mitochondria (10 mg protein/ml) were solubilized in buffer containing 0.25 M sucrose, 10 mM Tris–HCl pH 7.0, 1 mM EDTA and 0.5% NP-40. The resulting particulate fraction was washed once with the same buffer and designated P2. Western blot analysis of P2 was performed as described above. P2, or naked DNA extracted from P2, was suspended in 10 µl of buffer containing 0.25 M sucrose, 10 mM Tris–HCl pH 7.5 and 2.5 mM CaCl2 and then digested with micrococcal nuclease (Takara) at 4°C for 10 min. The reaction was terminated by adding 10 µl of buffer containing 100 mM Tris–HCl pH 8.0, 40 mM NaCl, 2 mM EDTA, 2% SDS and 50 µg/ml proteinase K. After 15 min further incubation at 37°C, the reaction mixture was electrophoresed and mtDNA was detected by Southern blotting.
Acknowledgments
ACKNOWLEDGEMENTS
Language assistance was provided by M. Ohara. This work was supported in part by Grants in Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports and Culture of Japan.
REFERENCES
- Bogenhagen D. and Clayton, D.A. (1978) Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J. Mol. Biol., 119, 49–68. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X. and Stillman, B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F.X. and Stillman, B. (1992) DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem., 267, 3368–3374. [PubMed] [Google Scholar]
- Farr C.L., Wang, Y. and Kaguni, L.S. (1999) Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. J. Biol. Chem., 274, 14779–14785. [DOI] [PubMed] [Google Scholar]
- Fisher R.P. and Clayton, D.A. (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol., 8, 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.P., Lisowsky, T. and Breen, G.A. (1991) A rapid, efficient method for purifying DNA-binding proteins. J. Biol. Chem., 266, 9153–9160. [PubMed] [Google Scholar]
- Gasnier F., Rousson, R., Lerme, F., Vaganay, E., Louisot, P. and Gateau-Roesch, O. (1993) Use of Percoll gradients for isolation of human placenta mitochondria suitable for investigating outer membrane proteins. Anal. Biochem., 212, 173–178. [DOI] [PubMed] [Google Scholar]
- Goto A., Matsushima, Y., Kadowaki, T. and Kitagawa, Y. (2001) Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem. J., 354, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Nishida, J., Iyama, A., Nakabeppu, Y., Furuichi, M., Fujiwara, T., Sekiguchi, M. and Takeshige, K. (1995) Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J. Biol. Chem., 270, 14659–14665. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Robberson, D.L. and Vinograd, J. (1971) A novel closed-circular mitochondrial DNA with properties of replicating intermediates. Proc. Natl Acad. Sci. USA, 68, 2252–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson N.G., Wang, J., Wilhelmsson, H., Oldfors, A., Rustin, P., Lewandoski, M., Barsh, G.S. and Clayton, D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- Maier D., Farr, C.L., Poeck, B., Alahari, A., Vogel, M., Fischer, S., Kaguni, L.S. and Schneuwly, S. (2001) Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol. Biol. Cell, 12, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T.L. and Chae, C.B. (1993) Functional complementarity between the HMG-like yeast mitochondrial histone HM and bacterial histone-like protein HU. J. Biol. Chem., 268, 12758–12763. [PubMed] [Google Scholar]
- Mignotte B., Marsault, J. and Barat-Gueride, M. (1988) Effects of Xenopus laevis mitochondrial single-stranded DNA-binding protein on the activity of DNA polymerase γ. Eur. J. Biochem., 174, 479–484. [DOI] [PubMed] [Google Scholar]
- Miyakawa I., Sando, N., Kawano, S., Nakamura, S. and Kuroiwa, T. (1987) Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci., 88, 431–439. [DOI] [PubMed] [Google Scholar]
- Miyako K., Kai, Y., Irie, T., Takeshige, K. and Kang, D. (1997) The content of intracellular mitochondrial DNA is decreased by 1-methyl-4-phenylpyridinium ion (MPP+). J. Biol. Chem., 272, 9605–9608. [DOI] [PubMed] [Google Scholar]
- Ohno T., Umeda, S., Hamasaki, N. and Kang, D. (2000) Binding of human mitochondrial transcription factor A, an HMG box protein, to a four-way DNA junction. Biochem. Biophys. Res. Commun., 271, 492–498. [DOI] [PubMed] [Google Scholar]
- Ohsato T., Muta, T., Fukuoh, A., Shinagawa, H., Hamasaki, N. and Kang, D. (1999) R-loop in the replication origin of human mitochondrial DNA is resolved by RecG, a Holliday junction-specific helicase. Biochem. Biophys. Res. Commun., 255, 1–5. [DOI] [PubMed] [Google Scholar]
- Parisi M.A., Xu, B. and Clayton, D.A. (1993) A human mitochondrial transcription activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco P.A. and van Tuyle, G.C. (1985) Purification and general properties of the DNA-binding protein (P16) from rat liver mitochondria. J. Cell Biol., 100, 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay R.R. and Singer, T.P. (1986) Energy-dependent uptake of N-methyl-4-phenylpyridinium, the neurotoxic metabolite of 1-methyl-4-phenyl-1,2,3, 6-tetrahydropyridine, by mitochondria. J. Biol. Chem., 261, 7585–7587. [PubMed] [Google Scholar]
- Shadel G.S. and Clayton, D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- Shen E.L. and Bogenhagen, D.F. (2001) Developmentally regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res., 29, 2822–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink J.N. et al. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature Genet., 28, 223–231. [DOI] [PubMed] [Google Scholar]
- Umeda S., Muta, T., Ohsato, T., Takamatsu, C., Hamasaki, N. and Kang, D. (2000) The D-loop structure of human mtDNA is destabilized directly by 1-methyl-4-phenylpyridinium ion (MPP+), a parkinsonism-causing toxin. Eur. J. Biochem., 267, 200–206. [DOI] [PubMed] [Google Scholar]
- van Dyck E., Foury, F., Stillman, B. and Brill, S.J. (1992) A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J., 11, 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]