Abstract
Advances in proteogenomic technologies have revealed hundreds to thousands of translated small open reading frames (sORFs) that encode microproteins in genomes across evolutionary space. While many microproteins have now been shown to play critical roles in biology and human disease, a majority of recently identified microproteins have little or no experimental evidence regarding their functionality. Computational tools have some limitations for analysis of short, poorly conserved microprotein sequences, so additional approaches are needed to determine the role of each member of this recently discovered polypeptide class. A currently underexplored avenue in the study of microproteins is structure prediction and determination, which delivers a depth of functional information. In this review, we provide a brief overview of microprotein discovery methods, then examine examples of microprotein structures (and, conversely, intrinsic disorder) that have been experimentally determined using crystallography, cryo-electron microscopy, and NMR, which provide insight into their molecular functions and mechanisms. Additionally, we discuss examples of predicted microprotein structures that have provided insight or context regarding their function. Analysis of microprotein structure at the angstrom level, and confirmation of predicted structures, therefore, has potential to identify translated microproteins that are of biological importance and to provide molecular mechanism for their in vivo roles.
Keywords: sORF, Microprotein, Structure, Mass Spectrometry – LC-MS/MS, Genome
1 ∣. Introduction:
Small open reading frames (sORFs, also termed smORFs) below 100 codons were excluded by the FANTOM genome annotation consortium to filter out the high rate of false positive sORFs that were detected under this size in eukaryotic long noncoding RNAs[1,2]. A similar small size cutoff of 50 codons was applied for prokaryotic gene annotation[3]. These cutoffs were expedient, since the number of known genes pales in comparison to the number of background ORF-like sequences within a genome, most of which are not expressed[4], but resulted in systematic under-detection of functional sORFs. Many expressed sORFs have now been discovered: recent studies have converged on hundreds of previously unannotated sORFs in bacteria[5,6] and thousands in human[7-9], and multiple CRISPR screens have suggested that hundreds of human sORFs are required for cell survival and proliferation[10,11]. The emerging relevance of sORFs to infectious disease[12], the microbiome[13,14], and human disease[10,15] opens the possibility of new therapeutic strategies, and, as such, consortium efforts to enter translated sORFs into the genome annotation are underway[16].
Early discoveries of functional sORF-encoded polypeptides, such as humanin[17] in human, tarsal-less/polished rice in Drosophila[18-20] and SgrT[21] in bacteria, occurred individually. As a result, the global nature of sORF translation was not recognized until the seminal demonstration of ubiquitous translating ribosome occupancy outside canonical reading frames by Ingolia et al.[22] and subsequent confirmation of the presence of a large number of unannotated sORF translation products with mass spectrometry[23,24]. The products of sORF translation have been termed small proteins[25], microproteins[26-28], micropeptides[29], sORF-encoded polypeptides (SEPs)[24] and, evocatively, ghost proteins[30]; we will utilize the term microprotein throughout this review. In addition, longer, non-annotated proteins, in some cases referred to as alternative proteins, particularly when they overlap canonical proteins[23], have also been identified, but they will not be specifically discussed herein. For the purpose of this review, our definition of a eukaryotic microprotein will extend to previously unannotated proteins below 130 amino acids, as many previously undetected ORFs of this length have been reported in human cells. Prokaryotic microproteins are typically categorized as less than or equal to 50 amino acids in length[31]; however, our definition in this work will extend to 70 amino acids since many unannotated microproteins of this size have been detected in multiple bacterial species.
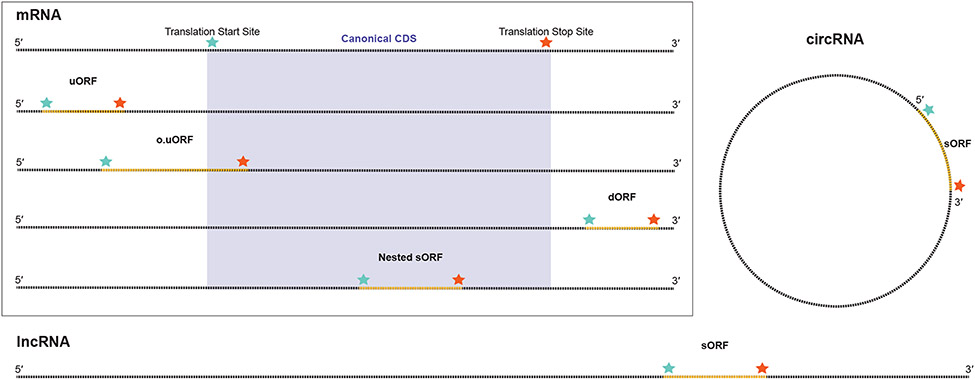
Multiple classes and regions of RNA, both coding and noncoding, have been shown to harbor sORFs in prokaryotes and eukaryotes (Figure 1). Functional sORFs have been discovered in small and long noncoding RNAs (ncRNAs and lncRNAs)[32,33], antisense lncRNAs[34,35], microRNA (miRNA) precursors[36], and circular RNAs[37] in bacteria, plants and other eukaryotes. Interestingly, an increasing number of genes have been shown to exert functions both at the level of the RNA and of the encoded microprotein, such as sgrST[38], azuCR[39], Spot42/SpfP[40], and some miRNAs[41,42]. Additional classes of sORFs have been identified in multicistronic mRNAs alongside canonical protein coding sequences (CDS) in both prokaryotes and, surprisingly, eukaryotes. sORFs in 5′ untranslated regions (UTRs) relative to an annotated CDS are referred to as upstream ORFs (uORFs)[43]. Importantly, while eukaryotic uORFs have long been regarded as cis-translational regulators that generally decrease translation efficiency of the downstream CDS[44], in some instances, uORFs encode microproteins with independent cellular functions in trans, such as MIEF1-MP[45], which regulates mitochondrial protein translation, and ASDURF[46], which is a previously unidentified component of the prefoldin-like module of the PAQosome. Some sORFs that initiate in the 5′ leader extend into and overlap the CDS in an alternative reading frame, and can be termed overlapping uORFs (o.uORFs), such as human alt-RPL36[47], which overlaps ribosomal protein L36 and regulates the phospholipid transporter TMEM24. It is important to note that, because they are translated in a different reading frame, o.uORF polypeptide amino acid sequence is completely different from that of the downstream, overlapping annotated protein. At the other end of the mRNA, the 3′ UTR has also been found to encode microproteins from downstream ORFs (dORF), which may also regulate CDS translation[48]. An emerging class of frameshifted sORFs occur internally within a protein CDS. These nested sORFs lie completely within the main ORF with translation initiating downstream of the main ORF start codon, and translation terminating upstream of the main ORF stop codon. Nested sORFs can occur in the +2 or +3 (same-strand, frameshifted) reading frames (Figure 2), such as E. coli GndA[49] and human alt-FUS[50]. Surprisingly, these findings point to the fact that mammalian mRNAs may be multicistronic or dual coding[51]. While prokaryotic organisms are known to express polycistronic mRNA transcripts termed operons, and compact viral genomes have long been known to contain overlapping open reading frames, eukaryotic transcripts have long been thought to be monocistronic as a result of the scanning model of translation initiation[52,53]. Importantly, microproteins and longer alternative proteins encoded in each of these classes of sORFs have been shown to be functional[3,25,54-58]. In summary, coding and noncoding regions of both prokaryotic and eukaryotic genomes encode functional sORFs in loci that are denser and more complex than previously presumed.
FIGURE 1.
Small open reading frames (sORFs) and RNA. Box: Within mRNA that encodes canonical protein coding sequences (CDSs), sORFs can appear in the 5′ UTR (upstreamORF [uORF]), initiating in the 5′ UTR and extending into the CDS in an alternative reading frame (upstream overlapping ORF [u.oORF]), in the 3′ UTR (downstreamORF [dORF]), or nested within the CDS in an alternative reading frame. sORFs can also be found in long noncoding RNA (lncRNA, bottom) and circular RNA (circRNA, right), as well as additional classes of RNA not pictured.
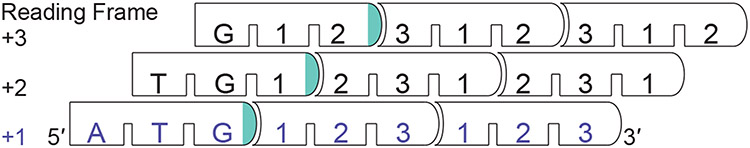
FIGURE 2.
Alternative Reading Frames for Same-Strand Overlapping (Nested) sORFs. The +1 reading frame corresponds to the canonical coding sequence and is always the frame of reference. Frameshifted translation in the +2 or +3 reading frames generates protein products with completely different amino acid sequences because the codon identities are changed in alternative reading frames.
2 ∣. Microprotein Discovery
2.1 ∣. Computation
Accurate annotation of sORFs using computational tools is challenging not only due to their short lengths that impede statistical analyses[24], but also because they exhibit intermediate conservation relative to longer genes, which has been interpreted as evidence for the de novo evolution of some microproteins[59]. Notwithstanding these challenges, algorithms and machine learning strategies are currently being developed to better find sORFs within genomes. Some computational efforts rely on phylogeny, nucleotide and amino acid homology, and secondary structure to identify unannotated sORFs with sequence or structural similarities to canonical proteins; examples include PhyloCSF[60] and miPFinder[61]. Additional dimensions of predictive information, including the presence of a ribosome binding site upstream of bacterial sORF start codons[62] or a Kozak consensus sequence surrounding a eukaryotic sORF start codon[63], have been applied to sORF prediction. Ambitiously, OpenProt predicts all AUG-initiated sORFs and alternative ORFs (alt-ORFs) within all known mRNAs for several organisms, and curates experimental evidence (or lack thereof) for their expression[64,65]. Finally, deep forest and deep learning models have been applied to sORF prediction, with application to individual microbial genomes, as well as the microbiome and metagenomes[14,66]. These methods have highlighted new sORFs in intergenic regions, noncoding RNAs and in multicistronic/dual coding mRNAs.
2.2 ∣. Ribosome Profiling
Deep sequencing of the protected mRNA footprints of actively translating ribosomes (ribosome profiling or Ribo-seq) has been reviewed[67] and provides a powerful technology for detection of translated sORFs. Ribo-seq is carried out by isolating translating ribosomes associated with mRNA transcripts, using either elongation inhibitors like cycloheximide[22,68,69] or rapid freezing[68,70,71]. Because translating 80S ribosomes protect bound mRNA fragments from digestion by RNase, sequencing the ribosome-protected fragments (RPFs) reports on translated regions of mRNA. Furthermore, the codon-by-codon elongation of 80S ribosome gives RPFs a characteristic 3-nucleotide periodic distribution, which can be used to infer the reading frame and confidently differentiate translated ORFs from noise[72]. Furthermore, translation efficiency can be assessed by comparing the frequency of ribosome footprint reads to mRNA transcript levels[73]. Rigorous data analysis, high-resolution datasets, and analysis of replicates are essential for calling sORF translation using Ribo-seq, because their short lengths and translation by monosomes lead to lower signal-to-noise in sORF-mapped reads relative to longer canonical protein coding sequences[8,74].
While Ribo-seq is powerful in profiling the footprints of elongating ribosomes and identifying novel coding regions, elongation inhibitors like cycloheximide are not well-suited to deconvolute some translation initiation sites, especially for ORFs with multiple start sites or overlapping reading frames[75]. As a result, a specialized method called translation initiation sequencing (TI-seq) has been developed for inhibition and profiling of the footprints of initiating ribosomes that leverage molecules like puromycin, harringtonine and lactimidomycin in eukaryotes[68,76-78], and retapamulin[6], tetracycline[79] and Onc112[5] in prokaryotes. The enrichment of ribosome footprints at canonical and non-canonical start codons in TI-seq datasets generates peaks at the beginning of putative sORFs as well as canonical protein coding sequences. This allows deconvolution of sORF translation initiation from larger main ORFs in multicistronic mRNAs, and is especially important for detection of nested and out of frame sORFs. TI-Seq can also be combined with Ribo-Seq to call translated ORFs with higher confidence[80].
2.3 ∣. Mass Spectrometry
Mass spectrometry proteomics is able to detect translational products of sORFs directly in biological samples using either bottom-up (from peptide fragments) or top-down (intact precursor) modalities[81,82]. However, specialized sample preparation and computational methods must be applied for high-sensitivity detection of small, unannotated microproteins. For example, a standard bottom-up proteomics experiment begins with isolation of the proteome, during which small molecules and proteolytic fragments are typically removed by SDS-PAGE or filter-aided sample preparation[83]. Furthermore, most peptide and protein identification from proteomics data is accomplished via spectral matching against the annotated proteome database[84]. For these reasons, sORF-encoded polypeptides are both de-enriched from proteomic samples, and absent from databases, and therefore cannot be detected with standard proteomic workflows and searches.
Multiple recent reviews and protocols describing microprotein identification via proteomics are available[81,82,85,86], so we provide a brief overview highlighting only the key concerns here. Microprotein discovery methods are built on the same technologies used for standard shotgun proteomics, with several modifications (Figure 3). First, because sORF-encoded microproteins are small, most are identified by only a single proteotypic or fingerprint tryptic fragment in a typical proteomics experiment[24]. A major factor complicating detection of microproteins is coelution and/or cofragmentation of the one or two detectable tryptic peptides derived from a given microprotein with abundant tryptic and/or proteolytic fragments of larger proteins. Resulting ion suppression and/or complex spectra preclude detection and/or identification of the microprotein fragment[87,88], regardless of its abundance; this consideration is less severe for larger, canonical proteins, which generate many tryptic peptides and thus detection of any individual fragment is not required. Therefore, the first critical step of any sORF proteomic experiment is to achieve proteome extraction in the absence of proteolysis of canonical proteins (e.g., via boiling in acidic solution or application of protease inhibitors[85]) to minimize sample complexity, followed by or concomitant with enrichment of the small proteome and exclusion of large proteins. Small protein enrichment can be achieved via multiple chemical and biophysical methods, such as solid phase extraction, peptide gels, GELFrEE resolution, and organic solvent or surfactant extraction[89-93]. When they have been compared head-to-head, these methods have typically been shown to offer comparable numbers, but non-overlapping sets, of microproteins detected[89-92]. Depending on the experimental goals, the size selection approach for microprotein proteomics can therefore be optimally chosen: for example, for the deepest coverage, multiple methods should be employed on replicate samples and the results combined; for a rapid, robust and economical approach, organic solvent or surfactant extraction may prove attractive.
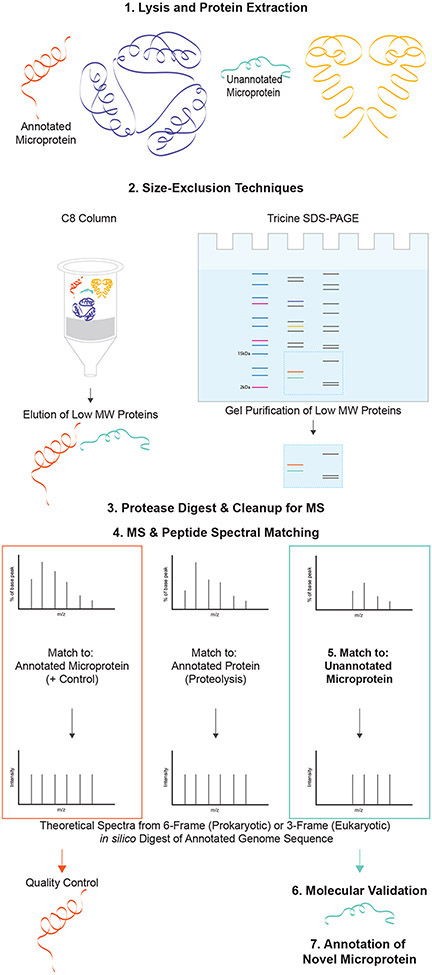
FIGURE 3.
Mass Spectrometry Workflow for Detection of Unannotated Microproteins. To search for novel microproteins in a sample of interest, low molecular weight proteins are isolated from total protein after cell lysis. Size-exclusion techniques include, but are not limited to, solid-phase extraction and polyacrylamide gel electrophoresis techniques. Low molecular weight protein is digested with a protease, producing a sample of uniform peptide length appropriate for mass spectrometric (MS) analysis. Experimental spectra are generated and matched to theoretical spectra from a custom database using proteomics software. Detection of annotated microproteins known to be expressed in the system of interest can serve as a positive control for success of small protein enrichment and known small proteome coverage, but these spectra are otherwise computationally excluded. Peptides deriving from proteolysis of canonical proteins before size-exclusion are computationally identified and excluded from consideration. High scoring experimental spectra without any matches to known microproteins can be subjected to further molecular validation, leading to annotation of novel microproteins.
Subsequent to small proteome isolation, most microprotein studies to date have employed bottom-up proteomic analysis, in which microproteins are enzymatically digested into peptide fragments (typically with trypsin, though multienzyme digests have been shown to improve small proteome coverage[94]), followed by liquid chromatography-tandem mass spectrometry, often with multi-dimensional separation[24]. This experiment provides thousands of raw peptide fragmentation spectra corresponding both to known canonical small proteins and microproteins, which must then be identified and distinguished. This is typically accomplished via peptide-spectral matching against expanded databases comprising the canonical proteome as well as candidate sORF sequences[85,95]. For eukaryotes, databases can be derived from three-frame transcriptome translations[24], ribosome profiling-derived translatomes[15,96,97], or publicly available noncanonical ORF databases such as OpenProt[64] and sORFs.org[98]; six-frame genomic translation can be employed for prokaryotes[49,95,99]. Peptide-spectral matching against any of these databases affords identifications of both canonical small proteins and unannotated microproteins. It is important to note that discrimination of false-positive identifications that arise from searching expanded databases is critical[100,101]; at the same time, the expansion of the decoy database also decreases sensitivity for true positive matches[102,103], so, ideally, the smallest database containing all known proteins and unannotated microproteins (and any common artefactual contaminants[104]) should be used. An alternative approach is to employ permissive false discovery rates, followed by either manual inspection of fragmentation spectra or a secondary algorithm like PepQuery to exclude false positive spectra better explained by peptides arising from canonical, mutant or post-translationally modified proteins[105]. After exclusion of peptides matching (or near-matching) annotated proteins, the resulting list of identifications represent candidate unannotated microproteins, which can be computationally mapped to the sORFs that encode them and experimentally validated[85].
Top-down proteomics is emerging as an alternative modality for microprotein discovery[81,106,107]. This technology has particular importance for identification of microprotein proteoforms, a term that refers to all post-transcriptionally and post-translationally processed protein products arising from a single gene[108,109]. For example, microproteins and other noncanonical proteins can exhibit multiple proteoforms as a result of alternative splicing[110], N-terminal proteolytic processing[111], phosphorylation[112,113], and other post-translational modifications[108], and most of this variability is obscured in bottom-up proteomic analyses as a result of incomplete sequence coverage and inability to distinguish whether modifications on different tryptic fragments occur on the same, or mutually exclusive, proteoforms[108]. Top-down analysis has identified novel microprotein proteoforms in microorganisms[81], and its expanded adoption should accelerate the identification of microprotein N- and C- termini as well as functional modification states in the future.
Mass spectrometry typically detects one to two orders of magnitude fewer microproteins than ribosome profiling[114]. This may be due to the abovementioned challenge in detecting single microprotein-derived fingerprint peptides; the relative insensitivity of mass spectrometry to some classes of microproteins, including membrane-localized, positively charged, and low-abundance species[49]; the instability of some sORF translation products[115]; reduced sensitivity for true-positive detections as a result of expanded decoy databases applied for stringent false discovery rate estimation[102]; or all of these factors. Nonetheless, mass spectrometry offers several advantages. First, enrichment strategies, such as membrane fractionation and chemical labeling, can enable identification of microproteins that are refractory to shotgun analysis of whole-cell tryptic digests, thus beginning to address one of the major limitations of microprotein proteomics while at the same time affording functional information about microproteins (e.g., chemical reactivity, subcellular localization) that is inaccessible to sequencing methods[86]. Second, without specialized analysis pipelines, ribosome profiling with elongation inhibitors is refractory to confident detection of sORFs that overlap canonical protein coding sequences in alternative reading frames, due to the requirement for three-nucleotide periodicity for ORF calling. In contrast, mass spectrometry can readily detect and identify microproteins derived from overlapping ORFs, which can represent as much as 30% of microproteins identified in a proteomic experiment[24]. Given the complementary nature of genomics, ribosome profiling and mass spectrometry, it is likely that the combination of these methods offers the greatest power for large-scale, high-confidence microprotein identification.
3 ∣. Microprotein Structure and Function
Dozens of human microproteins, and many more in model organisms, have now been assigned function at the molecular, cellular, and/or organismal level[116]. CRISPR screens have implicated hundreds of sORFs in cell survival and proliferation[10,11]. Experimental approaches are yielding insights into the roles of microproteins in biological processes and disease, which have been extensively reviewed[56,58,117]. Recently emerging trends in microprotein function include roles in immunity and inflammation[15,118-120], mitochondrial functions and energetics[35,45,121-123], adiposity[124,125], microbial carbon metabolism[38-40], and cancer initiation and progression[10,15,58,126-128], among others. Nonetheless, the vast majority of recently discovered microproteins remain entirely uncharacterized in mechanistic detail. This is in large part because bioinformatic predictions of sORF function are challenging—even when they exhibit signatures of conservation in multiple species, microproteins tend to lack primary sequence homology to proteins of known function[24,59]. While three-dimensional structure prediction and elucidation are likely to provide important insights into microprotein functions, structures of microproteins have not yet been examined on scale. However, the number of experimentally determined structures of microproteins, in isolation or in complex with their effectors, is growing, and general trends have begun to emerge, which we will describe in this section. First, we discuss a subclass of single-pass alpha-helical transmembrane microproteins, many of which are evolutionarily novel, and some of which bind to and regulate important transporters. Next, we consider examples of microproteins with solved or predicted structures and the potential relevance to their functions. Last, we will examine several intrinsically disordered microproteins that undergo regulatory protein-protein interactions.
3.1 ∣. Alpha-helical transmembrane microproteins
Intergenic regions of eukaryotic genomes are rich in A/T residues relative to genes, which are G/C rich[129]. When microproteins are expressed from “noncoding” regions, they therefore tend to contain predicted transmembrane helices arising from the preponderance of T/U residues within codons that correspond to hydrophobic and aromatic amino acids[129]. This intergenic sequence bias therefore affects the amino acid composition of evolutionarily young, species-specific microproteins, that arise de novo from previously noncoding regions of the genome[59]. A recent study demonstrated that C-terminal hydrophobic patches tend to target evolutionarily young microproteins to the BAG6 membrane protein triage complex, resulting either in membrane insertion or, if mislocalized or improperly folded, proteasomal degradation[115]. Interestingly, species-specific transmembrane microproteins that exhibit low expression can nonetheless contribute fitness advantage to cells[129], and examples have been shown to function in processes such as yeast mating[130]. Not all membrane-associated microproteins are evolutionarily novel; a large and growing number of well-characterized, conserved transmembrane microproteins are predicted to contain transmembrane helices, such as the lysosomal membrane-localized polypeptide regulator of mTORC1, SPAR[131], the plasma membrane localized micropeptide Myomixer[132], which is required for myoblast fusion during skeletal muscle development, and Kastor and Polluks, mammalian microproteins required for sperm motility[133]. The class of alpha-helical transmembrane microproteins is therefore large, and of outsize biological importance. We turn our attention in this section to those membrane-associated microproteins that have been subjected to experimental structure determination.
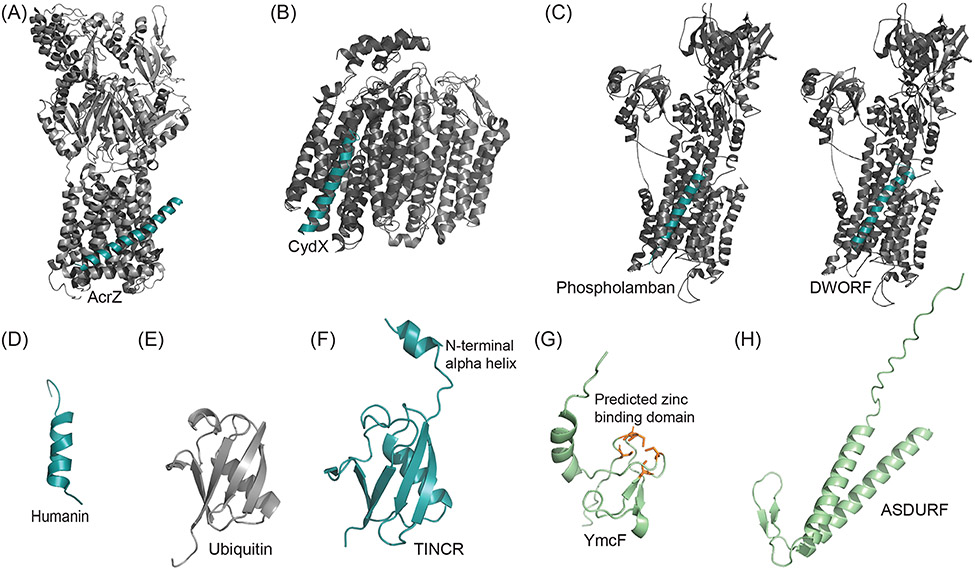
AcrZ, previously named YbhT, was reported in a seminal study identifying unannotated small protein genes in E. coli utilizing computational tools that incorporate ribosome binding site prediction[62]. AcrZ is a 49-amino acid microprotein that is conserved in many Gram-negative bacteria and localizes to the E. coli inner and outer membranes by virtue of an N-terminal transmembrane helix[62]. AcrZ binds to the AcrB subunit of the AcrAB/tolC multidrug efflux pump, increasing the efficiency of transport of (and, thus, resistance of E. coli to) a subset of its substrates[134]. Multiple structures of AcrZ in complex with the AcrB homotrimer have been solved, including crystal structures of detergent-solubilized complexes[135], as well as a cryo-electron microscopy (cryo-EM) structure of the complex reconstituted in lipid discs[136] (Figure 4A). AcrZ binds to a transmembrane groove within each molecule of AcrB. The cryo-EM structure revealed that AcrZ exhibits a profound bend between positions 10-15, conferred by a helix-breaking proline residue[136]. Mutagenesis studies revealed that the proline is required for interaction of AcrZ with AcrB. At the same time, proline, or an equally helix-breaking glycine residue, can be moved to any position within the AcrZ interaction motif while retaining its association with AcrB. Several of these mutations that retain AcrB binding also recapitulate the selective drug transport-promoting phenotype of wild-type AcrZ. While the precise effects of AcrZ binding on cargo occupancy and transport are not fully clear, allosteric modulation of binding sites in AcrB is evident by comparing the AcrB vs. AcrBZ structures. Furthermore, AcrZ promotes cardiolipin association with AcrB, likely contributing to allosteric modulation of cargo binding pockets in the transporter. Taken together, these results indicate that the bend in the transmembrane helical shape of AcrZ, and not its amino acid sequence, is essential for interaction and modulation of AcrB.
FIGURE 4.
Experimentally Determined Microprotein Structures. (A) Crystal structure of AcrB (grayscale) of the tolC efflux pump in complex with microprotein AcrZ (cyan). PDB: 5NC5. (B) Cryo-EM structure of bacterial microprotein CydX (cyan) in complex with transmembrane cytochrome bd-I oxidase (grayscale). PDB: 6RKO. (C) Crystal structure of SERCA1a calcium pump (grayscale) with bound single-pass transmembrane microprotein phospholamban (cyan), which downregulates SERCA activity. PDB: 4Y3U. Solid-state NMR structure of helix-loop-helix microprotein DWORF (cyan) modeled into SERCA1a calcium pump (grayscale) based on Venkateswara et al. 2022. PDB: 4Y3U, 7MPA. (D) NMR structure of wild-type humanin in 30% 2,2,2-trifluoroethanol (organic) solution. PBD: 1Y32. (E) Crystal structure of Ubiquitin monomer. PDB: 1AAR. (F) Crystal Structure of ubiquitin-like TINCR microprotein with additional N-terminal alpha helix. PDB: 7MRJ. (G) Predicted structure of bacterial microprotein YmcF generated with AlphaFold, obtained from UniProt[166] (green). Five cysteines (orange) in the YmcF sequence are predicted to form a zinc-finger domain common to RNA binding proteins. (H) Predicted structure of PAQosome binding microprotein ASDURF generated with AlphaFold, obtained from UniProt[166].
E. coli CydX was originally identified as YbgT, a predicted 37-amino acid microprotein encoded downstream of the cytochrome bd oxidase operon genes cydA and cydB[62]. Cytochrome bd oxidases operate as terminal electron acceptors in the electron transport chain under hypoxic conditions due to their high oxygen affinity[137]. The two canonical subunits, CydA and CydB, form a pseudosymmetric heterodimer, of which the CydA subunit contains all three heme residues responsible for reduction of molecular oxygen to water, as well as the Q loop that is responsible for binding an electron donor quinol. CydX is a single-pass alpha helical transmembrane protein that copurifies with the CydAB complex and is required for the assembly, stability, and/or activity of cytochrome bd oxidase in multiple species[137-139]. Several atomic structures of cytochrome bd oxidases have revealed the role of CydX homologs in the complex (Figure 4B). First, the presence of an unannotated, CydX homolog, CydS, was serendipitously discovered in a crystal structure of cytochrome bd oxidase purified from the gram-positive bacterium Geobacillus thermodenitrificans[140]. CydS forms an alpha helix that binds between helices 5 and 6 of CydA, leading the authors to speculate that it may stabilize the heme cofactor when the Q loop undergoes dynamic movement during catalysis[140]. A subsequent cryo-EM structure of the E. coli cytochrome BD oxidase revealed CydX bound to CydA between helices 1 and 6, again suggesting a structural role[141]. Interestingly, the E. coli CydAB unexpectedly revealed the presence of another single-pass transmembrane microprotein, CydH, which is encoded in the ynhF gene that is not located within the cytochrome bd oxidase operon. CydH binds between transmembrane helices 1 and 8 of CydA, on the opposite face of CydA relative to CydX[141]. CydH is proposed to occlude the proposed oxygen-conducting channel from the Geobacillus complex structure, which has been replaced with a hydrophobic channel that traverses CydB directly to the heme d site. The CydH oxygen channel rearrangement was proposed to be required due to the swapped positions of two heme cofactors in the E. coli enzyme relative to the Geobacillus structure, and, accordingly, CydH homologs are found in Proteobacteria. Overall, cytochrome bd oxidase is a unique system in which microproteins are required for activity, structure and stability of a critical complex of proteins.
In another well-characterized example, a class of microproteins (also called micropeptides[142]) termed “regulins” regulate the activity of the sarco/endoplasmic reticulum (SR/ER) calcium ATPase (SERCA)[143]. During muscle contraction, including the contraction of the heart and calcium-dependent signaling processes, calcium is released from the SR/ER into the cytosol; then, to terminate signaling or contraction, calcium is pumped back into the SR/ER against its concentration gradient using the energy of ATP hydrolysis by SERCA. Regulins colocalize with SERCA in the SR/ER membrane, and each micropeptide is expressed in the same, specific tissue as the SERCA isoform that it regulates. The first known regulins, phospholamban[144] and sarcolipin[145], were identified as inhibitors of SERCA in cardiac and skeletal muscle, respectively. Structural analysis of these canonical regulins, both of which are <100 amino acids, reveals that they are small, single-pass membrane proteins bearing a single transmembrane alpha-helix. The crystal structure of the SERCA-sarcolipin complex reveals that the micropeptide binds in a transmembrane groove in the SERCA channel between helices 2, 6 and 9, where it allosterically alters the conformation of SERCA to decrease its apparent calcium affinity[146]. Phospholamban binds to the same regulatory groove[147] (Figure 4C). One seminal discovery of novel SERCA regulating micropeptides came from a study in Drosophila[148]. In this work, Couso and colleagues analyzed putative lncRNAs associated with polysomes, suggesting that they are translated. Of these lncRNAs, one contained an sORF encoding a peptide predicted to be homologous to phospholamban and sarcolipin, which was accordingly given the name sarcolamban. Sarcolamban may have arisen via duplication of an ancestral phospholamban/sarcolipin gene in insects, which subsequently diverged to the sarcolamban sequence. Sarcolamban was demonstrated to bind SERCA in flies and its deletion caused heart arrythmias, consistent with a role in regulating SERCA. Docking the predicted structure of sarcolamban onto SERCA was consistent with a similar binding mode as that observed for phospholamban and sarcolipin. Just as importantly, additional novel regulins have also been discovered in mammals. In analyses of mammalian lncRNAs to identify potential micropeptides expressed in skeletal muscle and other tissues lacking known regulin expression, translated sORFs were identified that encode the novel SERCA binding micropeptides myoregulin[149], endoregulin, and another-regulin[150], all of which bind to the same transmembrane groove of SERCA, exhibit similar inhibition of SERCA to phospholamban, and are predicted to have similar single-pass transmembrane alpha-helical structures. Interestingly, an unannotated, SERCA-activating micropeptide, DWORF, was identified in yet another long noncoding RNA in mouse[151]. DWORF is expressed in skeletal muscle, and ectopic over-expression of DWORF in heart tissue enhances contractility and reverses heart failure in a model of heart failure[29]. However, the mechanism by which DWORF activates SERCA was unclear, since it is predicted to bear a similar alpha-helical transmembrane domain and binds to the same SERCA groove as previously characterized regulins, which are all inhibitory. Some evidence from fluorescence resonance energy transfer suggests that DWORF binding can directly activate SERCA[152]. A recent NMR structural study demonstrated that the alpha helix of DWORF is kinked at a unique proline residue, creating a significant bend in the transmembrane region without disrupting its binding to SERCA[153] (Figure 4C). Mutating this proline residue diminished the bend angle between the two alpha helical regions of DWORF, and not only prevented its activation of SERCA, but converted it into a SERCA inhibitor[153]. Therefore, activation of SERCA by DWORF appears to require its proline-induced kink, and, by extension, inhibition of SERCA by phospholamban, sarcolipin, myoregulin, endoregulin and another-regulin may be hypothesized to require binding of their uninterrupted transmembrane helices to the regulatory groove of SERCA. It is also fascinating to note the parallels between DWORF and AcrZ (see above), both of which utilize kinked transmembrane alpha-helices to allosterically regulate the membrane transporters SERCA and AcrB, respectively.
3.2 ∣. Humanin and its disorder-to-order transition
Humanin is a secreted 24-amino acid polypeptide found in human serum that protects neurons from cell death in the presence of familial early onset-Alzheimer’s disease-associated mutants of amyloid precursor protein[154-156]. Interestingly, the humanin coding sequence was mapped to a polyadenylated cDNA that was expressed in the surviving brain tissue of an Alzheimer’s disease patient, and is derived from the mitochondrial 16S ribosomal RNA (rRNA)[156]. Given that another mitochondrial peptide, MOTS-C, is encoded in a region overlapping the mitochondrial 12S rRNA[124], this raises the intriguing possibility that the mitochondrial rRNA genes may be polycistronic, though the molecular mechanisms by which microprotein-encoding transcripts are generated or processed are not yet defined[156]. Humanin’s neuroprotective effects have been proposed to occur through multiple intracellular and cell-surface interaction partners, including BAX[157], IGFBP3[158], FPRL1[159], and CNTF Receptor α/WSX-1/gp130[160], though the relative contributions of these pathways to its in vivo activity remain to be determined[156]. A circular dichroism and NMR study of humanin revealed that it does not adopt a stable secondary structure in aqueous solution, although through-space interactions consistent with turns at the N- and C-termini of the peptide were observed[161]. In contrast, in 30% organic solvent, humanin forms an alpha helix spanning residues G5 to L18[161] (Figure 4D). This suggests that humanin may fold in hydrophobic environments such as cell membranes or in complex with interaction partners. Testing this hypothesis could provide deeper insight into its localization and associations with functional interaction partner(s).
3.3 ∣. Ubiquitin-like microproteins
Several groups recently reported the discovery of ubiquitin-like microproteins. In one example, the ubiquitin pseudogene UBBP4 was reported to be translated[162]. Interestingly, UBBP4 encodes three ubiquitin variants within two independent open reading frames, and mass spectrometric evidence uniquely identifying all three have been previously obtained. The UBBP4 ubiquitin-like proteins exhibit high sequence similarity to canonical ubiquitin, with 1 (variant Ubbp4A1), 4 (Ubbp4B1 or UbKEKS), or 8 amino acid substitutions (Ubbp4A2). Ubbp4A2 and UbKEKS retain a functional C-terminal diglycine motif and can be covalently conjugated to high molecular weight cellular proteins, while UbbpA1 was predominantly observed as a monomer. Despite being ~700-fold less abundant than canonical ubiquitin, UbKEKS modifies a specific subset of cellular proteins including lamins, and, rather than promoting proteasomal degradation, may be important for regulating target protein localization and/or function.
In 2020 the TINCR RNA, which was previously classified as noncoding, was shown to encode an 87-amino acid microprotein with 85% sequence homology to ubiquitin. The microprotein translated from TINCR RNA, termed pTINCR[163,164] or TUBL[165], was predicted to adopt a ubiquitin-like fold[166] (Figure 4D). This prediction was confirmed in a recent crystal structure of pTINCR, which revealed an overall ubiquitin-like fold with a positively charged N-terminal domain hypothesized to enable interaction with other biomolecules[164] (Figure 4E). Due to the lack of a C-terminal diglycine motif, pTINCR is a type II ubiquitin-like protein that associates with ubiquitin-binding proteins rather than being covalently attached to proteins. pTINCR is expressed in skin, and mice lacking pTINCR exhibit a mild delay in wound healing[165]. Importantly, two reports have identified pTINCR as a tumor suppressor in cutaneous squamous cell carcinoma and other epithelial cancers[163,164]. pTINCR is upregulated after DNA damage-induced p53 activation, and it is frequently lost or mutated in squamous cell carcinoma[163,164]. It normally promotes differentiation of keratinocytes and other epithelial cell types via its interaction with SUMOylated Cdc42[163]. Consequently, mouse embryonic stem cell-derived teratomas overexpressing pTINCR exhibit decreased growth and increased keratin deposition consistent with involvement in differentiation of skin cells[163]. Along the same lines, pTINCR overexpression inhibits the proliferation of squamous cell carcinoma cells in culture and in xenografts. Additionally, mice heterozygous for Xpc that lack pTINCR are DNA damage repair-deficient and exhibit increased formation of invasive skin papillomas and squamous cell carcinomas relative to Xpc heterozygous/pTINCR wild-type mice upon UV exposure[164]. Overall, pTINCR is a type II ubiquitin-like microprotein that is required for keratinocyte differentiation and acts as a tumor suppressor in squamous cell carcinoma.
3.4 ∣. Microproteins with predicted structures
With the advent of three-dimensional macromolecular structure prediction tools such as Rosetta[167], I-TASSER[168], Phyre[169], and, most recently, AlphaFold[170], many recently discovered, now-annotated microproteins have been subjected to computational structure prediction, and these structural models are publicly available[171,172]. For microproteins that remain unannotated, the same computational tools can be used to generate predicted structures. Importantly, tools for predicting protein functions (for example, gene ontology (GO) terms) from structural models[173-175], which can outperform sequence homology-based functional prediction[174], hold promise for application to generate hypotheses about the functions of microproteins; however, the caveat that microproteins exhibit limited sequence and, presumably, structural homology to canonical proteins must be considered. On a more granular level, one-by-one analysis of predicted microprotein structural homology has been informative. For example, analysis of the recently identified E. coli cold-shock microprotein YmcF using iTasser led to the hypothesis that YmcF may adopt a folded structure consisting of an alpha helix and 2-3 beta strands separated by a turn, homologous to a zinc-binding domain of aspartate transcarbamoylase[99] (Figure 4G). While no functional data for YmcF yet exists, this predicted structural model, if correct, may have implications in the cold shock response, which requires RNA binding proteins—some of which coordinate zinc[176]—to chaperone RNA secondary structures that become hyper-stable at low temperature[177-179]. In another example, plant microProteins are specifically defined as proteins predicted to fold into single domains that bind to and generally antagonize the functions of their effectors, such as transcription factors[28,61,180].
Critically, predicted microprotein structures and functions will require experimental validation. In a key example, a translated uORF encoding a 96-amino acid microprotein within the 5′ UTR of the human ASNSD1 gene was reported by Oyama et al. in 2007[181] and in subsequent proteomic analyses[23,24], leading to the annotation of the microprotein as ASDURF (ASNSD1 upstream open reading frame). As discussed above, evidence is accumulating that uORF microproteins can function in trans[11,45,182,183]. Remarkably, Coulombe and colleagues recently implicated ASDURF as the “missing” subunit of a chaperone complex termed the PAQosome[46]. Proximity biotinylation and pull-down experiments with PAQosome subunits revealed ASDURF as an interaction partner, and in vitro reconstitution assays suggested that it is an integral member of a PAQosome subcomplex. The PAQosome is a recently discovered chaperone that is essential for assembling complicated macromolecular complexes in the cell, including RNA polymerases, components of the spliceosome, and protein phosphatases[184]. The PAQosome consists of two modules, one of which is termed the prefoldin-like (PFDL) module[184]. The PFDL module shares some subunits and putative structural homology to prefoldin, another cellular chaperone required for folding cytoskeletal proteins and other clients[185]. Prefoldin and the PFDL module are both hexameric, consisting of three alpha- and three beta-prefoldin subunits, which both contain an alpha-helical coiled-coil separated by either one (beta) or two (alpha) hairpins; however, only five of the six PFDL subunits (three alpha and two beta) had been identified[46]. Tertiary structure modeling with Phyre suggested that ASDURF is a beta-prefoldin bearing a single beta hairpin and coiled-coil (Figure 4H), consistent with its potential identification as the undiscovered beta subunit of the PFDL module of the PAQosome – suggesting it had been missed because it was not part of the proteome annotation at the time of the PAQosome’s discovery[46]. Many additional interesting questions are raised by the ASDURF microprotein: Why is it encoded in an upstream ORF within the ASNSD1 gene? Does its 5′ UTR location confer stress responsiveness via translational regulation, as suggested by Cloutier et al.[46]? Is its function or regulation related to the downstream ASNSD1 protein, per the model of Chen, Weissman and colleagues that co-encoded microproteins and proteins tend to function in the same pathways[11]? Regardless, while the structural model requires experimental validation, it appears that ASDURF is a particularly compelling example of a microprotein for which structure prediction informs its interactions and likely function.
3.5 ∣. Intrinsically disordered microproteins
Microproteins are much shorter than annotated proteins, and they tend to exhibit limited conservation to protein domains of known function[24,59]. As a result, it is challenging to perform bioinformatic analyses, for example of predicted structure or intrinsic disorder, of microproteins with confidence, particularly because many of these predictive algorithms rely, at least in part, on homology to structures of known, larger proteins on which they are trained[170,186]. Nonetheless, some studies have suggested that microproteins may be enriched in intrinsic disorder relative to canonical proteins[128,187,188] (though an alternative analysis suggests that evolutionarily young microproteins are de-enriched in intrinsic disorder[189]), which, if true, suggests that some microproteins could carry out cellular functions associated with intrinsically disordered proteins, such as regulating signaling and other processes by binding to protein partners via short linear interaction motifs (SLIMs)[190]. In this section we discuss two human microproteins that have been experimentally confirmed to be predominantly intrinsically disordered.
MRI (Modulator of retroviral infection) was first identified in a cDNA library screen for host proteins that could complement resistance to retroviral infection of human cells[191], but it remained annotated as a predicted or uncharacterized protein-coding gene (C7ORF49) in the early 2010s. While the long isoform of MRI (MRI-1 hereafter) is 157 amino acids long and therefore not a microprotein, a 2013 peptidomics study identified an unannotated, sORF encoded isoform (MRI-2) of 69 amino acids[24]. Follow-up work demonstrated that the long MRI-1 and short MRI-2 proteins could interact with a complex of proteins essential for the non-homologous end joining pathway (NHEJ)[192], which is essential for repairing DNA double strand breaks in G1 phase of the cell cycle, as well as for B and T cell receptor gene diversification via V(D)J recombination. Specifically, MRI-1 interacts with the double-strand break binding adaptor proteins Ku70/80 (Ku) and DNA-PKcs (DNA-dependent protein kinase catalytic subunit), while MRI-2 binds to Ku[192]. Both of these MRI isoforms contain an N-terminal Ku-binding motif, explaining their association with Ku, while MRI-1 also contains a C-terminal XLF-like motif (XLM) that associates with additional, distinct NHEJ factors[193,194]. The XLM of MRI-1 is absent in the frameshifted, truncated MRI-2 isoform. One study suggests that MRI inhibits aberrant NHEJ at telomeres during S phase[195], while two studies to date are consistent with a positive role for MRI in NHEJ during most phases of the cell cycle[192,194], suggesting that the activity of MRI may be context-dependent. Purified MRI-2 was shown to promote NHEJ in vitro[192], WHILE abrogating all isoforms via knockout of the MRI gene in vivo and in pre-B cells increases sensitivity to ionizing radiation and inhibits NHEJ when coupled with knockout of the NHEJ “sentinel” gene XLF[194]. Purified MRI-1 was shown to be predominantly intrinsically disordered via hydrogen-deuterium exchange[194]; while MRI-2 was not directly investigated in this study, it is likely to have a similar degree of intrinsic disorder because these proteins share substantial sequence identity until the frameshift that truncates MRI-2. Interestingly, the N-terminal and C-terminal motifs of MRI-1 alone can nucleate separate complexes of NHEJ factors, and MRI-1 can recruit NHEJ factors to chromatin in the presence of DNA double strand breaks[194]. It is interesting to speculate MRI-2 may therefore be able to serve the same nucleating function in NHEJ via its Ku-binding motif even in the absence of the C-terminal XLM. Sleckman and colleagues proposed that MRI-1 serves as an adaptor protein for NHEJ, promoting stable association of active NHEJ complexes at sites of double strand breaks as a result of its (1) intrinsic disorder, (2) independent linear interaction motifs, and (3) its potential to multimerize[194]. While better understanding of the contributions of individual MRI isoforms to their function in vivo is required, MRI-1 and MRI-2 appear to be examples of intrinsically disordered (micro)proteins that promote assembly of a functional protein interaction network.
Another example of an experimentally validated, intrinsically disordered microprotein is NBDY. NBDY is a 68-amino acid microprotein expressed from a previously misannotated lncRNA (LOC550643)[24,27]. NBDY associates with members of the cytoplasmic mRNA decapping complex[27]. The interaction partners of NBDY, EDC4 and DCP1A, are coactivators required for allosteric activation of DCP2, which catalyzes the first step in 5′-to-3′ mRNA decay (removal of the 7-methylguanosine cap), thus regulating the stability of thousands of specific mRNA substrates [196,197]. Genetic ablation or silencing of NBDY predominantly stabilizes of DCP2 substrates, consistent with the requirement of NBDY for their effective decapping, including transcripts encoding proteins involved in immune responses – a pathway previously reported to be regulated by DCP2[198,199]. However, at the same time, a number of DCP2 substrates are destabilized by NBDY ablation, suggesting that the microprotein may act as a specificity factor for recruitment of mRNA targets to the decapping complex[198]. In particular, in the presence of NBDY, DCP2 substrate mRNAs with shorter 5′UTRs decay more rapidly, suggesting that there may be a requirement for NBDY for efficient recognition of transcripts with short leader sequences by DCP2[198]. While the molecular mechanism by which NBDY regulates the mRNA decapping complex is not yet known, mRNA decapping proteins have previously been reported to associate via SLIMs within disordered regions[200], and it is likely that NBDY participates in this network. NMR experiments indicated that NBDY is largely intrinsically disordered in solution[113], consistent with its ability to phase-separate in the presence of RNA to form liquid droplets in vitro. Within the intrinsically disordered NBDY sequence, two independent SLIMs interact with the WD40 domain of EDC4 and the EVH1 domain of DCP1A[198]. The interaction between EDC4 and NBDY appears to be more important for NBDY function in mutagenesis experiments[198], but, given the relatively low affinity of NBDY for EDC4 (KD ~ 1 micromolar)[198], the interaction with DCP1A could speculatively be important for increasing avidity of NBDY for the mRNA decapping complex, retaining it at interaction sites. Importantly, NBDY also partially localizes to and regulates phase-separated RNA granules termed P-bodies in cells[27,113], consistent with a role for intrinsically disordered microproteins in biological phase separation. NBDY is phosphorylated downstream of EGFR and cyclin-dependent kinase signaling, and this phosphorylation is required for dissociation of P-bodies – likely via electrostatic repulsion of negatively charged P-body components that promotes liquid-phase remixing and cell proliferation[113]. Taken together, NBDY’s intrinsic disorder enables its SLIM-mediated protein-protein interactions, phase separation and regulation of P-bodies, providing a well-defined example of the functional significance of intrinsic disorder in a microprotein.
4 ∣. Conclusion
As microproteins are increasingly linked with roles in human health and disease, elucidating their numbers and biological roles will be ever more essential. Regarding the complete annotation of microproteins, while there are still inconsistencies in the specific sORF loci identified across ribosome profiling studies, most recent studies detect comparable numbers of translated sORFs in a given organism. This developing consensus suggests that meta-analysis of ribosome profiling data has the potential to resolve complete sORF translatomes in the near future[16]. As this effort advances, large-scale CRISPR screens and other methods can be (re-)employed to identify functional sORFs on scale. However, it is important to note that most sORF functional screens to date have focused on cell proliferation/survival[10,11], protein-protein interactions[188], and/or conservation[72], thus potentially screening out sORFs with roles beyond these readouts. For example, microproteins with clear involvement in yeast mating[130] and cellular responses to stress[35,49,57,99] have been reported, but can be species-specific, nonessential and may not undergo long-lived interactions with other proteins, and thus would not appear as hits in most functional screens to date. Thus, alternative avenues to identify microproteins with potential functions are needed. Given the exquisite link between protein three-dimensional structure and function, investigation of microprotein structure holds tremendous promise to address this need. The advent of AlphaFold[170], combined with the rapidly increasing number of solved microprotein structures and experimentally characterized intrinsically disordered microproteins, including those described above, are already contributing to the improved power of structural prediction to generate functional hypotheses about uncharacterized microproteins. Experimental structural investigations are also providing critical mechanistic insights into how microproteins exert their functions, for example in allosteric regulation of target proteins. Combined with insights into disease-associated microprotein mutations and dysregulation, structural and mechanistic information may also pave the way to determining whether microproteins and/or their binding partners are druggable in the future.
Acknowledgments
This work was supported by a Mark Foundation for Cancer Research Emerging Leader Award, a Paul G. Allen Frontiers Group Distinguished Investigator Award, and a Sloan Research Fellowship (FG-2022-18417) (to S.A.S.), a Yale University fellowship associated with the NIGMS Chemistry-Biology Interface Training Program (5T32GM067543), and a Roberts Fellowship from the Yale University Department of Chemistry (to J.J.M.).
Abbreviations:
- sORF
small open reading frame
- SEPs
sORF-encoded polypeptides
- lncRNA
long noncoding RNA
- miRNA
microRNA
- UTR
untranslated region
- uORF
upstream ORF
- o.uORF
overlapping uORF
- dORF
downstream ORF
- alt-ORF
alternative ORF
- Ribo-seq
ribosome profiling
- RPF
ribosome-protected fragment
- TI-seq
translation initiation sequencing
- cryo-EM
cryo-electron microscopy
- SR/ER
sarco/endoplasmic reticulum
- SERCA
calcium ATPase
- rRNA
ribosomal RNA
- SLIMs
short linear interaction motifs
- XLM
XLF-like motif
- NHEJ
non-homologous end joining
- GO
gene ontology
- PFDL
prefoldin-like
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- [1].Dinger ME, Pang KC, Mercer TR, Mattick JS, Differentiating Protein-Coding and Noncoding RNA: Challenges and Ambiguities. PLoS Comput Biol 2008, 4, e1000176-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Okazaki Y, Furuno M, Kasukawa T, Adachi J, et al. , Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- [3].Storz G, Wolf YI, Ramamurthi KS, Small proteins can no longer be ignored. Annu Rev Biochem 2014, 83, 753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Basrai MA, Hieter P, Boeke JD, Small open reading frames: beautiful needles in the haystack. Genome Res 1997, 7, 768–771. [DOI] [PubMed] [Google Scholar]
- [5].Jeremy W, Fuad M, R BA, Gisela S, Identifying Small Proteins by Ribosome Profiling with Stalled Initiation Complexes. mBio 2019, 10, 10.1128/mbio.02819-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Meydan S, Marks J, Klepacki D, Sharma V, et al. , Retapamulin-Assisted Ribosome Profiling Reveals the Alternative Bacterial Proteome. Mol Cell 2019, 74, 481–493.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, et al. , The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [DOI] [PubMed] [Google Scholar]
- [8].Martinez TF, Chu Q, Donaldson C, Tan D, et al. , Accurate annotation of human protein-coding small open reading frames. Nat Chem Biol 2020, 16, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chothani SP, Adami E, Widjaja AA, Langley SR, et al. , A high-resolution map of human RNA translation. Mol Cell 2022, 82, 2885–2899.e8. [DOI] [PubMed] [Google Scholar]
- [10].Prensner JR, Enache OM, Luria V, Krug K, et al. , Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat Biotechnol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen J, Brunner AD, Cogan JZ, Nunez JK, et al. , Pervasive functional translation of noncanonical human open reading frames. Science 2020, 367, 1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Irina L, Kyle M, Xinhao S, J HA, et al. , Discovery of Unannotated Small Open Reading Frames in Streptococcus pneumoniae D39 Involved in Quorum Sensing and Virulence Using Ribosome Profiling. mBio 2022, 13, e01247–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sberro H, Fremin BJ, Zlitni S, Edfors F, et al. , Large-Scale Analyses of Human Microbiomes Reveal Thousands of Small, Novel Genes. Cell 2019, 178, 1245–1259.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Durrant MG, Bhatt AS, Automated Prediction and Annotation of Small Open Reading Frames in Microbial Genomes. Cell Host Microbe 2021, 29, 121–131.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ouspenskaia T, Law T, Clauser KR, Klaeger S, et al. , Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat Biotechnol 2022, 40, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mudge JM, Ruiz-Orera J, Prensner JR, Brunet MA, et al. , Standardized annotation of translated open reading frames. Nat Biotechnol 2022, 40, 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hashimoto Y, Niikura T, Tajima H, Yasukawa T, et al. , A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Aβ. Proc Natl Acad Sci U S A 2001, 98, 6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pueyo JI, Couso JP, Tarsal-less peptides control Notch signalling through the Shavenbaby transcription factor. Dev Biol 2011, 355, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP, Peptides Encoded by Short ORFs Control Development and Define a New Eukaryotic Gene Family. PLoS Biol 2007, 5, e106-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kondo T, Plaza S, Zanet J, Benrabah E, et al. , Small Peptides Switch the Transcriptional Activity of Shavenbaby During Drosophila Embryogenesis. Science 2010, 329, 336–339. [DOI] [PubMed] [Google Scholar]
- [21].Wadler CS, Vanderpool CK, A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 2007, 104, 20454–20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS, Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vanderperre B, Lucier JF, Bissonnette C, Motard J, et al. , Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One 2013, 8, e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, et al. , Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 2013, 9, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hemm MR, Weaver J, Storz G, Escherichia coli Small Proteome . EcoSal Plus 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bhati KK, Kruusvee V, Straub D, Nalini Chandran AK, et al. , Global analysis of cereal microProteins suggests diverse roles in crop development and environmental adaptation. G3: Genes, Genomes, Genetics 2020, 10, 3709–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].D’Lima NG, Ma J, Winkler L, Chu Q, et al. , A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 2017, 13, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eguen T, Straub D, Graeff M, Wenkel S, MicroProteins: small size-big impact. Trends Plant Sci 2015, 20, 477–482. [DOI] [PubMed] [Google Scholar]
- [29].Makarewich CA, Munir AZ, Schiattarella GG, Bezprozvannaya S, et al. , The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. Elife 2018, 7, e38319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cardon T, Fournier I, Salzet M, Shedding Light on the Ghost Proteome. Trends Biochem Sci 2021, 46, 239–250. [DOI] [PubMed] [Google Scholar]
- [31].Orr MW, Mao Y, Storz G, Qian SB, Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 2020, 48, 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou B, Yang H, Yang C, Bao Y lu, et al. , Translation of noncoding RNAs and cancer. Cancer Lett 2021, 497, 89–99. [DOI] [PubMed] [Google Scholar]
- [33].Patraquim P, Magny EG, Pueyo JI, Platero AI, Couso JP, Translation and natural selection of micropeptides from long non-canonical RNAs. Nat Commun 2022, 13, 6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Na Z, Dai X, Zheng S, Baserga SJ, et al. , Mapping subcellular localizations of unannotated microproteins and alternative proteins with MicroID. Mol Cell 2022, 82, 2900–2911.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chu Q, Martinez TF, Novak SW, Donaldson CJ, et al. , Regulation of the ER stress response by a mitochondrial microprotein. Nat Commun 2019, 10, 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dozier C, Plaza S, Functions of animal microRNA-encoded peptides: the race is on! EMBO Rep 2022, 23, 2021–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang M, Zhao K, Xu X, Yang Y, et al. , A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 2018, 9, 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].R LC, Seongjin P, Jingyi F, K VC, The Small Protein SgrT Controls Transport Activity of the Glucose-Specific Phosphotransferase System. J Bacteriol 2017, 199, 10.1128/jb.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Raina M, Aoyama JJ, Bhatt S, Paul BJ, et al. , Dual-function AzuCR RNA modulates carbon metabolism. Proceedings of the National Academy of Sciences 2022, 119, e2117930119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aoyama JJ, Raina M, Zhong A, Storz G, Dual-function Spot 42 RNA encodes a 15–amino acid protein that regulates the CRP transcription factor. Proceedings of the National Academy of Sciences 2022, 119, e2119866119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lauressergues D, Ormancey M, Guillotin B, San Clemente H, et al. , Characterization of plant microRNA-encoded peptides (miPEPs) reveals molecular mechanisms from the translation to activity and specificity. Cell Rep 2022, 38, 110339. [DOI] [PubMed] [Google Scholar]
- [42].Dozier C, Plaza S, Functions of animal microRNA-encoded peptides: the race is on! EMBO Rep 2022, 23, e54789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang H, Wang Y, Lu J, Function and Evolution of Upstream ORFs in Eukaryotes. Trends Biochem Sci 2019, 44, 782–794. [DOI] [PubMed] [Google Scholar]
- [44].Calvo SE, Pagliarini DJ, Mootha VK, Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proceedings of the National Academy of Sciences 2009, 106, 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rathore A, Chu Q, Tan D, Martinez TF, et al. , MIEF1 Microprotein Regulates Mitochondrial Translation. Biochemistry 2018, 57, 5564–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cloutier P, Poitras C, Faubert D, Bouchard A, et al. , Upstream ORF-Encoded ASDURF Is a Novel Prefoldin-like Subunit of the PAQosome. J Proteome Res 2020, 19, 18–27. [DOI] [PubMed] [Google Scholar]
- [47].Cao X, Khitun A, Luo Y, Na Z, et al. , Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24. Nat Commun 2021, 12, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wu Q, Wright M, Gogol MM, Bradford WD, et al. , Translation of small downstream ORFs enhances translation of canonical main open reading frames. EMBO J 2020, 39, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yuan P, D’Lima NG, Slavoff SA, Comparative Membrane Proteomics Reveals a Nonannotated E. coli Heat Shock Protein. Biochemistry 2018, 57, 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Brunet MA, Jacques JF, Nassari S, Tyzack GE, et al. , The FUS gene is dual-coding with both proteins contributing to FUS-mediated toxicity. EMBO Rep 2021, 22, e50640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Brunet MA, Levesque SA, Hunting DJ, Cohen AA, Roucou X, Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res 2018, 28, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kozak M, Initiation of translation in prokaryotes and eukaryotes. Gene 1999, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- [53].Wright BW, Molloy MP, Jaschke PR, Overlapping genes in natural and engineered genomes. Nat Rev Genet 2022, 23, 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zanet J, Chanut-Delalande H, Plaza S, Payre F, Small Peptides as Newcomers in the Control of Drosophila Development. Curr Top Dev Biol 2016, 117, 199–219. [DOI] [PubMed] [Google Scholar]
- [55].Garai P, Blanc-Potard A, Uncovering small membrane proteins in pathogenic bacteria: Regulatory functions and therapeutic potential. Mol Microbiol 2020, 114, 710–720. [DOI] [PubMed] [Google Scholar]
- [56].Makarewich CA, The hidden world of membrane microproteins. Exp Cell Res 2020, 388, 111853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Khitun A, Ness TJ, Slavoff SA, Small open reading frames and cellular stress responses. Mol Omics 2019, 15, 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Merino-Valverde I, Greco E, Abad M, The microproteome of cancer: From invisibility to relevance. Exp Cell Res 2020, 392, 111997. [DOI] [PubMed] [Google Scholar]
- [59].Carvunis AR, Rolland T, Wapinski I, Calderwood MA, et al. , Proto-genes and de novo gene birth. Nature 2012, 487, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lin MF, Jungreis I, Kellis M, PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Straub D, Wenkel S, Cross-Species Genome-Wide Identification of Evolutionary Conserved MicroProteins. Genome Biol Evol 2017, 9, 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE, Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol 2008, 70, 1487–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Camargo AP, Sourkov V, Pereira GAG, Carazzolle MF, RNAsamba: Neural network-based assessment of the protein-coding potential of RNA sequences. NAR Genom Bioinform 2020, 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Brunet MA, Lucier JF, Levesque M, Leblanc S, et al. , OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res 2021, 49, D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Brunet MA, Lekehal AM, Roucou X, How to Illuminate the Dark Proteome Using the Multi-omic OpenProt Resource. Curr Protoc Bioinformatics 2020, 71, e103. [DOI] [PubMed] [Google Scholar]
- [66].Miravet-Verde S, Ferrar T, Espadas-García G, Mazzolini R, et al. , Unraveling the hidden universe of small proteins in bacterial genomes. Mol Syst Biol 2019, 15, e8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Brar GA, Weissman JS, Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 2015, 16, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS, The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 2012, 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ingolia NT, Lareau LF, Weissman JS, Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Webster MW, Chen YH, Stowell JAW, Alhusaini N, et al. , mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol Cell 2018, 70, 1089–1100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wu CCC, Zinshteyn B, Wehner KA, Green R, High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol Cell 2019, 73, 959–970.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, et al. , Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014, 33, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Thoreen CC, Chantranupong L, Keys HR, Wang T, et al. , A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Heyer EE, Moore MJ, Redefining the Translational Status of 80S Monosomes. Cell 2016, 164, 757–769. [DOI] [PubMed] [Google Scholar]
- [75].Lee S, Liu B, Lee S, Huang SX, et al. , Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A 2012, 109, E2424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, et al. , Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 2010, 6, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gao X, Wan J, Liu B, Ma M, et al. , Quantitative profiling of initiating ribosomes in vivo. Nat Methods 2015, 12, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Fritsch C, Herrmann A, Nothnagel M, Szafranski K, et al. , Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res 2012, 22, 2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nakahigashi K, Takai Y, Kimura M, Abe N, et al. , Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Research 2016, 23, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhang P, He D, Xu Y, Hou J, et al. , Genome-wide identification and differential analysis of translational initiation. Nat Commun 2017, 8, 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cassidy L, Kaulich PT, Maaß S, Bartel J, et al. , Bottom-up and top-down proteomic approaches for the identification, characterization, and quantification of the low molecular weight proteome with focus on short open reading frame-encoded peptides. Proteomics 2021, 21, 2100008. [DOI] [PubMed] [Google Scholar]
- [82].Fabre B, Combier J-P, Plaza S, Recent advances in mass spectrometry–based peptidomics workflows to identify short-open-reading-frame-encoded peptides and explore their functions. Curr Opin Chem Biol 2021, 60, 122–130. [DOI] [PubMed] [Google Scholar]
- [83].Wiśniewski JR, Zougman A, Nagaraj N, Mann M, Universal sample preparation method for proteome analysis. Nat Methods 2009, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- [84].Yates JR, Eng JK, Clauser KR, Burlingame AL, Search of sequence databases with uninterpreted high-energy collision-induced dissociation spectra of peptides. J Am Soc Mass Spectrom 1996, 7, 1089–1098. [DOI] [PubMed] [Google Scholar]
- [85].Khitun A, Slavoff SA, Proteomic Detection and Validation of Translated Small Open Reading Frames. Curr Protoc Chem Biol 2019, 11, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chen Y, Cao X, Loh KH, Slavoff SA, Chemical labeling and proteomics for characterization of unannotated small and alternative open reading frame-encoded polypeptides. Biochem Soc Trans 2023, BST20221074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Annesley TM, Ion Suppression in Mass Spectrometry. Clin Chem 2003, 49, 1041–1044. [DOI] [PubMed] [Google Scholar]
- [88].Ting YS, Egertson JD, Payne SH, Kim S, et al. , Peptide-Centric Proteome Analysis: An Alternative Strategy for the Analysis of Tandem Mass Spectrometry Data. Molecular & Cellular Proteomics 2015, 14, 2301–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ma J, Diedrich JK, Jungreis I, Donaldson C, et al. , Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. Anal Chem 2016, 88, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ma J, Ward CC, Jungreis I, Slavoff SA, et al. , Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J Proteome Res 2014, 13, 1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kaulich PT, Cassidy L, Weidenbach K, Schmitz RA, Tholey A, Complementarity of Different SDS-PAGE Gel Staining Methods for the Identification of Short Open Reading Frame-Encoded Peptides. Proteomics 2020, 20, 2000084. [DOI] [PubMed] [Google Scholar]
- [92].Cassidy L, Kaulich PT, Tholey A, Depletion of High-Molecular-Mass Proteins for the Identification of Small Proteins and Short Open Reading Frame Encoded Peptides in Cellular Proteomes. J Proteome Res 2019, 18, 1725–1734. [DOI] [PubMed] [Google Scholar]
- [93].Fijalkowski I, Peeters MKR, Van Damme P, Small Protein Enrichment Improves Proteomics Detection of sORF Encoded Polypeptides. Front Genet 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Kaulich PT, Cassidy L, Bartel J, Schmitz RA, Tholey A, Multi-protease Approach for the Improved Identification and Molecular Characterization of Small Proteins and Short Open Reading Frame-Encoded Peptides. J Proteome Res 2021, 20, 2895–2903. [DOI] [PubMed] [Google Scholar]
- [95].Jaffe JD, Berg HC, Church GM, Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 2004, 4, 59–77. [DOI] [PubMed] [Google Scholar]
- [96].Menschaert G, Van Criekinge W, Notelaers T, Koch A, et al. , Deep Proteome Coverage Based on Ribosome Profiling Aids Mass Spectrometry-based Protein and Peptide Discovery and Provides Evidence of Alternative Translation Products and Near-cognate Translation Initiation Events. Molecular & Cellular Proteomics 2013, 12, 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Koch A, Gawron D, Steyaert S, Ndah E, et al. , A proteogenomics approach integrating proteomics and ribosome profiling increases the efficiency of protein identification and enables the discovery of alternative translation start sites. Proteomics 2014, 14, 2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Olexiouk V, Crappe J, Verbruggen S, Verhegen K, et al. , sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 2016, 44, D324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].D’Lima NG, Khitun A, Rosenbloom AD, Yuan P, et al. , Comparative Proteomics Enables Identification of Nonannotated Cold Shock Proteins in E. coli. J Proteome Res 2017, 16, 3722–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhang K, Fu Y, Zeng W-F, He K, et al. , A note on the false discovery rate of novel peptides in proteogenomics. Bioinformatics 2015, 31, 3249–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li H, Joh YS, Kim H, Paek E, et al. , Evaluating the effect of database inflation in proteogenomic search on sensitive and reliable peptide identification. BMC Genomics 2016, 17, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cardon T, Salzet M, Franck J, Fournier I, Nuclei of HeLa cells interactomes unravel a network of ghost proteins involved in proteins translation. Biochimica et Biophysica Acta (BBA) - General Subjects 2019, 1863, 1458–1470. [DOI] [PubMed] [Google Scholar]
- [103].Hadjeras L, Heiniger B, Maaß S, Scheuer R, et al. , Unraveling the small proteome of the plant symbiont Sinorhizobium meliloti by ribosome profiling and proteogenomics. microLife 2023, 4, uqad012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, et al. , The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 2013, 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wen B, Wang X, Zhang B, PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res 2019, 29, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Cassidy L, Helbig AO, Kaulich PT, Weidenbach K, et al. , Multidimensional separation schemes enhance the identification and molecular characterization of low molecular weight proteomes and short open reading frame-encoded peptides in top-down proteomics. J Proteomics 2021, 230, 103988. [DOI] [PubMed] [Google Scholar]
- [107].Delcourt V, Franck J, Leblanc E, Narducci F, et al. , Combined Mass Spectrometry Imaging and Top-down Microproteomics Reveals Evidence of a Hidden Proteome in Ovarian Cancer. EBioMedicine 2017, 21, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Cassidy L, Kaulich PT, Tholey A, Proteoforms expand the world of microproteins and short open reading frame-encoded peptides. iScience 2023, 26, 106069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Aebersold R, Agar JN, Amster IJ, Baker MS, et al. , How many human proteoforms are there? Nat Chem Biol 2018, 14, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A, A Human Short Open Reading Frame (sORF)-encoded Polypeptide That Stimulates DNA End Joining. Journal of Biological Chemistry 2014, 289, 10950–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Pauli A, Norris ML, Valen E, Chew G-L, et al. , Toddler: An Embryonic Signal That Promotes Cell Movement via Apelin Receptors. Science 2014, 343, 1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Samandi S, Roy AV, Delcourt V, Lucier J-F, et al. , Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. Elife 2017, 6, e27860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Na Z, Luo Y, Cui DS, Khitun A, et al. , Phosphorylation of a Human Microprotein Promotes Dissociation of Biomolecular Condensates. J Am Chem Soc 2021, 143, 12675–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Bogaert A, Fijalkowska D, Staes A, Van de Steene T, et al. , Limited Evidence for Protein Products of Noncoding Transcripts in the HEK293T Cellular Cytosol. Molecular & Cellular Proteomics 2022, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kesner JS, Chen Z, Shi P, Aparicio AO, et al. , Noncoding translation mitigation. Nature 2023, 617, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wright BW, Yi Z, Weissman JS, Chen J, The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol 2022, 32, 243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Schlesinger D, Elsässer SJ, Revisiting sORFs: overcoming challenges to identify and characterize functional microproteins. FEBS J 2022, 289, 53–74. [DOI] [PubMed] [Google Scholar]
- [118].Jackson R, Kroehling L, Khitun A, Bailis W, et al. , The translation of non-canonical open reading frames controls mucosal immunity. Nature 2018, 564, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Niu L, Lou F, Sun Y, Sun L, et al. , A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci Adv 2023, 6, eaaz2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Lee CQE, Kerouanton B, Chothani S, Zhang S, et al. , Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat Commun 2021, 12, 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zhang S, Reljic B, Liang C, Kerouanton B, et al. , Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat Commun 2020, 11, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Makarewich CA, Baskin KK, Munir AZ, Bezprozvannaya S, et al. , MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep 2018, 23, 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Stein CS, Jadiya P, Zhang X, McLendon JM, et al. , Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep 2018, 23, 3710–3720.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Lee C, Zeng J, Drew BG, Sallam T, et al. , The Mitochondrial-Derived Peptide MOTS-c Promotes Metabolic Homeostasis and Reduces Obesity and Insulin Resistance. Cell Metab 2015, 21, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Martinez TF, Lyons-Abbott S, Bookout AL, De Souza EV, et al. , Profiling mouse brown and white adipocytes to identify metabolically relevant small ORFs and functional microproteins. Cell Metab 2023, 35, 166–183.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Polycarpou-Schwarz M, Gross M, Mestdagh P, Schott J, et al. , The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 2018, 37, 4750–4768. [DOI] [PubMed] [Google Scholar]
- [127].Huang JZ, Chen M, Chen, Gao XC, et al. , A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell 2017, 68, 171–184 e6. [DOI] [PubMed] [Google Scholar]
- [128].Erady C, Boxall A, Puntambekar S, Suhas Jagannathan N, et al. , Pan-cancer analysis of transcripts encoding novel open-reading frames (nORFs) and their potential biological functions. NPJ Genom Med 2021, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Vakirlis N, Acar O, Hsu B, Castilho Coelho N, et al. , De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences. Nat Commun 2020, 11, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wacholder A, Parikh SB, Coelho NC, Acar O, et al. , A vast evolutionarily transient translatome contributes to phenotype and fitness. Cell Syst 2023, 14, 363–381.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Matsumoto A, Pasut A, Matsumoto M, Yamashita R, et al. , mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [DOI] [PubMed] [Google Scholar]
- [132].Bi P, Ramirez-Martinez A, Li H, Cannavino J, et al. , Control of muscle formation by the fusogenic micropeptide myomixer. Science 2017, 356, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Mise S, Matsumoto A, Shimada K, Hosaka T, et al. , Kastor and Polluks polypeptides encoded by a single gene locus cooperatively regulate VDAC and spermatogenesis. Nat Commun 2022, 13, 1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G, Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proceedings of the National Academy of Sciences 2012, 109, 16696–16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Du D, Wang Z, James NR, Voss JE, et al. , Structure of the AcrAB–TolC multidrug efflux pump. Nature 2014, 509, 512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Du D, Neuberger A, Orr MW, Newman CE, et al. , Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment. Structure 2020, 28, 625–634.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].E VC, Shantanu B, J AR, P BE, et al. , The Escherichia coli CydX Protein Is a Member of the CydAB Cytochrome bd Oxidase Complex and Is Required for Cytochrome bd Oxidase Activity. J Bacteriol 2013, 195, 3640–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Sun Y-H, de Jong M, den Hartigh A, Roux C, et al. , The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front Cell Infect Microbiol 2012, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Hoeser J, Hong S, Gehmann G, Gennis RB, Friedrich T, Subunit CydX of Escherichia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett 2014, 588, 1537–1541. [DOI] [PubMed] [Google Scholar]
- [140].Safarian S, Rajendran C, Müller H, Preu J, et al. , Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science 2016, 352, 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Safarian S, Hahn A, Mills DJ, Radloff M, et al. , Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science 2019, 366, 100–104. [DOI] [PubMed] [Google Scholar]
- [142].Makarewich CA, Olson EN, Mining for Micropeptides. Trends Cell Biol 2017, 27, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Singh DR, Dalton MP, Cho EE, Pribadi MP, et al. , Newly Discovered Micropeptide Regulators of SERCA Form Oligomers but Bind to the Pump as Monomers. J Mol Biol 2019, 431, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Tada M, Ohmori F, Yamada M, Abe H, Mechanism of the stimulation of Ca2+-dependent ATPase of cardiac sarcoplasmic reticulum by adenosine 3‘:5‘-monophosphate-dependent protein kinase. Role of the 22,000-dalton protein. Journal of Biological Chemistry 1979, 254, 319–326. [PubMed] [Google Scholar]
- [145].Odermatt A, Taschner PEM, Scherer SW, Beatty B, et al. , Characterization of the Gene Encoding Human Sarcolipin (SLN), a Proteolipid Associated with SERCA1: Absence of Structural Mutations in Five Patients with Brody Disease. Genomics 1997, 45, 541–553. [DOI] [PubMed] [Google Scholar]
- [146].Toyoshima C, Iwasawa S, Ogawa H, Hirata A, et al. , Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature 2013, 495, 260–264. [DOI] [PubMed] [Google Scholar]
- [147].Traaseth NJ, Ha KN, Verardi R, Shi L, et al. , Structural and Dynamic Basis of Phospholamban and Sarcolipin Inhibition of Ca2+-ATPase. Biochemistry 2008, 47, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Magny EG, Pueyo JI, Pearl FMG, Cespedes MA, et al. , Conserved Regulation of Cardiac Calcium Uptake by Peptides Encoded in Small Open Reading Frames. Science 2013, 341, 1116–1120. [DOI] [PubMed] [Google Scholar]
- [149].Anderson DM, Anderson KM, Chang C-L, Makarewich CA, et al. , A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Anderson DM, Makarewich CA, Anderson KM, Shelton JM, et al. , Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci Signal 2016, 9, ra119–ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Nelson BR, Makarewich CA, Anderson DM, Winders BR, et al. , A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Fisher ME, Bovo E, Aguayo-Ortiz R, Cho EE, et al. , Dwarf open reading frame (DWORF) is a direct activator of the sarcoplasmic reticulum calcium pump SERCA. Elife 2021, 10, e65545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Reddy UV, Weber DK, Wang S, Larsen EK, et al. , A kink in DWORF helical structure controls the activation of the sarcoplasmic reticulum Ca2+-ATPase. Structure 2022, 30, 360–370.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Hashimoto Y, Niikura T, Tajima H, Yasukawa T, et al. , A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Aβ. Proceedings of the National Academy of Sciences 2001, 98, 6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Hashimoto Y, Ito Y, Niikura T, Shao Z, et al. , Mechanisms of Neuroprotection by a Novel Rescue Factor Humanin from Swedish Mutant Amyloid Precursor Protein. Biochem Biophys Res Commun 2001, 283, 460–468. [DOI] [PubMed] [Google Scholar]
- [156].Lee C, Yen K, Cohen P, Humanin: a harbinger of mitochondrial-derived peptides? Trends in Endocrinology & Metabolism 2013, 24, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Guo B, Zhai D, Cabezas E, Welsh K, et al. , Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 2003, 423, 456–461. [DOI] [PubMed] [Google Scholar]
- [158].Ikonen M, Liu B, Hashimoto Y, Ma L, et al. , Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proceedings of the National Academy of Sciences 2003, 100, 13042–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ying G, Iribarren P, Zhou Y, Gong W, et al. , Humanin, a Newly Identified Neuroprotective Factor, Uses the G Protein-Coupled Formylpeptide Receptor-Like-1 as a Functional Receptor1. The Journal of Immunology 2004, 172, 7078–7085. [DOI] [PubMed] [Google Scholar]
- [160].Hashimoto Y, Kurita M, Aiso S, Nishimoto I, Matsuoka M, Humanin Inhibits Neuronal Cell Death by Interacting with a Cytokine Receptor Complex or Complexes Involving CNTF Receptor α/WSX-1/gp130. Mol Biol Cell 2009, 20, 2864–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Benaki D, Zikos C, Evangelou A, Livaniou E, et al. , Solution structure of humanin, a peptide against Alzheimer’s disease-related neurotoxicity. Biochem Biophys Res Commun 2005, 329, 152–160. [DOI] [PubMed] [Google Scholar]
- [162].Dubois M-L, Meller A, Samandi S, Brunelle M, et al. , UBB pseudogene 4 encodes functional ubiquitin variants. Nat Commun 2020, 11, 1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Boix O, Martinez M, Vidal S, Giménez-Alejandre M, et al. , pTINCR microprotein promotes epithelial differentiation and suppresses tumor growth through CDC42 SUMOylation and activation. Nat Commun 2022, 13, 6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Morgado-Palacin L, Brown JA, Martinez TF, Garcia-Pedrero JM, et al. , The TINCR ubiquitin-like microprotein is a tumor suppressor in squamous cell carcinoma. Nat Commun 2023, 14, 1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Nita A, Matsumoto A, Tang R, Shiraishi C, et al. , A ubiquitin-like protein encoded by the “noncoding” RNA TINCR promotes keratinocyte proliferation and wound healing. PLoS Genet 2021, 17, e1009686-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Eckhart L, Lachner J, Tschachler E, Rice RH, TINCR is not a non-coding RNA but encodes a protein component of cornified epidermal keratinocytes. Exp Dermatol 2020, 29, 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, et al. , Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat Methods 2020, 17, 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Zhang Y, I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE, The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015, 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Jumper J, Evans R, Pritzel A, Green T, et al. , Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Varadi M, Anyango S, Deshpande M, Nair S, et al. , AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 2022, 50, D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Consortium, T.U., UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023, 51, D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Roy A, Kucukural A, Zhang Y, I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010, 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Gligorijević V, Renfrew PD, Kosciolek T, Leman JK, et al. , Structure-based protein function prediction using graph convolutional networks. Nat Commun 2021, 12, 3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Bernhofer M, Dallago C, Karl T, Satagopam V, et al. , PredictProtein - Predicting Protein Structure and Function for 29 Years. Nucleic Acids Res 2021, 49, W535–W540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Hall TMT, Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 2005, 15, 367–373. [DOI] [PubMed] [Google Scholar]
- [177].Zhang Y, Burkhardt DH, Rouskin S, Li G-W, et al. , A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol Cell 2018, 70, 274–286.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Rennella E, Sára T, Juen M, Wunderlich C, et al. , RNA binding and chaperone activity of the E. coli cold-shock protein CspA. Nucleic Acids Res 2017, 45, 4255–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Jiang W, Hou Y, Inouye M, CspA, the Major Cold-shock Protein of Escherichia coli, Is an RNA Chaperone. Journal of Biological Chemistry 1997, 272, 196–202. [DOI] [PubMed] [Google Scholar]
- [180].Bhati KK, Kruusvee V, Straub D, Chandran AKN, et al. , Global Analysis of Cereal microProteins Suggests Diverse Roles in Crop Development and Environmental Adaptation. G3 Genes∣Genomes∣Genetics 2020, 10, 3709–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, et al. , Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol Cell Proteomics 2007, 6, 1000–1006. [DOI] [PubMed] [Google Scholar]
- [182].Huang N, Li F, Zhang M, Zhou H, et al. , An Upstream Open Reading Frame in Phosphatase and Tensin Homolog Encodes a Circuit Breaker of Lactate Metabolism. Cell Metab 2021, 33, 128–144.e9. [DOI] [PubMed] [Google Scholar]
- [183].Jayaram DR, Frost S, Argov C, Liju VB, et al. , Unraveling the hidden role of a uORF-encoded peptide as a kinase inhibitor of PKCs. Proceedings of the National Academy of Sciences 2021, 118, e2018899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Coulombe B, Cloutier P, Pinard M, Forget D, et al. , The PAQosome, a novel molecular chaperoning machine for assembly of human protein complexes and networks. The FASEB Journal 2020, 34, 1. [Google Scholar]
- [185].Liang J, Xia L, Oyang L, Lin J, et al. , The functions and mechanisms of prefoldin complex and prefoldin-subunits. Cell Biosci 2020, 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Katuwawala A, Kurgan L, Comparative Assessment of Intrinsic Disorder Predictions with a Focus on Protein and Nucleic Acid-Binding Proteins. Biomolecules 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Mackowiak SD, Zauber H, Bielow C, Thiel D, et al. , Extensive identification and analysis of conserved small ORFs in animals. Genome Biol 2015, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Sandmann C-L, Schulz JF, Ruiz-Orera J, Kirchner M, et al. , Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames. Mol Cell 2023, 83, 994–1011.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Vakirlis N, Vance Z, Duggan KM, McLysaght A, De novo birth of functional microproteins in the human lineage. Cell Rep 2022, 41, 111808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].van der Lee R, Buljan M, Lang B, Weatheritt RJ, et al. , Classification of Intrinsically Disordered Regions and Proteins. Chem Rev 2014, 114, 6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Agarwal S, Harada J, Schreifels J, Lech P, et al. , Isolation, characterization, and genetic complementation of a cellular mutant resistant to retroviral infection. Proceedings of the National Academy of Sciences 2006, 103, 15933–15938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A, A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J Biol Chem 2014, 289, 10950–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Grundy GJ, Rulten SL, Arribas-Bosacoma R, Davidson K, et al. , The Ku-binding motif is a conserved module for recruitment and stimulation of non-homologous end-joining proteins. Nat Commun 2016, 7, 11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Hung PJ, Johnson B, Chen B-R, Byrum AK, et al. , MRI Is a DNA Damage Response Adaptor during Classical Non-homologous End Joining. Mol Cell 2018, 71, 332–342.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Arnoult N, Correia A, Ma J, Merlo A, et al. , Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 2017, 549, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Luo Y, Schofield JA, Simon MD, Slavoff SA, Global Profiling of Cellular Substrates of Human Dcp2. Biochemistry 2020, 59, 4176–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Luo Y, Schofield JA, Na Z, Hann T, et al. , Discovery of cellular substrates of human RNA-decapping enzyme DCP2 using a stapled bicyclic peptide inhibitor. Cell Chem Biol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Na Z, Luo Y, Schofield JA, Smelyansky S, et al. , The NBDY Microprotein Regulates Cellular RNA Decapping. Biochemistry 2020, 59, 4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Li Y, Dai J, Song M, Fitzgerald-Bocarsly P, Kiledjian M, Dcp2 Decapping Protein Modulates mRNA Stability of the Critical Interferon Regulatory Factor (IRF) IRF-7. Mol Cell Biol 2012, 32, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Jonas S, Izaurralde E, The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev 2013, 27, 2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.