Abstract
Previously characterized Ras subfamily proteins have been found to be predominantly associated with the plasma membrane where they function in signal transduction pathways to convey extracellular signals to intracellular targets. Here, we provide evidence that the Dictyostelium Ras subfamily protein RasB has a novel subcellular localization and function. The protein is predominantly localized in the nucleus during most of the cell cycle. Furthermore, during mitosis and cytokinesis RasB assumes a diffuse cellular localization despite the fact that the nuclear membrane stays intact. The linkage between the position of RasB in the cell and division suggests that it may have a role in nuclear division. Consistent with this idea, rasB– cells exhibit severe growth defects and cells overexpressing an activated version of RasB are multinucleate.
INTRODUCTION
Ras subfamily proteins function as simple molecular switches in the intracellular transmission of a variety of extracellular signals (Reuther and Der, 2000; Takai et al., 2001). The two states of the switch are generated by the binding of guanine nucleoside triphosphate (GTP) or guanine nucleoside diphosphate (GDP), producing active and inactive states, respectively. The reversible conversion between states involves the exchange of GDP for GTP and the subsequent hydrolysis of GTP to GDP (Bourne et al., 1990; Hall, 1990). H-Ras and K-Ras have recently been shown to occupy different locations on the plasma membrane resulting from different membrane localization signals in the C-terminal hypervariable region of the proteins (reviewed in Magee and Marshall, 1999). In addition, K-Ras, but not H-Ras, has been found to be an essential gene in mice (Johnson et al., 1997), suggesting that individual Ras subfamily proteins may have some distinct functions (reviewed in Shields et al., 2000).
Dictyostelium contains six partially characterized ras subfamily genes (rasD, rasG, rasS, rasC, rasB and rap), the encoded proteins of which show at least 52% amino acid sequence identity to H-Ras (reviewed in Chubb and Insall, 2000). Gene deletion studies have indicated that each of the Dictyostelium Ras subfamily proteins thus far characterized regulate distinct downstream pathways. For example, disruption of the rasG gene resulted in multiple effects including a cytokinesis defect, a growth defect, reduced cell polarity and cell motility, and an altered F-actin cytoskeleton (Tuxworth et al., 1997). The cytokinesis defect of rasG– cells is likely to result from incomplete cytokinesis, as cells are unable to sever cleavage furrows to complete cell division. In contrast, RasC is involved in the regulation of the early aggregation stage of Dictyostelium development (Lim et al., 2001). rasC– gene deletion strains are unable to initiate aggregation upon starvation and cAMP-induced adenylyl cyclase and Akt/PKB activation are greatly reduced, suggesting that RasC regulates G protein-coupled cAMP receptor activation. rasS gene deletion strains exhibit severe growth defects resulting from an inability of cells to conduct pinocytosis and phagocytosis, while at the same time they exhibit increased rates of random cell motility, suggesting that RasS regulates the distribution of F-actin polymerization between the two opposing processes of endocytosis and movement (Chubb et al., 2000). Finally, RasD is required for the phototaxis and thermotaxis of slugs during late development (Wilkins et al., 2000). To begin to understand the function of RasB, we examined the subcellular localization of RasB, disrupted the rasB gene and examined a strain expressing an activated version of the gene (Delehanty, 1998). Our results are consistent with the idea that RasB performs a function different from those reported for other Dictyostelium Ras subfamily proteins and for Ras proteins in other organisms.
RESULTS AND DISCUSSION
RasB subcellular localization
To examine the subcellular localization of RasB, we generated specific antibodies using a peptide corresponding to the hypervariable C-terminus of RasB (amino acids 173–194) as an antigen, and these antibodies were affinity purified using a RasB peptide-affinity column (Khosla et al., 2000). These purified antibodies did not cross-react with other Dictyostelium Ras subfamily proteins (Khosla et al., 2000) and only a single band of the predicted size of the RasB protein was detected on western blots of cell lysates, indicating the antibodies are RasB specific (see Figure 2). Following adsorption against the RasB peptide antigen, antibodies no longer interacted with any proteins in western blots (see supplementary figure 1 available as Supplementary data at EMBO reports Online). Indirect immunofluorescence on fixed vegetative Dictyostelium cells showed clearly that RasB co-localized with 4′-6-diamidine-2-phenylindole (DAPI) stain in the nucleus (Figure 1A). This is the first Ras subfamily protein of any species to be described that exhibits this novel localization.Since RasB contains a CLIL C-terminus motif and a basic hypervariable domain (Daniel et al., 1993), which are membrane localization signals in the mammalian Ras subfamily proteins (reviewed in Prior and Hancock, 2001), we considered the possibility that the protein may be localized in the nuclear membrane. Double fluorescence experiments were conducted using the RasB antibody and an antibody specific for a nuclear pore protein, mAb17 (Fukuzawa et al., 1997), to delineate the nuclear membrane. RasB did appear to localize with the nuclear pore protein, but was also found within the nucleus (Figure 1B). To estimate the proportion of RasB associated with the nuclear membrane as opposed to the nucleosol, cells were lysed mechanically and fractionated by differential centrifugation. Microscopic examination and DAPI staining showed the lysates to be 99% free of intact cells and nuclei. RasB was predominantly, if not exclusively, located in the 100 000 g pellet fraction containing membranes and cytoskeletal matrix components (Figure 2), indicating that the RasB observed in the nucleosolic space (Figure 1) is predominantly associated with the nuclear matrix.
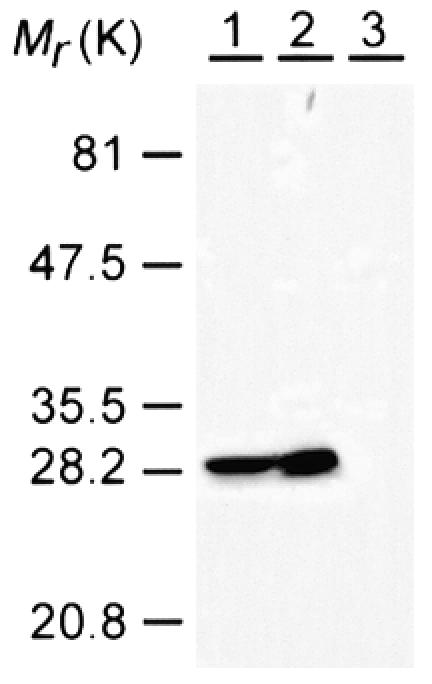
Fig. 2. RasB is associated with the nuclear membrane and nuclear matrix. Thirty micrograms of total cellular protein (lane 1, control) and volumes of the 100 000 g pellet (lane 2) and 100 000 g supernant fractions (lane 3) that were equivalent to the 30 µg of total cellular protein were separated by SDS–PAGE, transferred to Hybond-P polyvinylidene difluoride membrane (Amersham Pharmacia Biotech) and probed with the RasB antibody. Mr, relative molecular mass.
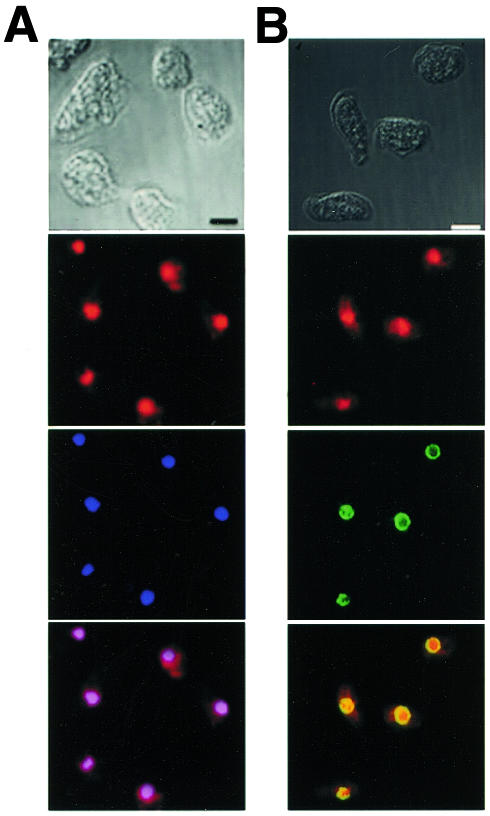
Fig. 1. RasB is localized predominantly in the nucleus of vegetative Dictyostelium cells. (A) Indirect immunofluorescence using the RasB antibody (red) and DAPI stain (blue). The merged image verifies strong co-localization (violet). (B) Indirect immunofluorescence using RasB antibody (red) and an antibody against the nuclear pore complex protein (mAb17) (green). The merged image reveals partial co-localization (yellow). Nomarski images are shown. Scale bar, 5 µm.
Examination of a large population of vegetative cells immunostained with the anti-RasB antibody revealed that a small proportion (∼1%) exhibited diffuse fluorescence throughout the entire cell and that nuclei within these cells contained condensed chromosomes, as determined by the smaller yet more intensely stained areas of DAPI fluorescence. This suggested the possibility that RasB is not associated with the nucleus during mitosis. To investigate this idea, we examined RasB localization during the various mitotic phases and cytokinesis in cells that had been cell-cycle synchronized by a cold-shock method (Maeda, 1986). Triple staining experiments with anti-α-Tubulin antibodies (Piperno and Fuller, 1985), anti-RasB antibodies and DAPI stain were then used to determine the localization of RasB, the mitotic spindle and the chromosomes (Figure 3). Without exception, cells in interphase exhibited the RasB nuclear localization pattern described above (Figure 3A). However, beginning at metaphase and extending through to late telophase, RasB was no longer associated with the nucleus (Figure 3C–F). These results show that RasB left the nucleus at or just prior to the onset of mitosis (Figure 3B) and re-entered the nucleus at or slightly after the end of mitosis (Figure 3G).
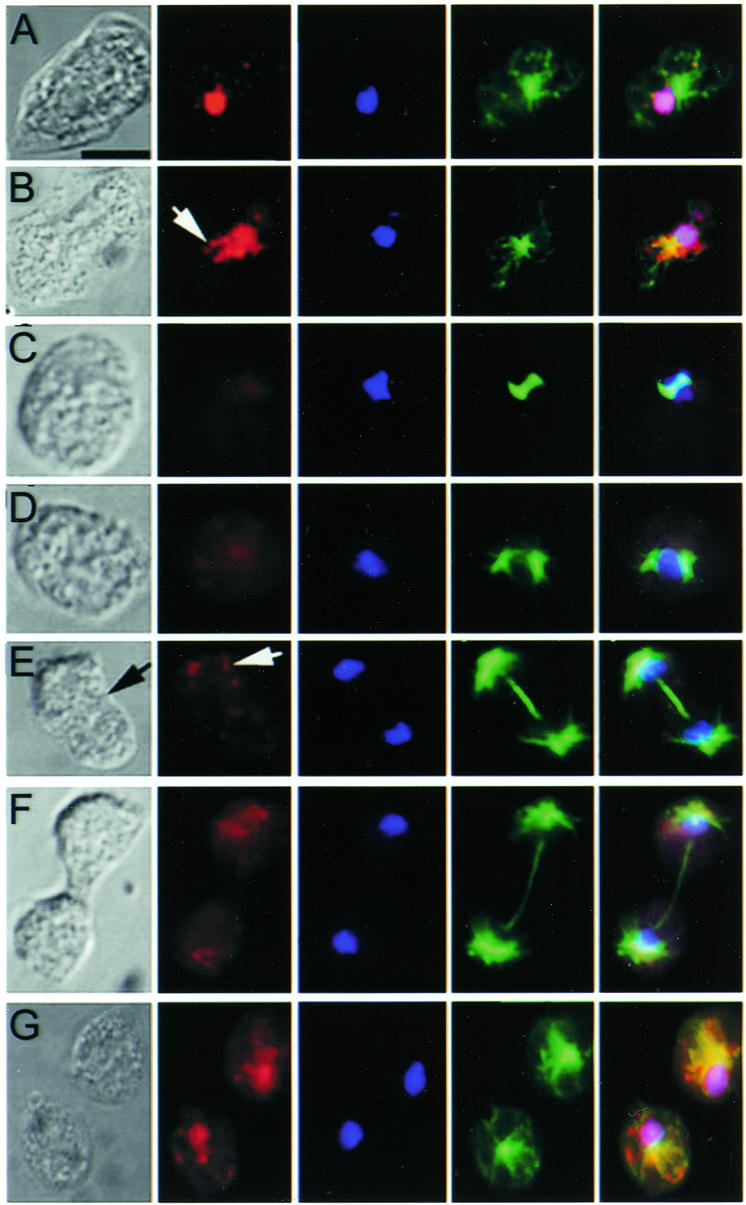
Fig. 3. RasB cellular localization is regulated during the cell cycle. Vegetative cells were synchronized, fixed and immunostained with RasB antibody (red), DAPI stain (blue) or α-tubulin antibody (green). Representative cells in the major mitotic stages are shown in temporal order: interphase (A), prophase (B), metaphase (C), anaphase (D), early telophase (E), late telophase (F) and separated daughter cells (G). Strong co-localization of RasB and DAPI is shown by the violet color in the merged images. Perinuclear punctuate patches of RasB are indicated by white arrows. The black arrow in the Nomarski image (E) indicates the beginning of cleavage furrow formation. Scale bar, 5 µm.
The Dictyostelium nuclear membrane is maintained during mitosis (Moens, 1976), but it becomes highly fenestrated. Thus RasB could passively diffuse out of the nucleus, but relocalization to the nucleus requires a different mechanism. Punctuate patches of RasB were observed in a perinuclear location (Figure 3, white arrows) just prior to or at the beginning of prophase (Figure 3B) and at telophase (Figure 3E). This distribution pattern may reflect the active shuttling of RasB in and out of the nucleus since punctuate protein patterns have been attributed previously to trafficking of Ras proteins from one cellular location to another (Choy et al., 1999). In order to directly monitor the passage of RasB in and out of the nucleus in living cells, we attempted to express a green fluorescent protein tagged RasB construct. However, this construct proved to be toxic to cells. A similar construct with the –CLIL putative membrane attachment site deleted was not expressed. Western blots of cell extracts collected during division of the cell cycle-synchronized populations of Ax2 cells did not reveal changes in the levels of RasB protein during mitosis (see supplementary figure 2), indicating that the disappearance of RasB from the nucleus was not accompanied by protein degradation.
RasB protein-deficient cell strains
To determine whether RasB plays a functional role in the regulation of the cell cycle, we generated Dictyostelium strains in which the rasB gene was disrupted. This was accomplished by inserting the Blasticidin S resistance gene into the rasB promoter region just upstream of the start codon by homologous recombination (Manstein et al., 1989). In these cells, RasB was not detectable by western blot analysis (Figure 4A) or immunofluorescence (Figure 4B). DAPI staining revealed that cells were mononucleate and contained morphologically normal nuclei. However, these cells exhibited a severe growth defect (Figure 4C), a result consistent with the idea that RasB plays an important role in progression through the cell cycle. Blasticidin-resistant rasB– transformant strains proved to be unstable and revertants, which began to produce RasB and exhibited increased growth rates, arose within populations of the clonal isolates. Attempts to generate more stable rasB null strains by inserting the blastocidin resistance gene within the coding region of the rasB gene or by deleting the promoter region and/or part of the coding region were unsuccessful. This inability to produce stable rasB null strains raises the possibility that the rasB gene is essential and that cells with the rasB promoter disruption expressed a very low level of RasB, undetectable by western blot analysis or immunofluorescence, but sufficient to support slow growth.
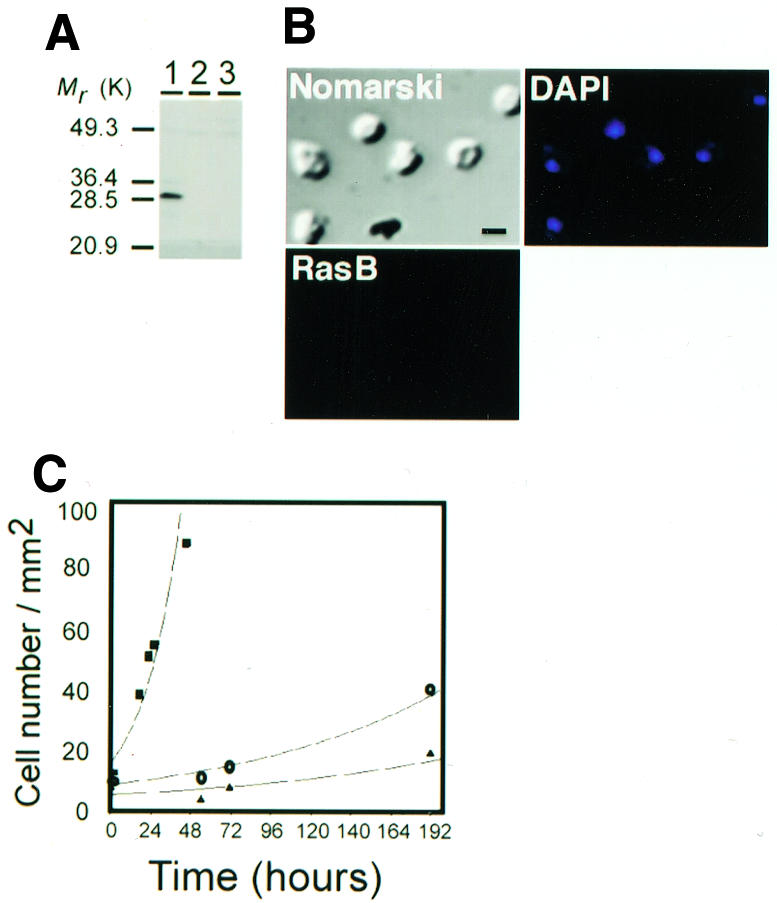
Fig. 4. Characteristics of RasB-deficient cells. (A) Western blot of total cell lysates from an Ax2 parental strain (lane 1) and two rasB gene disrupted strains (lanes 2 and 3) immunostained with the RasB antibody. Mr, relative molecular mass. (B) Immunofluorescence using the RasB antibody and DAPI stain (blue) on cells of a rasB gene disrupted strain. Scale bar, 5 µm. (C) Growth curve of Ax2 wild type cells (squares) and two rasB– gene disrupted strains (circles and triangles). Cell numbers were counted at the indicated times by microscopic examination of cells grown on tissue culture plates at 22°C.
Cells containing the activated rasB(G12T) gene
Next we examined Dictyostelium strains that had been transformed with a rasB gene containing the G12T mutation (Delehanty, 1998). Mammalian Ras proteins containing this substitution are constrained in the GTP bound form. The most dramatic effect of RasB(G12T) overexpression (Figure 5A) was that a significant proportion (30%) of the cell population became multinucleate (Figure 5C), suggesting that the coordination of karyokinesis and cytokinesis had been disrupted. The intensity of DAPI staining indicated that the DNA within the nuclei of multinucleate cells appeared normal, suggesting no apparent defect in chromosomal replication. Since rasG null cells also exhibit a defect in the regulation of cytokinesis (Tuxworth et al., 1997), it is possible that the multinucleate phenotype associated with overexpression of RasB(G12T) results from the interference with a RasG-regulated pathway, rather than a RasB regulated pathway. However, since there are considerable differences in the appearance of the two types of cells (Figure 5) (Tuxworth et al., 1997), we consider this possibility to be unlikely.
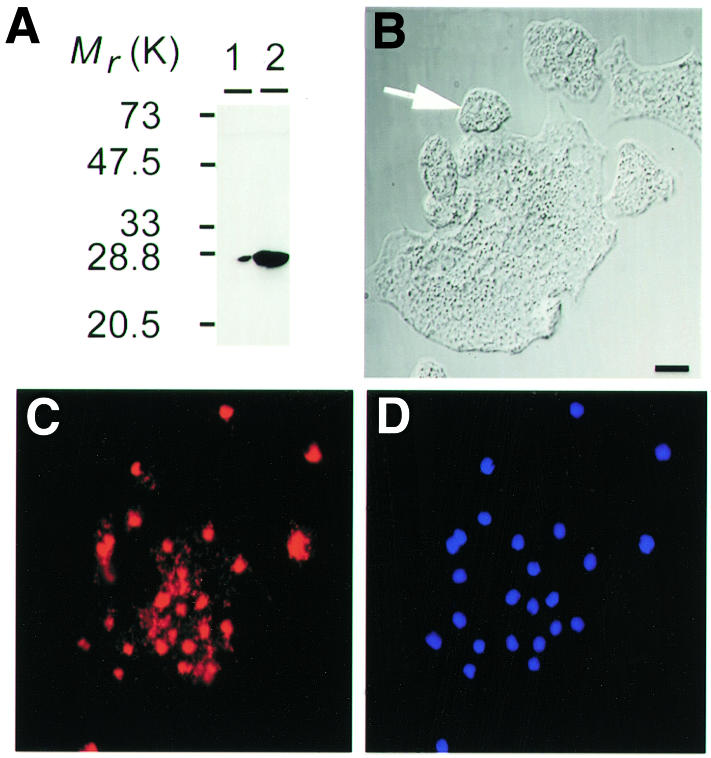
Fig. 5. Appearance of cells expressing the rasB(G12T) gene. (A) Western blots, immunostained with the RasB antibody, of cell lysates of cells grown in the presence or absence of folate. Indirect immunofluorescence images immunostained with RasB antibody (C, red) and DAPI stain (D, blue) of cells overexpressing RasB(G12T). The white arrow indicates wild-type sized cell. Nomarski images are shown in (B). Scale bar, 5 µm.
Recent evidence has implicated the Ras-related GTPase, Ran, in regulating microtubule spindle assembly during mitosis (reviewed in Dasso, 2001). Active Ran (GTP) is localized in the nucleus and inactive Ran (GDP) is localized in the cytoplasm during interphase. A Ran (GTP) gradient regulating the aster promoting activity components that are required for spindle assembly at chromosomes has been implicated in mitosis progression (Dasso, 2001). Although both Ran and RasB exhibit cell cycle-dependent nuclear localization, it is unlikely that RasB fulfills the functional role of a Ran protein in Dictyostelium during mitosis/cytokinesis, because Dictyostelium contains a Ran protein that exhibits high amino acid sequence identity to mammalian Ran (Bush and Cardelli, 1993). However, it is entirely possible that RasB is an upstream component in a Ran-regulated pathway, or vice versa, involved in mitosis/cytokinesis progression. Such a pathway would be a counterpart to ones in which Ras subfamily proteins at the plasma membrane activate downstream Rho GTPase-regulated pathways involved in cytoskeletal organization (Van Aelst and D’Souza-Schorey, 1997; Zohn et al., 1998).
In summary, our findings indicate a novel location for a Ras subfamily protein. The observations that RasB is localized in the nucleus except during mitosis/cytokinesis, that growth is dramatically reduced in the absence of detectable RasB and that cytokinesis is abnormal in strains expressing constitutively activated RasB are consistent with a direct role for the protein in cell-cycle regulation. Since the localization of some of the mammalian Ras subfamily proteins has not yet been determined, our findings raise the possibility that one of these proteins might exhibit a similar cell cycle linking nuclear localization and function.
METHODS
Dictyostelium cell growth
Dictyostelium discoideum Ax2 cells were grown axenically at 22°C in HL-5 medium (Watts and Ashworth, 1970), either on tissue culture plates (Gibco BRL) or in rotatory agitated suspension (150 r.p.m.). In addition, cell strains were clonally isolated by growth on rich nutrient agar plates in association with Klebsiella oxytoca at 22°C.
Immunofluorescence and DAPI staining
For tubulin, RasB and DAPI localization studies, cells attached to glass coverslips were fixed with 2.5% formaldehyde in 15 mM PIPES and 1 mM EGTA for 20 min at room temperature and then extracted for 5 min with methanol containing 1% formaldehyde at –20°C. The fixed cells were incubated with rabbit RasB antibody (1:50 dilution) and/or the mouse α-tubulin antibody (Piperno and Fuller, 1985) (1:50 dilution) for 40 min at room temperature. For RasB and nuclear pore protein co-localization studies, cells were fixed with methanol at room temperature for 10 min and incubated with mouse mAB17 (Fukuzawa et al., 1997) (1:300 dilution) and rabbit RasB antibody (1:50 dilution) for 40 min at room temperature. Cells were then incubated for 30 min at room temperature with the following secondary antibodies (Sigma): for RasB in Figure 1A, anti-rabbit IgG FITC conjugate (1:100 dilution) and for RasB in Figures 1B, 3 and 5, anti-rabbit IgG TRITC conjugate (1:33 dilution); for tubulin, anti-mouse IgG FITC conjugate (1:100 dilution); and for nuclear pore protein, anti-mouse IgM FITC conjugate (1:100 dilution). Where indicated, cells were incubated with DAPI stain (Sigma) (1 µM) for 10 min at room temperature. Between all incubation steps, cells were washed three times for 5 min with phosphate-buffered saline. Coverslips were mounted on slides using the ProLongR Antifade Kit (Molecular Probes) following the manufacturer’s instructions. Images visualized with Zeiss Axioplan 2 and Axiovert S100TV microscopes were captured with a SPOT (Diagnostic Instruments) camera using Northern Eclipse Image Analysis Software.
Cell-cycle synchronization
Cell-cycle synchronization was accomplished by the previously described cold-shock method (Maeda, 1986) with some modifications. Vegetative Ax2 cells were grown in the rich nutrient media, HL-5, at 22°C to a density of 5 × 106 cells/ml. Cells were then diluted into fresh media to a density of 1 × 106 cells/ml and incubated at 10°C for 20 h. Aliquots of cells were then attached to glass coverslips at a density of 4 × 104 cells/cm2 at 22°C to allow growth. Samples were fixed and immunostained every 15–30 min. Approximately 55% of the cell population consistently passed through mitosis between 2 and 3 h following the end of the cold-shock treatment.
Subcellular fractionation
Axenically grown cells were harvested by centrifugation at 1100 g for 5 min and washed twice with KK2 buffer (20 mM potassium phosphate pH 6.0). Cell pellets were resuspended in 1 ml lysis buffer (5 mM Tris–Cl pH 7.4, 5 µg/ml leupeptin, 1 µg/ml aprotinin, 2 µg/ml pepstatin, 10 µg/ml phenylmethylsulfonyl fluoride and 2 mM sodium bisulfite) and mechanically lysed using a Dounce homogenizer (50 strokes) at 4°C. Complete cellular lysis and nuclear membrane breakage were confirmed microscopically by the lack of intact cells and nuclei. Fractionation of membrane and cytoskeletal components from freely soluble cytoplasmic and nucleosolic proteins was accomplished by centrifugation at 100 000 g for 1 h at 4°C. The resultant membrane and cytoskeletal pellet was resuspended in a volume of lysis buffer equivalent to that of the supernatant fraction. Thirty micrograms of protein samples were boiled at 100°C for 5 min in 2% SDS, 5% 2-mercaptoethanol, 10 mM Tris–HCl pH 6.8, 10% glycerol (vol/vol) and 0.01% Bromophenol Blue and separated by discontinuous SDS–PAGE.
Supplementary data
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Michael Koonce and Masashi Fukuzawa for the kind gifts of the α-tubulin and nuclear-pore-protein (mAB17) antibodies, respectively, and Megan Delehanty for generating the rasB(G12T) cell strain. We thank Drs G.R. Ehrhardt, M. Gold and J.W. Schrader for comments on the manuscript. This work was supported by a CIHR research grant (to G.W.) and a Canadian MRC studentship (to B.W.S.).
REFERENCES
- Bourne H.R., Sanders, D.A. and McCormick, F. (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature, 348, 125–132. [DOI] [PubMed] [Google Scholar]
- Bush J. and Cardelli, J. (1993) Molecular cloning and DNA sequence of a Dictyostelium cDNA encoding a Ran/TC4 related GTP binding protein belonging to the ras superfamily. Nucleic Acids Res., 21, 1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E., Chiu, V.K., Silletti, J., Feoktistov, M., Morimoto, T., Michaelson, D., Ivanov, I.E. and Philips, M.R. (1999) Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell, 98, 69–80. [DOI] [PubMed] [Google Scholar]
- Chubb J.R. and Insall, R.H. (2000) Dictyostelium: an ideal organism for genetic dissection of Ras signalling networks. Biochim. Biophys. Acta, 1525, 262–271. [DOI] [PubMed] [Google Scholar]
- Chubb J.R., Wilkins, A., Thomas, G.M. and Insall, R.H. (2000) The Dictyostelium RasS protein is required for macropinocytosis, phagocytosis and the control of cell movement. J. Cell Sci., 113, 709–719. [DOI] [PubMed] [Google Scholar]
- Daniel J., Spiegelman, G.B. and Weeks, G. (1993) Characterization of a third ras gene, rasB, that is expressed throughout the growth and development of Dictyostelium discoideum. Oncogene, 8, 1041–1047. [PubMed] [Google Scholar]
- Dasso M. (2001) Running on Ran: nuclear transport and the mitotic spindle. Cell, 104, 321–324. [DOI] [PubMed] [Google Scholar]
- Delehanty M.C. (1998) The effect of overexpression of activated RasB, RasC and RasS on Dictyostelium discoideum. MSc dissertation, University of British Columbia, Vancouver, BC, Canada.
- Fukuzawa M., Hopper, N. and Williams, J. (1997) cudA: a Dictyostelium gene with pleiotropic effects on cellular differentiation and slug behaviour. Development, 124, 2719–2728. [DOI] [PubMed] [Google Scholar]
- Hall A. (1990) The cellular functions of small GTP-binding proteins. Science, 249, 635–640. [DOI] [PubMed] [Google Scholar]
- Johnson L. et al. (1997) K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Gene Dev., 11, 2468–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla M., Spiegelman, G.B., Insall, R. and Weeks, G. (2000) Functional overlap of the Dictyostelium RasG, RasD and RasB proteins. J. Cell Sci., 113, 1427–1434. [DOI] [PubMed] [Google Scholar]
- Lim C.J., Spiegelman, G.B. and Weeks, G. (2001) Dictyostelium RasC is required for activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J., 20, 4490–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M. (1986) Effects of low temperature on differentiation of Dictyostelium cells in the vegetative and preaggregation stages. Dev. Growth Differ., 28, 259–266. [DOI] [PubMed] [Google Scholar]
- Magee T. and Marshall, C. (1999) New insights into the interaction of Ras with the plasma membrane. Cell, 98, 9–12. [DOI] [PubMed] [Google Scholar]
- Manstein D.J., Titus, M.A., De Lozanne, A. and Spudich, J.A. (1989) Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J., 8, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P.B. (1976) Spindle and kinetochore of Dictyostelium discoideum. J. Cell Biol., 68, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G. and Fuller, M. (1985) Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol., 101, 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I.A. and Hancock, J.F. (2001) Compartmentalization of Ras proteins. J. Cell Sci., 114, 1603–1608. [DOI] [PubMed] [Google Scholar]
- Reuther G. and Der, C. (2000) The Ras branch of small GTPases: Ras family members don’t fall far from the tree. Curr. Opin. Cell Biol., 12, 157–165. [DOI] [PubMed] [Google Scholar]
- Shields J., Pruitt, K., McFall, A., Shaub, A. and Der, C. (2000) Understanding Ras: ‘it ain’t over ‘til it’s over’. Trends Cell Biol., 10, 147–154. [DOI] [PubMed] [Google Scholar]
- Takai Y., Sasaki, T. and Matozaki, T. (2001) Small GTP-binding proteins. Physiol. Rev., 81, 153–208. [DOI] [PubMed] [Google Scholar]
- Tuxworth R.I., Cheetham, J.L., Machesky, L.M., Spiegelman, G.B., Weeks, G. and Insall, R.H. (1997) Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol., 138, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L. and D’Souza-Schorey, C. (1997) Rho GTPases and signaling networks. Gene Dev., 11, 2295–2322. [DOI] [PubMed] [Google Scholar]
- Watts D.J. and Ashworth, J.M. (1970) Growth of myxamoebae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J., 119, 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins A., Khosla, M., Fraser, D.J., Spiegelman, G.B., Fisher, P.R., Weeks, G. and Insall, R.H. (2000) Dictyostelium RasD is required for normal phototaxis, but not differentiation. Gene Dev., 14, 1407–1413. [PMC free article] [PubMed] [Google Scholar]
- Zohn I.M., Campbell, S.L., Khosravi-Far, R., Rossman, K.L. and Der, C.J. (1998) Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene, 17, 1415–1438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


