Abstract
In mammalian cells, the expression level of the cyclin B1 gene plays a critical role in the progression through mitosis. Here we demonstrate that the transcriptional activity of the human cyclin B1 promoter, as well as the rate of gene transcription, is high during mitosis. Indeed, the cyclin B1 promoter maintains an open chromatin configuration at the mitotic stage. Consistent with this, we show that the cyclin B1 promoter is occupied and bound to NF-Y during mitosis in vivo. Our results provide the first example of RNA polymerase II-dependent transcription during mitosis in mammalian cells.
INTRODUCTION
Repression of transcription during mitosis has been known in mammalian cells for more than 30 years. It has been demonstrated that specific RNA polymerase II (pol II) factors, and several transcription factors, are displaced from the chromosomes during mitosis (Gottesfeld and Forbes, 1997). However, mitotic dispersal was not observed for some transcription factors such as the serum response factor p67SRF, and 10–20% of the TFIID complex (Gauthier-Rouviere et al., 1991; Segil et al., 1996).
In mammalian cells, the G2/M transition is controlled by the mitotic cyclins A and B1 (Jackman and Pines, 1997). The expression of cyclin B1 is cell-cycle regulated, peaking at the G2/M transition. It has been described that the transcriptional activity of the human cyclin B1 promoter is induced during the G2/M transition and repressed in G1 (Cogswell et al., 1995; Hwang et al., 1995; Piaggio et al., 1995; Farina et al., 1996; Katula et al., 1997). However, it has not been demonstrated yet whether the cyclin B1 promoter retains transcriptional activity during mitosis. Here we provide evidence for cyclin B1 transcription at the mitotic stage in mammalian cells. Indeed, we show that the cyclin B1 promoter is accessible to restriction endonucleases and interacts with transcription factors during mitosis in vivo. In addition, we demonstrate by formaldehyde cross-linking and immunoprecipitation assays that NF-Y is bound to the cyclin B1 promoter during mitosis. Our results show that the RNA pol II-transcribed gene cyclin B1 is actively transcribed during mitosis where the promoter retains open conformation and factor occupancy.
RESULTS
Transcriptional activity of the exogenous cyclin B1 promoter is high in the G2/M phases of the cell cycle
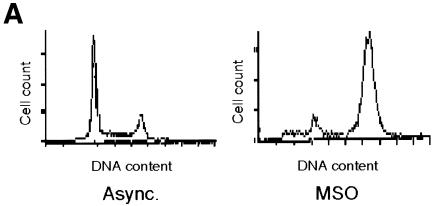
To investigate the transcriptional activity of the cyclin B1 promoter during mitosis, we stably transfected HeLa cells with a chloroamphenicol acetyltransferase (CAT) reporter gene driven by a human cyclin B1 promoter fragment (–150 to +182 bp) (Piaggio et al., 1995). We synchronized these cells by a double thymidine block. After release from the block, the cells were collected by mitotic shake-off (MSO). Flow cytometric analysis (Figure 1A) and mitotic index (see Supplementary data available at EMBO reports Online) of synchronized cells showed that in the MSO population, 92% of cells were mitotic. The levels of CAT mRNA in cycling and mitotic cells were analyzed by northern blot. The results showed that the activity of the exogenous cyclin B1 promoter fragment was high during mitosis of HeLa cells (9.2-fold higher than asynchronous cells), in agreement with the expression of the endogenous gene (Figure 1B). Since the half-life of CAT mRNA is longer than the length of mitosis in HeLa cells, it was impossible to determine whether the CAT mRNA levels detected in our MSO population were transcribed in G2 or in mitosis.
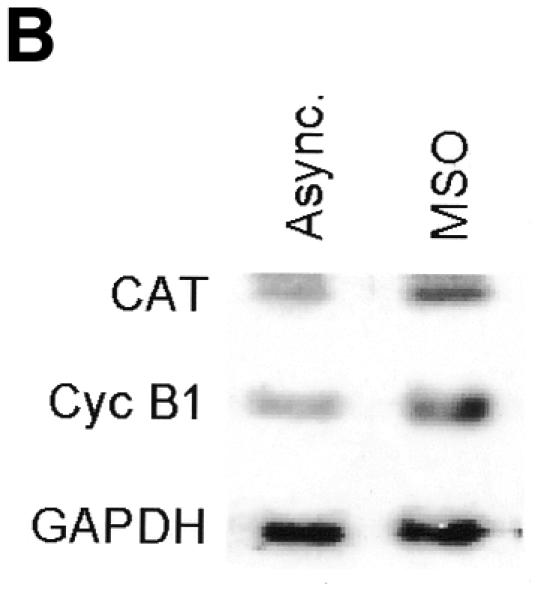
Fig. 1. Transcriptional activity of the exogenous cyclin B1 promoter is high in the G2/M phases of the cell cycle. (A) DNA distribution analysis of propidium iodide-stained asynchronous (Async.) and MSO HeLa cells. (B) Northern blot analysis of CAT mRNA levels in asynchronous (Async.) and MSO cells. HeLa cells stably transfected with the CAT reporter gene fused to the 332 bp cyclin B1 promoter were synchronized by a double thymidine block. The filter was hybridized with CAT, murine cyclin B1 and human GAPDH cDNAs.
The cyclin B1 promoter is accessible to restriction endonucleases in vivo
To analyze cyclin B1 promoter chromatin conformation during mitosis, we performed in vivo restriction site accessibility assays (Bhattacharyya et al., 1997) on a region spanning the promoter and the first intron of the cyclin B1 gene. Nuclear preparations from asynchronous cells and permeabilized mitotic cells were partially digested with BsaAI (Figure 2A) and Sau96I (Figure 2B), cutting at the E-box region and at the Sp1 site of the cyclin B1 promoter, respectively. DNA was then purified and fully digested with XbaI and NcoI generating fragments of 697 and 746 bp, respectively. The resultant fragments were detected by Southern blot analysis using a probe spanning 401 bp of the 5′ region of the cyclin B1 promoter. In mitotic cells the accessibility to BsaAI and Sau96I was comparable to that in asynchronous nuclei, demonstrating that the core promoter region is accessible to the endonucleases during mitosis. In contrast, accessibility to the enzymes that cut outside the core promoter was much lower even in the asynchronous nuclei, indicating an inaccessible chromatin structure in both tested regions (Figure 2C and D). Taken together, these results demonstrate that the cyclin B1 core promoter region is accessible to restriction enzymes, indicating that it maintains an open configuration at the mitotic stage.
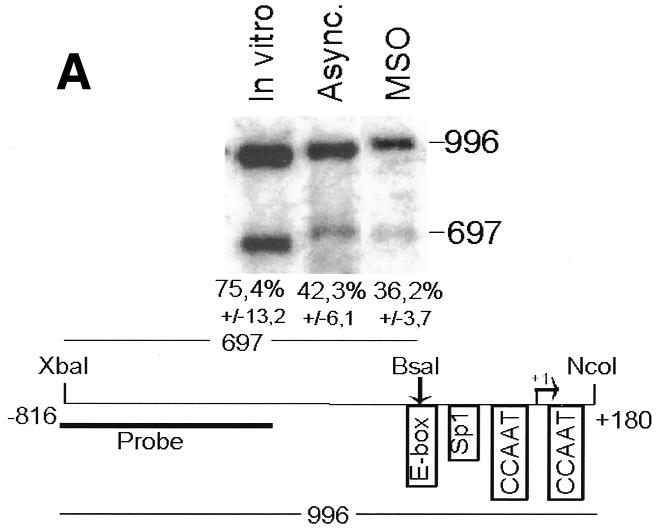
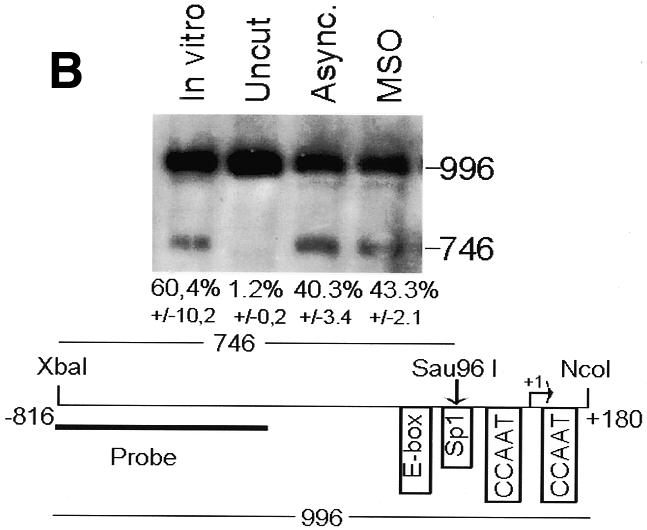
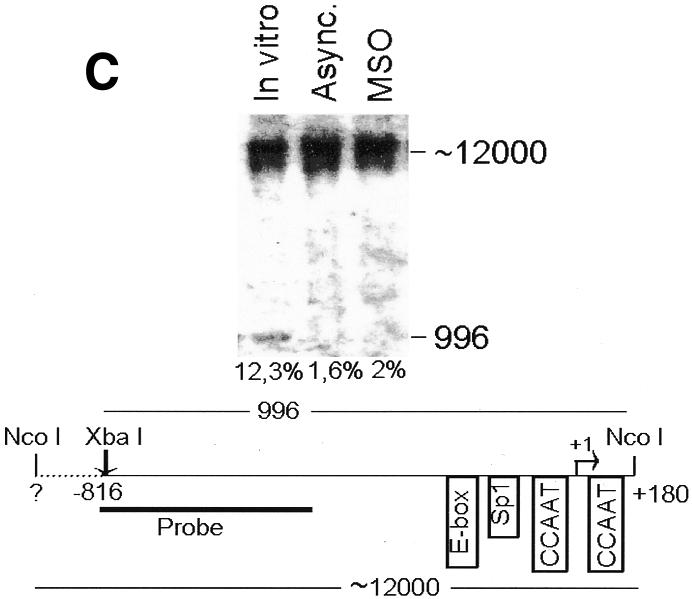
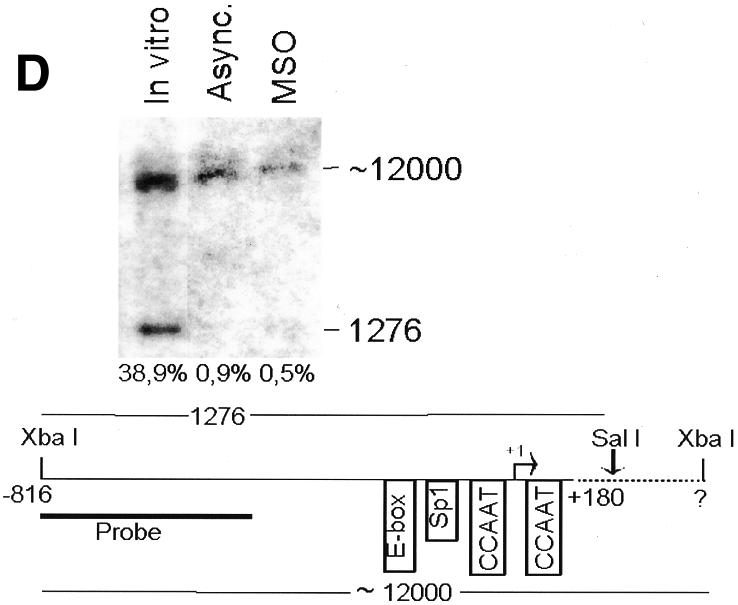
Fig. 2. The cyclin B1 promoter is accessible to restriction endonucleases in vivo. Nuclei isolated from asynchronous (Async.) (20 µg of DNA) and permeabilized MSO cells were partially digested with 100 U of BsaAI (A), 100 U of Sau96I (B), 200 U of XbaI (C) or 100 U of SalI (D). Purified DNA was fully digested with NcoI and XbaI when BsaAI was used for the partial digestion, and with NcoI when XbaI was used for the partial digestion. The probe used for the hybridization is shown at the bottom of each panel. Arrows mark the digested and undigested bands. The schematic diagram at the bottom of each panel indicates the position of the digested bands in the cyclin B1 promoter. The percent digestion is shown below each lane.
The cyclin B1 promoter interacts with transcription factors during mitosis in vivo
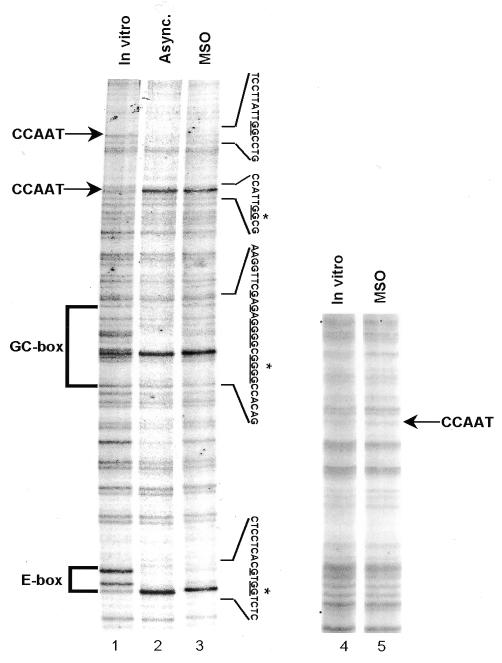
To test whether the open chromatin structure leads to transcription factor–DNA interactions, we performed in vivo genomic footprinting of the cyclin B1 promoter during mitosis. Asynchronous and mitotic cells were treated with the DNA alkylating reagent dimethyl sulfate (DMS), and methylated G residues were identified using the ligation-mediated polymerase chain reaction technique (LM–PCR) (Dey et al., 1992). In asynchronous cells, four regions on the cyclin B1 promoter non-coding strand were hypersensitive to or protected from DMS methylation: the E box (–124 to –119 bp), the GC box (–80 to –71 bp), the upstream CCAAT box (–17 to –12 bp) and the downstream CCAAT box (+15 to +20 bp) (Figure 3, lane 2). Genomic footprinting of mitotic cells showed the same methylation pattern as in cycling cells, thus indicating a persistent interaction of sequence-specific transcription factors with the cyclin B1 promoter at the mitotic stage (Figure 3, lane 3). It has been demonstrated that the hsp70 promoter is devoid of sequence-specific transcription factor interactions during mitosis in HeLa cells, while it shows protection of the main cis-acting elements in asynchronous cells (Martinez-Balbas et al., 1995). As a control, we performed genomic footprinting experiments on the hsp70 promoter (Martinez-Balbas et al., 1995). As expected, this promoter was devoid of sequence-specific transcription factor interactions in MSO cells (Figure 3, lane 5). Consistent with the restriction site accessibility data shown in Figure 2, these results demonstrate that, during mitosis, the cyclin B1 promoter is occupied by sequence-specific transcription factors.
Fig. 3. The cyclin B1 promoter interacts with transcription factors during mitosis in vivo. Genomic footprinting of the non-coding strand of the human cyclin B1 promoter from asynchronous (Async.) and MSO HeLa cells (lanes 2 and 3), and of the coding strand of the human hsp70 promoter from MSO HeLa cells (lane 5). As a control, the DMS reactivity for DNA purified from asynchronous HeLa cells is shown (In vitro). The changes in DMS reactivity found in asynchronous and MSO cells are noted at the corresponding G residues by an asterisk (hypersensitivity) and by underlines (protection).
NF-Y binds the cyclin B1 promoter during mitosis in vivo
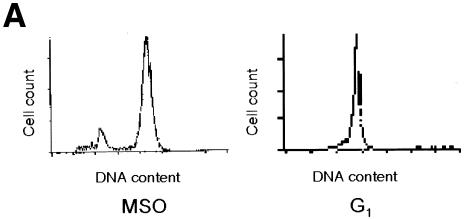
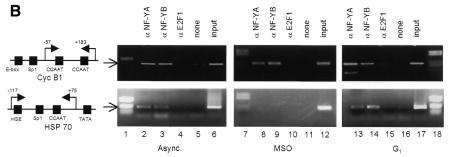
Genomic footprinting experiments demonstrate that the CCAAT boxes of the cyclin B1 promoter are occupied during mitosis in vivo. We have demonstrated previously that these two elements are key sequences for the cyclin B1 promoter activity and NF-Y binds these sequences in vitro (Farina et al., 1999). Thus, we asked whether NF-Y interacted with the cyclin B1 promoter during mitosis in vivo. Cycling and mitotic cells were treated with formaldehyde to cross-link proteins to DNA. After sonication, the cross-linked chromatin from equivalent numbers of asynchronous and mitotic cells was immunoprecipitated using anti-NF-YA and NF-YB antibodies. As negative controls, we included a reaction lacking the primary antibody and one containing anti-E2F1 antibody. E2F1 is a nuclear transcription factor that does not have a binding site on the cyclin B1 promoter. After immunoprecipitation, enrichment of the endogenous cyclin B1 promoter fragment in each sample was monitored by PCR amplification using primers amplifying the cyclin B1 promoter region from –57 to +183 bp. The results show that both anti-NF-Y immunoprecipitates from cycling and mitotic chromatin contained the cyclin B1 promoter (Figure 4B). To investigate the ability of NF-Y to bind the cyclin B1 promoter in G1, we performed chromatin immunoprecipitation experiments on G1 synchronized cells (Figure 4A). As shown in Figure 4B, anti-NF-Y immunoprecipitates from G1 chromatin contained the cyclin B1 promoter. To evaluate the specificity of the NF-Y interaction with the cyclin B1 promoter, we used the hsp70 promoter. Indeed, a CCAAT box is also required for the transcriptional activation of this promoter in proliferating cells, and NF-Y binds this sequence in vitro and in vivo (Imbriano et al., 2001). NF-Y antibodies immunoprecipitated the hsp70 promoter from asynchronous and G1 cells, but not from mitotic cells (Figure 4B). The non-specific antibody E2F1 was unable to immunoprecipitate the cyclin B1 or hsp70 promoters. Taken together, these results further provide in vivo evidence for a specific retention of NF-Y to the cyclin B1 promoter during mitosis.
Fig. 4. NF-Y binds to the cyclin B1 promoter in vivo during mitosis. (A) DNA distribution analysis of propidium iodide-stained MSO and G1 HeLa cells. (B) Formaldehyde cross-linked chromatin was prepared from the same number of asynchronous (Async.), mitotic and G1 HeLa cells. Cross-linked chromatin from each sample was incubated with antibodies to NF-Y (lanes 2, 3, 8, 9, 13 and 14) and E2F1 (lanes 4, 10 and 15), or in the absence of antibody (none). Immunoprecipitates from each antibody were aliquoted and then analyzed by PCR with specific primers for the cyclin B1 or hsp70 promoters. A sample representing 0.02% of the total input chromatin (input) was included in the PCR analysis.
Transcriptional activity of the endogenous cyclin B1 promoter is high during mitosis
To investigate whether the endogenous cyclin B1 gene is transcribed at the mitotic stage, we analyzed the rate of gene transcription by whole-cell run-on assays (Parsons and Spencer, 1997). In these experiments a constant cell number of asynchronous, MSO, nocodazole-treated and G1 phase cells have been employed. The RNA labeled during run-on transcription assays was hybridized to filters containing denatured double-stranded cDNA probes for B1, A, E and D1 cyclins. Transcription was detected for all tested genes in the asynchronous cell population. Transcription in mitotic cells was reduced for A, E and D1 cyclin genes, while the cyclin B1 gene showed a high level of transcription in mitotic cells (Figure 5). Interestingly, although not completely repressed, cyclin B1 gene transcription was down-regulated in the G1 phase. Taken together, our data demonstrate that the human cyclin B1 gene is actively transcribed during mitosis in HeLa cells.
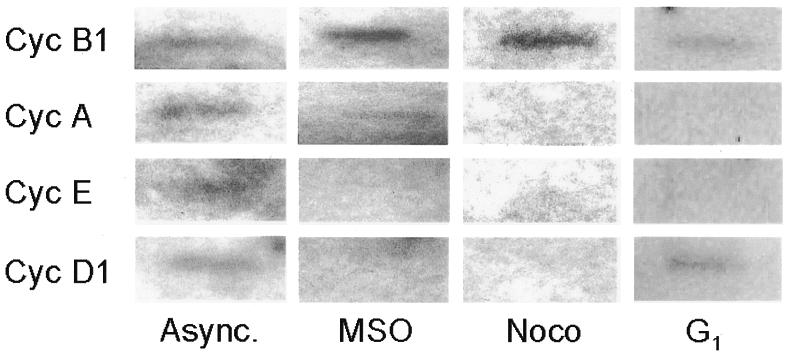
Fig. 5. Transcriptional activity of endogenous cyclin B1 promoter is high during mitosis. Asynchronous (Async.), mitotic and G1 phase HeLa cells were tested by a whole-cell run-on assay. Probes were cDNAs from the murine cyclin B1, A, E and D1 genes.
DISCUSSION
We investigated the occurrence of transcription during mitosis on an RNA pol II-transcribed gene. We have found that the human cyclin B1 gene is actively transcribed at the mitotic stage. This result is surprising, since it is widely accepted that transcription is repressed during mitosis in higher eukaryotes. Interestingly, in fission yeast the rate of RNA synthesis is maintained during passage through mitosis (Baum et al., 1998). In mammalian cells, until now, no RNA pol II-dependent transcription has been reported in mitotic cells, although there is evidence showing that 10–20% of the TFIID population remains associated with the condensed mitotic chromatin (Segil et al., 1996). Whether the transcription of the cyclin B1 gene occurs during all the four mitosis phases remains to be elucidated. The cyclin B1 protein is quickly degraded at the metaphase. Whenever a spindle checkpoint is imposed during metaphase, there is a reappearance of cyclin B1 protein due to a loss of cyclin B1 destruction (Clute and Pines, 1999). Our results suggest that transcription could also contribute to ensure adequate cyclin B1 levels, which could be immediately available whenever a spindle checkpoint is activated. Whether the mitotic transcription of the cyclin B1 gene is an event occurring in transformed cells and not in non-transformed cells remains to be elucidated.
Our results not only demonstrate that the cyclin B1 promoter is accessible to restriction endonucleases, but also show that this promoter is occupied by transcription factors during mitosis. In particular, we demonstrate that NF-Y binds the cyclin B1 promoter during mitosis. This result is in agreement with the evidence for retaining the TFIID on mitotic chromatin (Segil et al., 1996). Interestingly, the single-stranded DNA-binding proteins, heterogeneous nuclear ribonucleoprotein K and far upstream binding protein, which are both able to transactivate the c-myc gene in asynchronous cells, remain bound to the c-myc promoter during mitosis (Michelotti et al., 1997). The results presented here indicate strongly that the cyclin B1 gene transcription occurring during mitosis is due to a functional NF-Y complex bound on its promoter. Importantly, we did not find NF-Y binding to the hsp70 promoter during mitosis, indicating that specific sequences, other than the CCAAT boxes per se, are required for the recruitment of NF-Y on mitotic chromosomes. Several cell cycle-regulated promoters contain more than one CCAAT in a position similar to that of the cyclin B1 promoter. Thus, it is possible that mitotic recruitment of NF-Y might be common to other cell cycle-related promoters containing two or more CCAAT boxes.
Interestingly, NF-Y also binds the cyclin B1 promoter in the G1 phase. Since our ChIP experiments are not quantitative, we cannot exclude that NF-Y binds in G1 the cyclin B1 promoter with a lower affinity than in mitosis. However, this result is consistent with our genomic footprinting data demonstrating that the two CCAAT boxes of the cyclin B1 promoter bind proteins in the G1 phase (S.Sciortino, unpublished observations). Run-on experiments indicate that cyclin B1 transcription, although not completely repressed, is down-regulated in G1. One possibility is that the cell cycle-dependent transcription of the cyclin B1 gene is due to a recruitment of different protein complexes by NF-Y on the promoter in the different cell-cycle phases. We are currently investigating this issue.
In conclusion, although there is a general repression of transcription during mitosis in mammalian cells concomitant with dispersal of transcription factors from the mitotic chromatin, the cyclin B1 gene retains transcriptionally active complexes on its promoter and is actively transcribed at the mitotic stage.
METHODS
HeLa cells synchronization and transfections
For the double thymidine block, HeLa cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (Gibco BRL) to 60–70% confluence and incubated with 2.5 mM thymidine (Sigma). After 14 h, fresh medium was replaced for 8.5 h. The second block was performed with 2.5 mM thymidine for 14 h. Cells were incubated with fresh medium for 10 h and the mitotic cells were detached by tapping the flasks 10 times (MSO). These cells were replated, and after 3 h G1 phase synchronized cells were collected. For the nocodazole block the cells were treated for 12 h with 4 µg/ml of nocodazole (Sigma) and prometaphase cells were detached by tapping the flasks 10 times. For DNA distribution analysis of propidium iodide-stained cells, 104 events were analyzed by an Epics cytofluorimeter (Coulter) using computer-assisted analysis (see Supplementary data). For the mitotic index, cells were cytocentrifuged, fixed and stained with a 1 mg/ml solution of Hoechst 33258 dye (see Supplementary data). Stable DNA transfections were performed using the calcium-phosphate precipitation technique (Graham and van der Eb, 1973).
Northern blot analysis
Total RNA was extracted as described previously (Manfioletti and Schneider, 1988). The probes were cDNA fragments of the CAT, cyclin B1 and GAPDH genes.
Whole-cell run-on transcription assays
Run-on transcription assays were performed as described previously (Parsons and Spencer, 1997). Probes were cDNAs from the murine B1, A, E and D1 cyclin genes.
Endonuclease restriction accessibility assays
Nuclei from asynchronous cells were prepared and stored as described previously (Bhattacharyya et al., 1997). MSO cells were collected and an amount of cells corresponding to 10–20 µg of DNA was pelletted, permeabilized with 0.5% NP-40 and resuspended in endonuclease buffer. These cells were homogenized gently 10 times in a Dounce homogenizer and brought to a final volume of 100 µl with endonuclease buffer. All the samples were digested at 30°C for 20 min. The following enzymes were used: BsaAI, Sau96I and XbaI from New England Biolabs, and SalI from Boehringer Mannheim. Purified deproteinated HeLa genomic DNA was isolated, digested to completion with the same enzymes and used as controls. Digested samples, resolved on a 1% agarose gel, were hybridized with a probe spanning the first 401 bp of the 5′ region of the cyclin B1 promoter, and detected by autoradiography. Percent cut was calculated as the intensity of digested bands/intensity of the undigested plus digested bands × 100.
In vivo DNA footprinting
The in vivo DNA footprinting was performed using LM–PCR as described previously (Dey et al., 1992). For cyclin B1, the following oligonucleotides were used: first primer, TGGCATTGGCAACGCACAC (Tm = 54°C); second primer, ACTGGCTTCACTTGCTCTCCAGGTGCGCTG (Tm = 63°C); third primer, CATGGCTTCCTCTTCACCAGG (Tm = 69°C).
For hsp70, oligonucleotides described previously were used (Martinez-Balbas et al., 1995).
Formaldehyde cross-linking and chromatin immunoprecipitation
Formaldehyde cross-linking and chromatin immunoprecipitation were performed as described previously (Boyd et al., 1998). Immunoprecipitation was performed with protein G. The chromatin solution was precleared by adding protein G for 15 min at 4°C, aliquoted and incubated with 2 µg of affinity-purified rabbit polyclonal antibodies (α-NF-YB was a gift from R. Mantovani, α-NF-YA was a mix of an antibody from Rockland and a gift from R. Mantovani, and α-E2F1 was from Santa Cruz) for 3 h at 4°C with mild shaking. Before use, protein G was blocked with 1 µg/µl sheared herring sperm DNA and 1 µg/µl bovine serum albumin for 4 h at 4°C and then incubated with chromatin and antibodies overnight. Immunoprecipitates were eluted and, after reverse cross-linking, ethanol precipitated. Recovered material was treated with proteinase K, extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated. The pellets were resuspended in 30 µl of H2O and analyzed using PCR. The total input sample was resuspended in 100 µl of H2O and diluted 1:100 before PCR analysis. For PCR the following oligonucleotides were used: cyclin B1 (+183): GGCTTCCTCTTCACCAGGCA; cyclin B1 (−57): CGCGATCGCCCTGGAAAC; hsp70 (+75): AGCCTTGGGACAACGGGAG; hsp70 (−117): GGCGAAACCCCTGGAATATTCCCGA.
Supplementary data
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Nissan Bhattacharyya for technical advice and Maria Pia Gentileschi for oligonucleotide synthesis. The financial support of AIRC and CNR to G.P. is gratefully acknowledged. A.G., I.M. and G.F. are recipients of a FIRC fellowship. This work is dedicated to the memory of Franco Tatò.
REFERENCES
- Baum B., Nishitani, H., Yanow, S. and Nurse, P. (1998) Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J., 17, 5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya N., Dey, A., Minucci, S., Zimmer, A., John, S., Hager, G. and Ozato, K. (1997) Retinoid-induced chromatin structure alterations in the retinoic acid receptor β2 promoter. Mol. Cell. Biol., 17, 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd K.E., Wells, J., Gutman, J., Bartley, S.M. and Farnham, P.J. (1998) c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl Acad. Sci. USA, 95, 13887–13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P. and Pines, J. (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nature Cell. Biol., 1, 82–87. [DOI] [PubMed] [Google Scholar]
- Cogswell J.P., Godlevski, M.M., Bonham, M., Bisi, J. and Babiss, L. (1995) Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol. Cell. Biol., 15, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Thornton, A.M., Lonergan, M., Weissman, S.M., Chamberlain, J.W. and Ozato, K. (1992) Occupancy of upstream regulatory sites in vivo coincides with major histocompatibility complex class I gene expression in mouse tissues. Mol. Cell. Biol., 12, 3590–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A., Gaetano, C., Crescenzi, M., Puccini, F., Manni, I., Sacchi, A. and Piaggio, G. (1996) The inhibition of cyclin B1 gene transcription in quiescent NIH 3T3 cells is mediated by an E-box. Oncogene, 13, 1287–1296. [PubMed] [Google Scholar]
- Farina A., Manni, I., Fontemaggi, G., Tiainen, M., Cenciarelli, C., Bellorini, M., Mantovani, R., Sacchi, A. and Piaggio, G. (1999) Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene, 18, 2818–2827. [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C., Cavadore, J.C., Blanchard, J.M., Lamb, N.J. and Fernandez, A. (1991) p67SRF is a constitutive nuclear protein implicated in the modulation of genes required throughout the G1 period. Cell. Regulat., 2, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J.M. and Forbes, D.J. (1997) Mitotic repression of the transcriptional machinery. Trends Biochem. Sci., 22, 197–202. [DOI] [PubMed] [Google Scholar]
- Graham F.L. and van der Eb, A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–467. [DOI] [PubMed] [Google Scholar]
- Hwang A., Maity, A., McKenna, W.G. and Muschel, R.J. (1995) Cell cycle-dependent regulation of the cyclin B1 promoter. J. Biol. Chem., 270, 28419–28424. [DOI] [PubMed] [Google Scholar]
- Imbriano C., Bolognese, F., Gurtner, A., Piaggio, G. and Mantovani, R. (2001) HSP-CBF is an NF-Y-dependent coactivator of the heat shock promoters CCAAT-boxes. J. Biol. Chem., 13, 26332–26339. [DOI] [PubMed] [Google Scholar]
- Jackman M.R. and Pines, J. (1997) Cyclins and the G2/M transition. Cancer Surv., 29, 47–73. [PubMed] [Google Scholar]
- Katula K.S., Wright, K.L., Paul, H., Surman, D.R., Nuckolls, F.J., Smith, J.W., Ting, J.P., Yates, J. and Cogswell, J.P. (1997) Cyclin-dependent kinase activation and S-phase induction of the cyclin B1 gene are linked through the CCAAT elements. Cell Growth Differ., 8, 811–820. [PubMed] [Google Scholar]
- Manfioletti G. and Schneider, C. (1988) A new and fast method for preparing high quality λ DNA suitable for sequencing. Nucleic Acids Res., 16, 2873–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Dey, A., Rabindran, S.K., Ozato, K. and Wu, C. (1995) Displacement of sequence-specific transcription factors from mitotic chromatin. Cell, 83, 29–38. [DOI] [PubMed] [Google Scholar]
- Michelotti E.F., Sanford, S. and Levens, D. (1997) Marking of active genes on mitotic chromosomes. Nature, 388, 895–899. [DOI] [PubMed] [Google Scholar]
- Parsons G.G. and Spencer, C.A. (1997) Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol., 17, 5791–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggio G., Farina, A., Perrotti, D., Manni, I., Fuschi, P., Sacchi, A. and Gaetano, C. (1995) Structure and growth-dependent regulation of the human cyclin B1 promoter. Exp. Cell Res., 216, 396–402. [DOI] [PubMed] [Google Scholar]
- Segil N., Guermah, M., Hoffmann, A., Roeder, R.G. and Heintz, N. (1996) Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Gene Dev., 10, 2389–2400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.