Abstract
UV-induced DNA damage causes cells to repress RNA synthesis and to initiate nucleotide excision repair (NER). NER and transcription are intimately linked processes. Evidence has been presented that, in addition to damaged genes, undamaged loci are transcriptionally inhibited. We investigated whether RNA synthesis from undamaged genes is affected by the presence of UV damage elsewhere in the same nucleus, using a novel technique to UV irradiate only part of a nucleus. We show that the basal transcription/repair factor TFIIH is recruited to the damaged nuclear area, partially depleting the undamaged nuclear area. Remarkably, this sequestration has no effect on RNA synthesis. This result was obtained for cells that are able to carry out NER and for cells deficient in NER. We conclude that cross talk between NER and transcription occurs only over short distances in nuclei of living cells.
INTRODUCTION
Short-wave UV light induces intrastrand DNA cross-links between adjacent pyrimidines, giving rise to the cyclobutane pyrimidine dimer (CPD) and the (6-4)photoproduct (6-4PP). These photoproducts are potent inhibitors of transcription by RNA polymerase II (RNA pol II). The removal of these DNA lesions is somehow prioritized at the expense of ongoing transcription. This may reflect an active cellular response to circumvent, for instance, the potential risks that might arise due to the production of aberrant gene products. The molecular mechanism underlying inhibition and recovery of transcription after DNA damage is not understood well (Friedberg et al., 1995).
Nucleotide excision repair (NER) is a sophisticated DNA repair mechanism that enables cells to resolve a wide variety of structurally diverse DNA injuries, including UV-induced photoproducts. NER involves the action of some 30 different gene products. Many protein interactions and biochemical characteristics of this repair system have been established (De Laat et al., 1999). There appear to be at least two mechanistically distinct NER pathways, commonly referred to as global genome repair (GGR) and transcription-coupled repair (TCR). In GGR, the XPC–hHR23B complex recognizes the damaged DNA and subsequently recruits the repair machinery to process the lesion (Sugasawa et al., 1998). On the other hand in TCR, DNA lesions are detected via an unknown mechanism that depends on RNA pol II-driven transcription (Hanawalt, 2000). Both CPD and 6-4PP, if present in the template strand of a transcribed gene, very efficiently block elongating RNA pol II. The stalled polymerase itself probably functions as a signal for the NER factors to be recruited. In agreement, XPC–hHR23B is not involved in TCR (Van Hoffen et al., 1995), whereas at least two gene products with poorly understood molecular roles, the CSA and CSB proteins, are necessary for TCR but not for GGR (Venema et al., 1990).
In addition to the ability of UV photoproducts to pose a physical block for RNA pol II, there have been other observations that suggest a tight coupling between NER and transcription. The most striking example of such a link is the finding that the basal transcription factor TFIIH is required for NER, both GGR and TCR (Hoeijmakers et al., 1996). In NER-proficient yeast extracts, it was found that transcription of an undamaged plasmid was decreased in the presence of another UV-damaged plasmid. This inhibition was dependent on the presence of the yeast homologue of the human CSB protein, Rad26, and could be reversed by addition of TFIIH (You et al., 1998). This supports a straightforward model for transcription inhibition by DNA damage: upon DNA damage, TFIIH is recruited to sites of damage and, as a result, is not then available for transcription initiation, causing a decrease in overall transcriptional activity.
In order to understand the mechanism by which DNA damage inhibits RNA pol II-driven transcription, we have investigated whether RNA synthesis from undamaged genes is affected by the presence of UV damage elsewhere in the nucleus. We developed a simple technique that enables one to irradiate only a limited volume of cell nuclei with UV light. This allowed us to analyse the functional cross talk between NER and transcription in the intact cell, showing that both systems only interact over short distances in the nucleus.
RESULTS AND DISCUSSION
Induction and detection of local UV damage
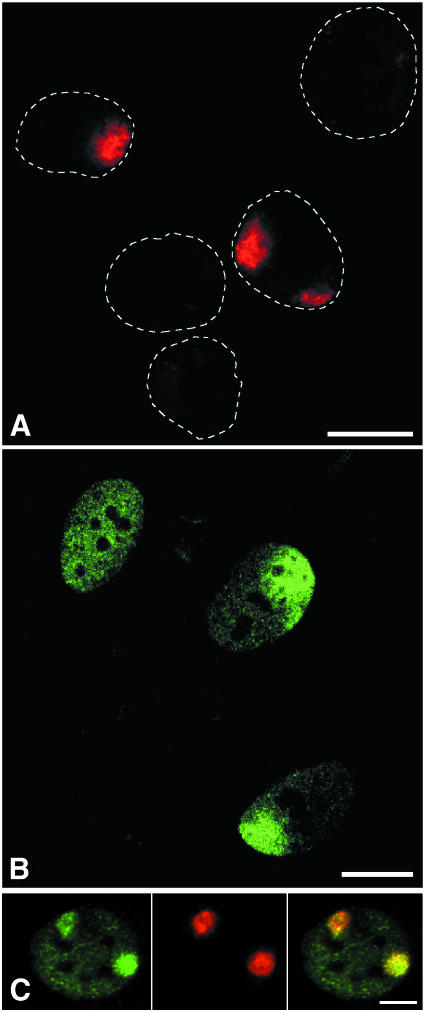
We have developed a method that enabled us to UV irradiate a confined area of individual cell nuclei. A polycarbonate filter containing pores of defined size (e.g. 3, 5, 8 or 10 µm in diameter) was used. Cells that were grown on coverslips were briefly rinsed with phosphate-buffered saline (PBS) and after removal of excess buffer the filter was carefully positioned on top of the cell monolayer. Filter-covered cells were immediately UV irradiated from above, after which the filter was removed. UV damage was then visualized by immunofluorescence labelling using the damage-specific monoclonal antibody TDM-2 (Mori et al., 1991). Figure 1A displays cells that were locally irradiated and subsequently immunostained for CPDs. This reveals DNA damage in discrete parts of cell nuclei. The number of nuclei that were locally damaged was consistent with the pore density of the filter (4 × 105 pores/cm2), resulting in a cell monolayer in which most nuclei were either unirradiated or locally damaged in a single spot. A small number of nuclei contained more than one damaged nuclear area. The diameter of UV-damaged spots was consistent with the diameter of the pores. Labelling in unirradiated parts of locally damaged nuclei was indistinguishable from background levels of unirradiated control cells in a separate experiment, indicating that the filter material efficiently absorbs 254 nm UV light. This technique allows one to locally UV irradiate large numbers of cells, while unirradiated control nuclei are present in the same monolayer of cells.
Fig. 1. (A) Detection of locally induced UV damage in cell nuclei. A UV-blocking polycarbonate filter containing pores of 5 µm in diameter was used to cover a monolayer of cells. The filter-covered cells were UV irradiated with 30 J/m2 and CPDs were subsequently detected by immunofluorescent labelling. Dotted lines denote the contours of individual cell nuclei. Two nuclei show labelling of CPDs in discrete nuclear areas. (B) Effect of local nuclear UV damage on the distribution of TFIIH. Human primary fibroblasts were locally UV irradiated with 100 J/m2 UV light, using a filter with 10 µm pores. Following irradiation, cells were grown for 1 h and immunolabelled against the p62 subunit of TFIIH. The top-left nucleus displays the characteristic labelling pattern of TFIIH in unirradiated cells, whereas the two remaining nuclei exhibit a TFIIH accumulation in UV-damaged nuclear areas, and a reduction in TFIIH signal outside these areas. A single confocal optical section is shown. (C) Colocalization of TFIIH (green) and CPDs (red). Human primary fibroblasts were locally irradiated with 30 J/m2 UV light, using a filter with 8 µm pores. Following irradiation, cells were grown for 30 min and dual labelled against both CPDs and the p62 subunit of TFIIH. Bars represent 10 µm.
TFIIH accumulates at sites of UV damage
TFIIH is indispensable for both transcription initiation and NER. Logically, this dual functionality has led to the suggestion that TFIIH could play a key role in orchestrating the down-regulation of transcription upon DNA damage-triggered onset of NER (Van Oosterwijk et al., 1996; Mullenders, 1998; You et al., 1998). Previous experiments by Houtsmuller et al. (1999) have shown that the ERCC1/XPF heterodimer, an endonuclease that makes the 5′ incision during NER, moves freely through undamaged cell nuclei and becomes immobilized for several minutes upon induction of UV damage. This shows that at least some NER factors are likely to be recruited to damaged sites in a distributive manner, rather than by processive scanning of large genomic segments. From that notion it is possible that TFIIH is targeted to sites of DNA damage in a similar fashion. We have tested whether this is the case.
Primary fibroblasts were locally UV irradiated and cultured for 1 h prior to fixation. Indeed, TFIIH had accumulated in large nuclear areas (Figure 1B). The size and number of these p62-enriched areas were consistent with filter specifications, and were also confirmed by applying filters with different pore size and pore density (data not shown). Moreover, double labelling shows that local UV damage and TFIIH indeed colocalize (Figure 1C). In addition to TFIIH accumulation in irradiated areas, TFIIH labelling intensity appeared to be partly depleted in undamaged parts of cell nuclei. This was the case even if a quarter to a third of the nuclear volume had been irradiated with 100 J/m2, resulting in an estimate number of lesions per nucleus that is ∼3-fold higher than the amount of lesions known to saturate the NER process and to inhibit >80% of transcription in cells that are uniformly UV irradiated (Mayne and Lehmann, 1982). These data show that recruitment of TFIIH from undamaged to damaged nuclear areas occurs and might thus affect transcription in undamaged genomic regions.
Local inhibition of transcription by local UV damage
In an in vitro yeast system, RNA pol II transcription of an undamaged template was significantly inhibited when NER became activated. Furthermore, this transcription inhibition could be restored by increasing the TFIIH concentration (You et al., 1998). Competition assays from human cell-free extracts have generated ambiguous results. Satoh and Hanawalt (1996) found no communication between transcription and NER in the presence of DNA damage. On the other hand, Vichi et al. (1997) did find damage-induced inhibition of transcription from an undamaged template, although the authors concluded that this coincided with sequestration of TATA box-binding protein rather than of TFIIH. We investigated whether local nuclear UV damage interfered with transcription in undamaged areas within the same intact nucleus.
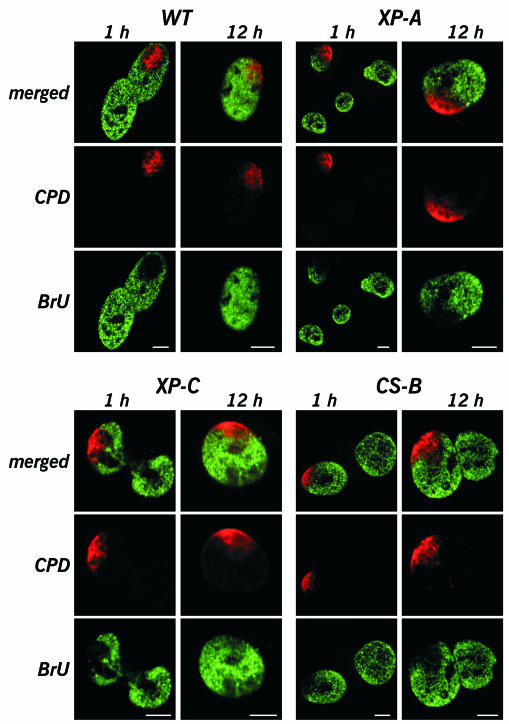
NER-proficient human primary fibroblasts were locally irradiated with 50 J/m2 UV light. Such a dose on a quarter to a third of the nuclear volume will generate a total number of lesions that is 30–100% larger than the number of lesions that is induced by 8–10 J/m2 uniform UV irradiation of cells. The latter condition is known to obliterate >80% of transcription 1 h after UV irradiation. After post-irradiation culturing for 1 h, cells were allowed to incorporate BrUTP into nascent RNA during run-on transcription (Jackson et al., 1993; Wansink et al., 1993). Subsequently, they were immunofluorescently dual labelled using anti-BrdU and anti-CPD antibodies. RNA synthesis was almost completely abolished within the UV-damaged nuclear area (Figure 2, WT). Surprisingly, cells maintained normal levels of transcriptional activity throughout undamaged regions of the nucleus. Inhibition of transcription in locally irradiated nuclei was fully confined to the UV-damaged area. The BrU signal outside the irradiated spot was comparable to RNA labelling intensities of nearby unirradiated control cells. If primary fibroblasts were allowed to grow for 12 h after local UV irradiation, CPD labelling was still clearly detectable (Figure 2, WT), but cells had largely restored transcription inside the damaged area. In accordance with previous data, it is anticipated that at this point in time the NER machinery will have repaired 6-4PPs and active genes. The bulk of the remaining damage will be CPDs in inactive chromatin, as these are known to be repaired much slower (Van Hoffen et al., 1995). Our results show that UV-induced DNA damage has a short-range inhibitory effect on transcription.
Fig. 2. Effect of local nuclear UV damage on transcription. Normal human primary fibroblasts (WT; NER-proficient) and immortalized primary fibroblasts from patients suffering from xeroderma pigmentosum group A (XP-A; no NER), group C (XP-C; no GGR) or Cockayne syndrome group B (CS-B; no TCR) were studied. Exponentially growing cells were locally UV irradiated with 50 J/m2 using filters containing 10 µm pores. After irradiation, cells were cultured for either 1 or 12 h. Subsequently, cells were allowed to incorporate BrUTP into nascent RNA during run-on transcription labelling. Nascent RNA (green) and CPDs (red) were dual labelled by immunostaining. Single confocal optical sections are shown. Bars represent 5 µm.
As NER and transcription are intimately linked processes, we investigated the effects of local UV irradiation in NER-deficient cell lines. XP-A cells are deficient in performing NER. Although RNA synthesis was efficiently inhibited in the damaged spot 1 h after local irradiation, BrU labelling intensities in undamaged nuclear compartments were comparable to labelling intensities of undamaged cells (Figure 2, XP-A). Twelve hours after induction of local damage, XP-A cells still exhibited high levels of transcription in undamaged nuclear areas, comparable to labelling intensities of undamaged control cells. Not surprisingly, due to the absence of NER in these cells, transcription in the damaged spot remained inhibited. Taken together, our results show that there is no long-distance functional cross talk between NER and transcription. Furthermore, cells harbour sufficient TFIIH to support normal levels of transcription and high levels of NER simultaneously.
We also examined the transcription response in XP-C and CS cells, deficient in GGR and TCR, respectively. The XPC protein is the earliest damage recognition factor during GGR (Sugasawa et al., 1998; Volker et al., 2001) and hence, the bulk of genomic DNA damage will remain devoid of NER complexes. Lesions are therefore likely to be available as a decoy for damage-binding transcription factors like TATA box-binding protein. Nonetheless, 1 h after local UV irradiation, transcription inhibition remained confined to the damaged nuclear area in XP-C cells (Figure 2, XP-C). The observation that XP-C cells reinitiated RNA synthesis in the UV-damaged area 12 h after local irradiation (Figure 2, XP-C) is consistent with the known ability of XP-C cells to recover inhibited transcription after UV damage (Mayne and Lehmann, 1982). CS cells are impaired in performing TCR (Venema et al., 1990) and are unable to recover transcription after inhibition by UV damage. Also in CS-B cells, BrU incorporation was not decreased in undamaged genomic regions after both 1 and 12 h of post-irradiation culturing (Figure 2, CS-B). As one would expect, transcription remained inhibited within the damaged spot.
Mechanism of UV damage-induced transcription inhibition
Several studies indicate that transcription from undamaged genes is affected by the presence of DNA damage elsewhere in the nucleus. For instance, mutations in the XPD subunit of TFIIH can lead to the repair-disorder XP (XP-D cells), or may result in XP combined with CS clinical features (XP-D/CS cells). Both XP-D and XP-D/CS cells appear equally inefficient in eliminating UV damage from transcribed genes, yet they differ in their ability to recover their UV-inhibited RNA synthesis. Whereas XP-D cells are able to regain normal transcription after irradiation with 2 J/m2 UVC, XP-D/CS cells fail to do so (Van Hoffen et al., 1999). XP-D/CS cells, despite their inefficient photoproduct removal, generate repair incisions with almost the same efficiency as normal cells (Berneburg et al., 2000). The incisions appear to be uncoupled from the NER process, as introduction of a UV-damaged plasmid in XP-D/CS cells triggered the induction of incisions in undamaged genomic DNA. Whatever the mechanism may be, these data indicate that XP-D/CS cells sense DNA damage and respond to it in trans. Our results show unambiguously that inhibition of RNA synthesis by UV irradiation occurs only in the close vicinity of damaged DNA, in both wild-type and CS cells (Figure 2). Stalling of elongating RNA pol II at lesions in transcribed templates will certainly contribute to the observed transcription inhibition. However, one cannot rule out that short-range sensing of DNA damage impedes transcription of a nearby undamaged gene, suggesting a local trans-effect. It has been shown that small numbers of RNA polymerases are tightly associated in transcription factories (Cook, 1999), suggesting some functional interaction between these polymerases. Hence, it is conceivable that stalling of for example one polymerase on a lesion could inhibit several polymerases within the same factory.
METHODS
Cell culture. Primary human fibroblasts (VH25) were grown in Ham’s F-10 medium containing 12% fetal calf serum (FCS), 1% glutamine, 100 IU/ml penicillin and 100 µg/ml streptomycin (Life Technologies). Cells were cultured in a 5% CO2 atmosphere at 37°C.
SV-40 immortalized primary fibroblasts (gifts from Dr W. Vermeulen) XP20MA (XP-C), XP12RO (XP-A) and CS1AN (CS-B) were grown in a 1:1 mixture of Ham’s F-10 and Dulbecco’s modified Eagle’s medium containing 10% FCS. Conditions and supplements were the same as for VH25. Cells were used at 50–80% confluency.
Local UV irradiation. Cells were rinsed in PBS. The PBS was then removed, leaving only a thin layer of buffer on top of the coverslip. An Isopore polycarbonate membrane filter (Millipore) containing pores of either 5 µm (4 × 105 pores/cm2) or 10 µm (105 pores/cm2) in diameter was placed on top of the cells. The coverslip with filter was irradiated from above using a TUV 15 W lamp (Philips) at a UVC fluency of 1.0 W/m2, as measured at 254 nm with an SHD 240/W detector connected to an IL 1700 radiometer (International Light). The filter was then removed and cells were either fixed or cultured for another period of time.
Immunofluorescent labelling. Nascent RNA was labelled with BrUTP during run-on transcription as described by Wansink et al. (1993).
Cells were fixed with 4% formaldehyde in PBS for 15 min at 4°C, permeabilized in 0.5% Triton X-100 (Serva) in PBS for 5 min, and incubated with 100 mM glycine in PBS for 10 min. Cells were rinsed in PB (130 mM KCl, 10 mM Na2HPO4 and 2.5 mM MgCl2 pH 7.4) and equilibrated in WB [PB containing 0.5% bovine serum albumen (BSA), 0.2% gelatin and 0.05% Tween 20; Sigma-Aldrich]. Antibody steps and washes were in WB. BrU-labelled RNA was labelled by a rat monoclonal anti-BrdU antibody (Seralab), and detected by FITC-conjugated donkey anti-rat antibody (Jackson ImmunoResearch Laboratories). TFIIH labelling was carried out using a mouse mAb against the p62 subunit of TFIIH (a gift from Dr J.M. Egly), and detection was by sheep anti-mouse Ig coupled to biotin, and FITC-conjugated streptavidin (Jackson).
Immunolabelling of CPDs was performed using mouse mAb TDM-2 (Mori et al., 1991). For this, the above steps were repeated, but prior to labelling DNA was denatured with 2 M HCl for 30 min at 37°C and blocked in 10% BSA in PB for 15 min. Detection was done using sheep anti-mouse Ig coupled to biotin (Amersham Pharmacia Biotech) and Cy3-conjugated streptavidin (Jackson). For double-labelling CPDs and TFIIH, a polyclonal anti-p62 antibody (Santa Cruz) was used, detected by goat anti-rabbit IgG coupled to FITC (Jackson). Samples were mounted in Vectashield.
Microscopy. Fluorescence microscopy was performed on a Leitz Aristoplan fluorescence microscope with a Zeiss Plan-Neofluar 63×/1.25 NA oil immersion objective and equipped with a CCD camera (Apogee).
Optical sections were recorded on an LSM510 confocal laser scanning microscope with a Zeiss Plan-Neofluar 100×/1.3 NA oil immersion objective. FITC and Cy3 were excited at 488 and 543 nm, respectively.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Drs W. Vermeulen and B. Van Steensel for support, H. Brugman for technical assistance and J. Mateos Langerak for advice on figure preparation. This work was supported by the Netherlands Organization for Scientific Research (NWO-ALW) grants 805-33-443-P and 805-44-007-P.
REFERENCES
- Berneburg M. et al. (2000) UV damage causes uncontrolled DNA breakage in cells from patients with combined features of XP-D and Cockayne syndrome. EMBO J., 19, 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.R. (1999) The organization of replication and transcription. Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- De Laat W.L., Jaspers, N.G. and Hoeijmakers, J.H.J. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker, G.C. and Siede, W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Hanawalt P.C. (2000) The bases for Cockayne syndrome. Nature, 405, 415–416. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H.J., Egly, J.-M. and Vermeulen, W. (1996) TFIIH: a key component in multiple DNA transactions. Curr. Opin. Genet. Dev., 6, 26–33. [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers, S., Nigg, A.L., Hoogstraten, D., Hoeijmakers, J.H.J. and Vermeulen, W. (1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science, 284, 958–961. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Hassan, A.B., Errington, R.J. and Cook, P.R. (1993) Visualization of focal sites of transcription within human nuclei. EMBO J., 12, 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne L.V. and Lehmann, A.R. (1982) Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res., 42, 1473–1478. [PubMed] [Google Scholar]
- Mori T., Nakane, M., Hattori, T., Matsunaga, T., Ihara, M. and Nikaido, O. (1991) Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol., 54, 225–232. [DOI] [PubMed] [Google Scholar]
- Mullenders L.H.F. (1998) Transcription response and nucleotide excision repair. Mutat. Res., 409, 59–64. [DOI] [PubMed] [Google Scholar]
- Satoh M.S. and Hanawalt, P.C. (1996) TFIIH-mediated nucleotide excision repair and initiation of mRNA transcription in an optimized cell-free DNA repair and RNA transcription assay. Nucleic Acids Res., 24, 3576–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng, J.M.Y., Masutani, C., Iwai, S., van der Spek, P.J., Eker, A.P.M., Hanaoka, F., Bootsma, D. and Hoeijmakers, J.H.J. (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell, 2, 223–232. [DOI] [PubMed] [Google Scholar]
- Van Hoffen A., Venema, J., Meschini, R., van Zeeland, A.A. and Mullenders, L.H.F. (1995) Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J., 14, 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoffen A., Kalle, W.H., de Jong-Versteeg, A., Lehmann, A.R., van Zeeland, A.A. and Mullenders, L.H.F. (1999) Cells from XP-D and XP-D-CS patients exhibit equally inefficient repair of UV-induced damage in transcribed genes but different capacity to recover UV-inhibited transcription. Nucleic Acids Res., 27, 2898–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterwijk M.F., Verstee, A., Filon, R., van Zeeland, A.A. and Mullenders, L.H.F. (1996) The sensitivity of Cockayne’s syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol. Cell. Biol., 16, 4436–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., Mullenders, L.H.F., Natarajan, A.T., van Zeeland, A.A. and Mayne, L.V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichi P., Coin, F., Renaud, J.P., Vermeulen, W., Hoeijmakers, J.H.J., Moras, D. and Egly, J.-M. (1997) Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J., 16, 7444–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M. et al. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wansink D.G., Schul, W., van der Kraan, I., van Steensel, B., van Driel, R. and de Jong, L. (1993) Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase-II in domains scattered throughout the nucleus. J. Cell Biol., 122, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z.Y., Feaver, W.J. and Friedberg, E.C. (1998) Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by holo-TFIIH and requirement for RAD26. Mol. Cell. Biol., 18, 2668–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]