Abstract
Our previous studies identified two iron-regulated cytoplasmic membrane proteins of 32 and 36 kDa expressed by both Staphylococcus epidermidis and Staphylococcus aureus. In this study we show by Triton X-114 phase partitioning and tritiated palmitic acid labelling that these proteins are lipoproteins which are anchored into the cytoplasmic membrane by their lipid-modified N termini. In common with those of some other gram-positive bacteria, these highly immunogenic lipoproteins were released from the bacterial cell into the culture supernatants, with release being promoted by growth of the bacteria under iron-restricted conditions. Immunoelectron microscopy with a monospecific rabbit antiserum to the 32-kDa S. epidermidis lipoprotein showed that the majority of the antigen was distributed throughout the staphylococcal cell wall. Only minor quantities were detected in the cytoplasmic membrane, and exposure of the lipoprotein on the bacterial surface was minimal. A monoclonal antibody raised to the 32-kDa lipoprotein of S. aureus was used in immunoblotting studies to investigate the conservation of this antigen among a variety of staphylococci. The monoclonal antibody reacted with polypeptides of 32 kDa in S. epidermidis and S. aureus and of 40 kDa in Staphylococcus hominis. No reactivity was detected with Staphylococcus lugdunensis, Staphylococcus cohni, or Staphylococcus haemolyticus. The gene encoding the 32-kDa lipoprotein from S. epidermidis has been isolated from a Lambda Zap II genomic DNA library and found to be a component of an iron-regulated operon encoding a novel ABC-type transporter. The operon contains three genes, designated sitA, -B, and -C, encoding an ATPase, a cytoplasmic membrane protein, and the 32-kDa lipoprotein, respectively. SitC shows significant homology both with a number of bacterial adhesins, including FimA of Streptococcus parasanguis and ScaA of Streptococcus gordonii, and with lipoproteins of a recently described family of ABC transporters with proven or putative metal ion transport functions. Although the solute specificity of this novel transporter has not yet been determined, we speculate that it may be involved in either siderophore- or transferrin-mediated iron uptake in S. epidermidis.
Staphylococci respond to iron deprivation in vitro and in vivo by increasing the expression of a number of iron-regulated proteins (24, 28, 36, 41). These include a 42-kDa cell wall-associated transferrin binding protein and two cytoplasmic membrane proteins of 32 and 36 kDa whose functions are at present unknown (28, 36). The association of the latter proteins with the cytoplasmic membrane is based on cell fractionation data, but the mechanism of anchorage of these proteins to the cytoplasmic membrane is also currently unclear. In other bacteria, some cytoplasmic membrane proteins are integral to the membrane and are held in place by hydrophobic membrane-spanning regions or hydrophobic N- or C-terminal anchors (31). Other membrane-associated proteins have been shown to be lipoproteins, and these are tethered to the outer surface of the membrane by their lipid-modified N termini (17, 38). Functionally, both groups of proteins may be components of ATP binding cassette (ABC)-type transporters involved in solute acquisition (13, 39). These multicomponent transporters have common structural and organizational features, and functions in addition to solute transport have recently been attributed to some ABC transporter lipoproteins. Studies with gram-positive bacteria show that the lipoproteins SarA and SsaB mediate adherence of Streptococcus gordonii and Streptococcus sanguis, respectively, to salivary pellicle components (15, 22, 29). FimA from Streptococcus parasanguis is involved in attachment of the bacterium to fibrin clots in a rat endocarditis model and to platelet fibrin matrix clots in vitro (14, 40). These functional studies and other antibody binding studies support the surface location of these streptococcal lipoproteins (20, 21, 40). The Enterococcus faecalis lipoprotein EfaA also shows sequence homology with streptococcal adhesins, including FimA, SsaB, and ScaA, indicating that EfaA may also be a surface-exposed adhesin (25).
In contrast to the case for other gram-positive organisms, ABC transporters in the staphylococci have been little studied. Proteins that may function as components of an ABC transporter involved in erythromycin resistance have been detected in Staphylococcus epidermidis (32), but this system has not been fully characterized and no homologies with such systems in other bacteria have yet been reported. ABC exporters are implicated in the secretion of the lantibiotics epidermin (13) and Pep5 (26) by S. epidermidis and of gallidermin (13) by Staphylococcus gallidermidis. DNA sequence analysis has also identified an ABC transporter of unknown function in Staphylococcus aureus (12). Currently the only well-studied staphylococcal lipoprotein is the β-lactamase of S. aureus (2, 30), which is found both in a membrane-bound form and in the bacterial culture supernatant (a common finding for other gram-positive lipoproteins [38]). There is currently no published information on transport systems involved in iron acquisition by the staphylococci.
Our ongoing interest in staphylococcal iron uptake mechanisms has led us to further characterize the role of the staphylococcal 32- and 36-kDa iron-regulated cytoplasmic membrane proteins as possible components of iron transport systems. This paper describes the identification of these iron-regulated proteins as lipoproteins and the molecular cloning of the 32-kDa lipoprotein from S. epidermidis, which sequence analysis reveals to be a component of a novel ABC-type transporter. Immunoelectron microscopy has also been used to confirm the cellular location of this lipoprotein in S. epidermidis, and immunoblotting has been used to investigate the conservation of these lipoproteins among other staphylococcal species.
MATERIALS AND METHODS
Staphylococcal strains and growth conditions.
S. aureus, S. epidermidis, Staphylococcus hominis, Staphylococcus cohni, Staphylococcus lugdunensis, Staphylococcus warneri, and Staphylococcus haemolyticus clinical isolates were obtained from the University and City Hospital NHS Trusts, Nottingham, United Kingdom. S. aureus BB (originally isolated from a case of bovine mastitis [8]) and 8325-4 were provided by J. P. Arbuthnott. Strains were maintained by regular subculture on horse blood agar.
For broth culture, strains were grown statically for 18 h at 37°C in RPMI 1640 tissue culture medium containing 2 mg of NaHCO3 per ml. Cultures were incubated in 5% CO2 in air, and where indicated the medium was supplemented with 20 μM Fe2(SO4)3 to produce iron-rich growth conditions.
Polyclonal and monoclonal antibody production.
Anti-S. aureus BB wall antibodies were raised in adult female BALB/c mice. The cell wall extract was prepared by digestion of whole bacterial cells, grown under iron-restricted conditions, with lysostaphin in the presence of 30% (wt/vol) raffinose (36, 41). Mice were bled 2 weeks after the third immunization. Spleens recovered from these mice were also used to generate hybridomas by fusion with the myeloma cell line NS0 by standard methods. Hybridomas secreting antistaphylococcal antibodies were selected by indirect enzyme-linked immunosorbent assay with S. aureus BB wall extract as the antigen, and antigen specificity was confirmed by immunoblotting.
Rabbit monospecific antisera raised against the S. epidermidis 32- and 36-kDa iron-regulated cytoplasmic membrane proteins were available from earlier studies (36, 41).
Immunoelectron microscopy.
S. epidermidis 901 was grown overnight in RPMI 1640. Bacteria were pelleted and fixed by resuspension in 1% (vol/vol) gluteraldehyde in phosphate-buffered saline, pH 7.4 (PBS) at room temperature for 2 h. Fixed bacteria were washed three times in PBS and dehydrated in a graded series of alcohol solutions (25 to 100% [vol/vol] in H2O). Bacterial pellets were then embedded in Lowicryl K4M resin (Agar Scientific, Essex, United Kingdom) by using UV light polymerization, and thin sections were prepared with a Reichert OMU3 ultramicrotome. Sections were transferred to carbon-coated copper grids, and nonspecific binding sites were blocked by incubation with 3% (wt/vol) bovine serum albumin (BSA) in PBS for 30 min at room temperature. Grids were then incubated with either a 1/10 dilution (in PBS plus 1% [wt/vol] BSA) of polyclonal monospecific rabbit antiserum to the S. epidermidis 32-kDa lipoprotein or a similar dilution of rabbit serum from a nonimmune animal for 1 h at room temperature. Grids were then washed in PBS and further incubated with a 1/20 dilution (in PBS plus 1% [wt/vol] BSA) of 15-nm-diameter-gold-conjugated protein A (Biocell) for 30 min at room temperature. Following washing in PBS, grids were poststained with 4% (wt/vol) uranyl acetate and lead citrate in H2O and examined in a Jeol Jem100C transmission electron microscope at 80 kV.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS PAGE) and immunoblotting.
All samples were solubilized by being boiled in Laemmli sample buffer (23) for 5 min. Polypeptides were separated by electrophoresis with a 4% (wt/vol) acrylamide stacking gel and a 10% (wt/vol) resolving gel in a Bio-Rad Mini Protean II gel apparatus as previously described (3). Where indicated, gels were stained with silver according to the method of Wray et al. (42). For immunoblotting, polypeptides were transferred to BioTrace NT membranes (Gelman); this was followed by blocking, incubation with primary antibody (a 1/500 dilution overnight for polyclonal antisera or a 1/2 dilution overnight for monoclonal antibodies) and conjugate (a 1/2,000 dilution of antirabbit, antirat, or antimouse peroxidase conjugates for 4 h), and detection of bound antibody as previously described (3).
Triton X-114 extraction.
Triton X-114 extractions were performed according to the method of Bordier (6) with minor modifications. Where appropriate, staphylococci were first digested with 100 μl of lysostaphin (80 μg/ml in PBS) for 15 min at 37°C, cooled on ice, and sonicated twice for 30 s at 8 mA on an MSE Soniprep sonicator fitted with a 3-mm-diameter probe. PBS (800 μl) and 10% (vol/vol) Triton X-114 (100 μl in PBS) were then added to each tube, and the tubes were then incubated at 4°C for 2 h. After incubation at 4°C, bacterial preparations were centrifuged (13,000 × g for 10 min at 4°C) to pellet insoluble debris. Supernatants were transferred to fresh tubes and incubated at 37°C for 30 min to allow phase separation. The detergent phase and associated proteins were then pelleted by centrifugation at room temperature, and the aqueous phase was removed and discarded. The detergent pellet was washed once in 1 ml of PBS at 4°C for 1 h and repelleted following incubation at 37°C for 30 min.
Lipoproteins present in culture supernatants were also extracted with Triton X-114. One hundred microliters of 10% (vol/vol) Triton X-114 in PBS was added to 5 ml of filter-sterilized culture supernatant, which was then processed as described above.
All Triton X-114 pellets were diluted 1:1 with H2O prior to solubilization for SDS-PAGE.
[3H]palmitic acid labelling of staphylococcal lipoproteins.
Staphylococci were grown in 3-ml volumes of RPMI 1640 in the presence of 0.3 MBq of 9,10-[3H](N)-palmitic acid (Dupont) for 24 h at 37°C. Bacteria were pelleted and lysed by digestion with lysostaphin, and whole cells were analyzed by SDS-PAGE. Alternatively, lipoproteins were extracted from lysostaphin-digested bacterial cells or filtered culture supernatants with Triton X-114 prior to analysis. Following SDS-PAGE, gels were soaked in Amplify (Amersham), dried, and exposed to X-OMAT AR film (Kodak) for 6 weeks.
Construction and screening of genomic DNA libraries.
Genomic DNA was prepared from staphylococci by the cetyltrimethylammonium bromide method described by Ausubel et al. (5). A genomic library of random EcoRI fragments of S. epidermidis 901 DNA was constructed in the phage vector Lambda Zap II (Stratagene) according to the manufacturer’s instructions. Recombinant phage were plated on Escherichia coli XLI-Blue, and plaques were transferred to nitrocellulose filters. Plaques were screened for expression of staphylococcal antigens by incubation with monospecific rabbit antisera raised to the 32- and 36-kDa S. epidermidis iron-regulated cytoplasmic membrane proteins (36) or with mouse anti-S. aureus BB cell wall antiserum. Bound antibody was detected by using appropriate species-specific peroxidase-conjugated antibodies, H2O2, and 4-chloro-1-naphthol (3). Reactive plaques were purified, and DNA inserts were recovered by in vivo excision into E. coli SOLR. Expression of staphylococcal antigens in E. coli SOLR was confirmed by immunoblotting whole bacterial cell lysates prepared from overnight broth cultures supplemented with 50 μg of ampicillin per ml. Recombinant plasmids were recovered by alkaline lysis minipreparation and subjected to restriction analysis. Inserts from positive clones were sequenced with an ABI automated DNA sequencer. Comparisons of the deduced protein primary sequences were performed on Seqnet. The OWL nonredundant protein database, searched by using NEW-SWEEP 2.00 (by Alan Bleasby), Tmbase and Tmpred (19), and PRINTS (4), was used to analyze protein sequences, and FASTA (9) and LASERGENE software (DNAstar Inc.) was used to analyze DNA sequences.
Southern and Northern blot analyses.
Staphylococcal chromosomal DNA was digested with EcoRI, electrophoresed, and transferred to a Hybond N+ membrane. The blot was incubated with a digoxigenin-labelled probe (Boehringer Mannheim) obtained by random priming of the 5.4-kb EcoRI fragment from pW32. Hybridization was performed at 42°C overnight, and blots were washed sequentially in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) SDS at room temperature and in 0.5× SSC–0.1% (wt/vol) SDS at 68°C according to the protocol of the manufacturer (Boehringer Mannheim). Bound probe was visualized with nitroblue tetrazolium salt color substrate (Boehringer Mannheim) according to the manufacturer’s instructions. A 2-kb NdeI-EcoRI fragment from pW32 containing most of the ABC operon was labelled with digoxigenin and used as a probe for Northern blot analysis. RNA was extracted from S. epidermidis 901 grown for 18 h under iron-rich or iron-restricted conditions in RPMI 1640 with a Qiagen Rneasy total RNA kit. Northern blotting was performed as described by Ausubel et al. (5). Digoxigenin-labelled RNA markers (Boehringer Mannheim) were electrophoresed on each gel.
Nucleotide sequence accession number.
The DNA sequence of the S. epidermidis ABC transporter is available in the GenBank database under accession no. X99127.
RESULTS
Growth of staphylococci under iron-restricted conditions in RPMI 1640.
In our previous studies on iron-regulated proteins of staphylococci, we used iron chelators such as ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) to restrict iron availability in standard broth media or pooled peritoneal dialysate fluid as an iron-restricted growth medium (28, 41). In the present studies we adopted a different approach and used a commercially available, intrinsically iron-restricted and defined growth medium, RPMI 1640, to investigate the phenotype of staphylococci in response to iron limitation. Our preliminary results indicated that all staphylococcal isolates tested grew readily in this medium, with or without supplementation with 20 μM Fe2(SO4)3. Qualitatively, the expression of iron-regulated proteins under iron-restricted conditions was essentially identical to that we had observed in our earlier studies (data not shown). Consequently, we used RPMI 1640 as a more convenient medium for growth of staphylococci for the present studies.
Extraction of iron-regulated staphylococcal cytoplasmic membrane proteins with Triton X-114.
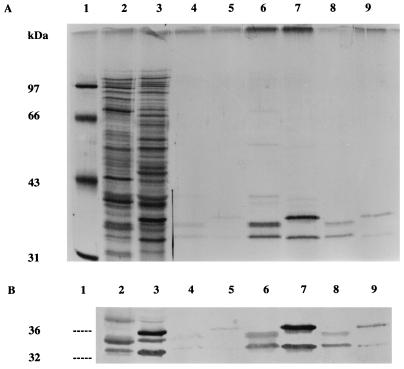
Triton X-114 phase partitioning provided preliminary evidence that the iron-regulated cytoplasmic membrane proteins in S. epidermidis and S. aureus may be lipoproteins. Extraction of lysed bacteria grown under iron-restricted conditions identified a subset of bacterial proteins partitioning into the detergent phase (Fig. 1A). Two major polypeptides of 32 and 36 kDa and of 32 and 35 kDa were detected in extracts of S. epidermidis and S. aureus, respectively. These polypeptides were not detected in the aqueous phase following detergent partitioning (data not shown). The relationship of these Triton X-114-extractable polypeptides to the previously described staphylococcal iron-regulated cytoplasmic membrane proteins was confirmed by immunoblotting with monospecific rabbit antisera (Fig. 1B). The minor differences between the molecular mass stated for one of the S. aureus proteins in the present study and those stated in our previous studies (36, 41) are attributable to minor variations in the electrophoresis technique and the source of molecular mass markers used.
FIG. 1.
SDS-PAGE (A) and immunoblots (B) of Triton X-114 extracts prepared from S. aureus BB and S. epidermidis 901 grown under iron-restricted growth conditions. Lanes: 1, molecular mass markers; 2 and 3, whole-cell polypeptide profiles; 4 and 5, Triton X-114 extracts of intact cells; 6 and 7, Triton X-114 extracts of lysostaphin-digested cells; lanes 8 and 9, Triton X-114 extracts of filtered culture supernatants; 2, 4, 6, and 8, S. aureus BB; 3, 5, 7, and 9, S. epidermidis 901. The gel in panel A was silver stained, and immunoblots were reacted with a pool of monospecific rabbit antisera to the 32- and 36-kDa iron-regulated cytoplasmic membrane proteins of S. epidermidis (36).
Only trace quantities of these proteins were extractable from lysed bacteria grown under iron-rich growth conditions (data not shown) or from intact iron-restricted cells (Fig. 1A), indicating their regulation by iron availability and the restricted exposure of detergent-soluble forms of these proteins on the surfaces of intact staphylococci.
Triton X-114 extraction of filtered S. epidermidis culture supernatants showed that both the 32- and 36-kDa polypeptides were also present in supernatants from iron-restricted cultures but that these polypeptides were not detectable in iron-rich culture supernatants (data not shown). In contrast, small quantities of Triton X-114-extractable polypeptides of 32 and 35 kDa were found in supernatants of iron-rich S. aureus cultures (data not shown), although larger amounts of these polypeptides were detected in iron-restricted supernatants (Fig. 1A).
[3H]Palmitic acid labelling studies.
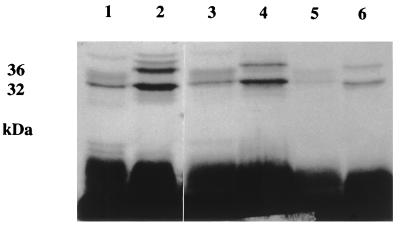
Confirmation of the identity of these major Triton X-114-extractable iron-regulated proteins as lipoproteins was provided by results of labelling studies using [3H]palmitic acid. The 32- and 36-kDa polypeptides of S. epidermidis and the 32- and 35-kDa polypeptides detected in detergent extracts of S. aureus were radiolabelled when the bacteria were grown in RPMI 1640 under iron-restricted conditions (Fig. 2). Labelled polypeptides with these molecular masses were also detected in Triton X-114 extracts of the respective filtered culture supernatants from both staphylococcal species (Fig. 2).
FIG. 2.
Autoradiograph showing [3H]palmitate labelling of Triton X-114-extractable lipoproteins of S. aureus BB and S. epidermidis 901. Lanes: 1 and 2, whole-cell [3H]palmitate-labelled profiles; 3 and 4, Triton X-114 extracts of [3H]palmitate-labelled lysostaphin-digested cells; 5 and 6, Triton X-114 extracts of [3H]palmitate-labelled filtered culture supernatants; 1, 3, and 5, S. aureus BB; 2, 4, and 6, S. epidermidis 901.
Antigenic relatedness between the staphylococcal lipoproteins.
A mouse monoclonal antibody raised against the S. aureus BB 32-kDa lipoprotein was used in immunoblotting studies to investigate the conservation of this antigen among a range of staphylococcal species. All bacteria were grown in RPMI 1640 under iron-restricted conditions. A polypeptide of the appropriate size was detected in Triton X-114 extracts of each of four S. epidermidis strains and all seven strains of S. aureus tested (data not shown). A 40-kDa polypeptide was detected in the single strain of S. hominis tested, but no reaction with this antibody was observed by immunoblotting with single strains of S. lugdunensis, S. cohni, and S. haemolyticus (data not shown).
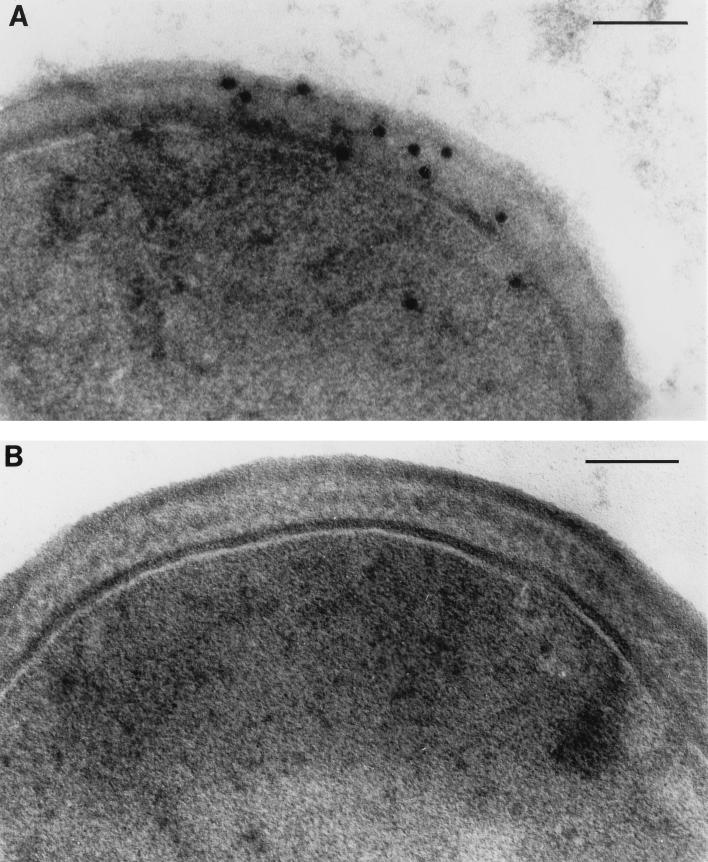
Localization of the 32-kDa lipoprotein in S. epidermidis by immunoelectron microscopy.
Immunogold labelling of thin sections of iron-restricted bacteria with a rabbit monospecific antibody to the S. epidermidis 32-kDa lipoprotein was used to further investigate the distribution of this antigen in the S. epidermidis cell. Figure 3A shows that the majority of the 32-kDa lipoprotein is distributed throughout the cell wall of S. epidermidis. Smaller quantities of this protein were located in the cytoplasmic membrane, and exposure of the antigen on the cell surface was minimal. Sections incubated with nonimmune rabbit serum showed minimal labelling with protein A-gold (Fig. 3B).
FIG. 3.
Electron micrographs of thin sections of S. epidermidis 901 cells reacted with monospecific antiserum to the 32-kDa S. epidermidis lipoprotein. (A) Sections reacted with monospecific rabbit anti-32-kDa lipoprotein antibody and protein A-gold conjugate; (B) sections reacted with nonimmune rabbit serum and protein A-gold conjugate. Bars, 1 μm.
Molecular cloning of the gene encoding the 32-kDa S. epidermidis lipoprotein.
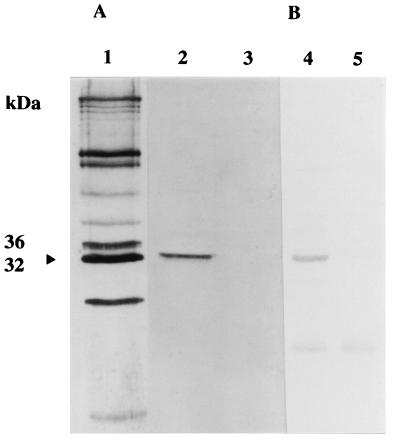
To further characterize the staphylococcal lipoproteins and investigate their potential functions, we sought to isolate the genes encoding these antigens from staphylococcal genomic DNA libraries. Initial antibody screening of an S. epidermidis 901 Lambda Zap II library with monospecific antisera to the 32- and 36-kDa iron-regulated cytoplasmic membrane lipoproteins failed to identify reactive plaques. Since our previous studies had indicated some antigenic cross-reactivity between iron-regulated proteins in different staphylococcal species (36), we rescreened the library with anti-S. aureus BB wall antibodies. This antiserum was raised against native staphylococcal antigen, in contrast to the denatured antigen used in the preparation of the monospecific anti-32- and anti-36-kDa rabbit sera. This second screen identified several plaques reactive with antistaphylococcal antibodies. Following plaque purification and in vivo excision of one of these plaques, a polypeptide of approximately 32 kDa was detected on immunoblots of an E. coli SOLR lysate probed with anti-S. aureus BB wall antiserum (Fig. 4A). The identity of the polypeptide as the S. epidermidis 32-kDa iron-regulated cytoplasmic membrane lipoprotein was confirmed by immunoblotting with monospecific anti-32-kDa antiserum (Fig. 4B). The plasmid encoding the 32-kDa lipoprotein was designated pW32.
FIG. 4.
Immunoblots of recombinant E. coli SOLR pW32 reacted with anti-S. aureus BB wall antiserum (A) or monospecific anti-32-kDa S. epidermidis lipoprotein antiserum (B). Lanes: 1, BB wall extract; 2 and 4, E. coli SOLRpW32 whole-cell extract; 3 and 5, E. coli SOLR whole-cell extract. The position of the 32-kDa lipoprotein antigen is indicated by the arrowhead.
Characterization of the cloned DNA fragment.
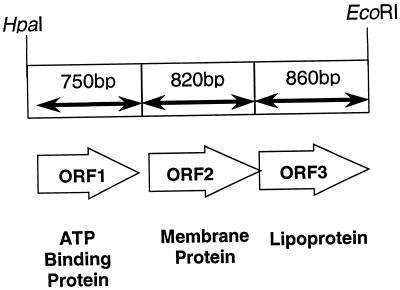
Restriction endonuclease mapping and subcloning of pW32 localized the gene encoding the 32-kDa lipoprotein to a 2.43-kb HpaI-EcoRI fragment. The plasmid containing this fragment was designated pW33. The entire DNA fragment was sequenced on both strands following synthesis of appropriate DNA primers, and three open reading frames encoding putative polypeptides of 28, 35, and 30 kDa were identified. We have designated the genes encoding these polypeptides sitA, -B, and -C, respectively (sit is for staphylococcal iron transport). The organization of these genes in pW33 is shown in Fig. 5.
FIG. 5.
Organization of the S. epidermidis ABC transporter operon.
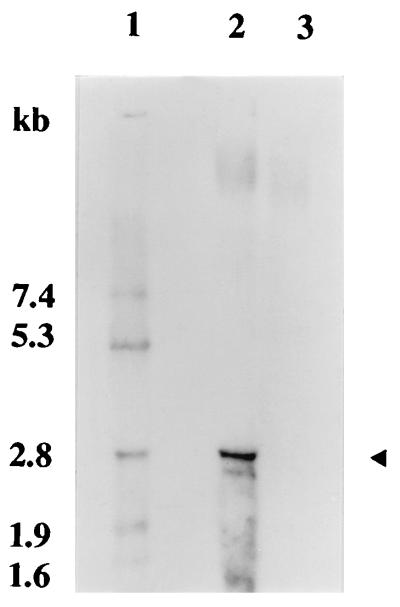
RNA transcript analysis by Northern blotting showed a single, iron-repressible RNA transcript of ∼2.7 kb, indicating that the operon containing sitA, -B, and -C is transcribed as a polycistronic mRNA (Fig. 6).
FIG. 6.
Northern blot showing transcript analysis of the S. epidermidis ABC transporter operon and differential transcription under iron-rich and iron-restricted growth conditions. Lanes: 1, molecular size markers; 2, RNA extracted from iron-restricted culture; 3, RNA extracted from iron-rich culture. The arrowhead indicates the position of the single 2.7-kb RNA transcript detected in S. epidermidis 901.
Southern blotting was used to investigate conservation of this operon among other staphylococcal strains. Probing with the 5.4-kb EcoRI insert from pW32 identified single copies of these genes in genomic DNAs from S. epidermidis 901 and 138; however, no signal was detected when DNA from S. aureus BB or 8325-4, S. lugdunensis, S. cohni, or S. warneri was similarly probed (data not shown). In addition, although these genes could be amplified by PCR with appropriate sequencing primer pairs and S. epidermidis genomic DNA as the template, no products could be obtained when DNAs from the other staphylococcal isolates were used (data not shown).
The cloned S. epidermidis DNA locus shows significant homology with bacterial ABC transporters.
Sequence comparisons indicated significant homologies at the DNA and protein levels between sitA, -B, and -C and their products and sequences of previously identified bacterial ABC transporters. Sequence comparisons yielding the highest homologies at the amino acid level are shown in Table 1.
TABLE 1.
Sequence homologies of the S. epidermidis ABC transporter operon proteins with similar proteins in databases
| Protein type (S. epidermidis protein) | Accession no. | % Identity | Organism(s) | Protein/functiona |
|---|---|---|---|---|
| ATP binding protein (SitA) | X99127 | 100 | S. epidermidis | ATP binding protein/Fe transport? |
| L11577 | 43 | Streptococcus gordonii | ScaA-associated ATP binding protein/? | |
| M26130 | 46 | Streptococcus parasanguis | FimA-associated ATP binding protein/? | |
| U55214 | 39 | Treponema pallidum | TroB/? | |
| U52850 | 38 | Erysipelothrix rhusiopathiae | EwlA-associated ATP binding protein/? | |
| L45003 | 34 | H. influenzae | FecE/iron(III) dicitrate transport | |
| L34630 | 33 | Synechocystis spp. | MntA/Mn transport | |
| Membrane protein (SitB) | X99127 | 100 | S. epidermidis | Membrane protein/Fe transport? |
| M26130 | 48 | Streptococcus parasanguis | FimA-associated membrane protein/? | |
| L11577 | 46 | Streptococcus gordonii | ScaA-associated membrane protein/? | |
| L45001 | 39 | H. influenzae | HI0359 membrane protein/? | |
| L34630 | 39 | Synechocystis spp. | MntB membrane protein/Mn transport | |
| U50597 | 38 | Y. pestis | YfeC membrane protein/Fe transport? | |
| Lipoprotein (SitC) | X99127 | 100 | S. epidermidis | Lipoprotein/Fe transport? |
| L19055 | 50 | Streptococcus pneumoniae | PapA lipoprotein/adhesion? | |
| U03756 | 49 | Enterococcus faecalis | EfaA/adhesion? | |
| M26130 | 50 | Streptococcus parasanguis | FimA/saliva binding protein | |
| U46542 | 48 | Streptococcus crista | ScbA/adhesion or transport? | |
| L11577 | 48 | Streptococcus gordonii | ScaA/coaggregation adhesion |
A question mark indicates that the function of the protein is presently unknown.
SitA shows significant homology with known or theoretical ATP binding proteins in a number of bacterial species, including Streptococcus gordonii and Streptococcus parasanguis. S. epidermidis SitA contains consensus ATP binding motifs, including a P loop (GPNGAGKS) (33). Secondary-structure analysis failed to identify potential membrane-spanning regions in this protein, suggesting a cytoplasmic location for the mature protein. Interestingly, SitA also shows homology with an ATP binding protein, FecE, involved in iron(III) dicitrate transport in the gram-negative pathogen Haemophilus influenzae.
SitB has significant homology with several ABC transporter membrane proteins, including those of Streptococcus gordonii and Streptococcus parasanguis. Other strong homologies include those with MntB, a protein involved in Mn2+ transport in Synechocystis spp., and YfeC, a putative iron transport protein in Yersinia pestis. Secondary-structure analysis identified seven hydrophobic transmembrane-spanning regions in the staphylococcal polypeptide, supporting its likely identity as a cytoplasmic membrane protein. This S. epidermidis membrane protein lacks an ATP binding motif, a feature considered to be characteristic of ABC importers rather than exporters (13).
SitC, the 32-kDa S. epidermidis lipoprotein, shows significant homology with other ABC transporter lipoproteins, including EfaA, ScaA, and FimA in Enterococcus faecalis, Streptococcus gordonii, and Streptococcus parasanguis, respectively. These proteins are all thought to play a role in bacterial adherence, and all contain recently described conserved motifs that define the family of bacterial adhesins designated ADHESINFAMILY or related periplasmic binding proteins (4). The staphylococcal lipoprotein has a consensus prelipoprotein signal peptide cleavage sequence (17) (ILAACG), with the cysteine residue at position 18, a finding which confirms our biochemical analysis, and also contains an ATP binding motif (GWFEKALDQAGKSTKDKN) (33).
DISCUSSION
The data presented here extend our previous observations on iron-regulated proteins in the staphylococci (36, 41). The iron-regulated cytoplasmic membrane proteins in S. epidermidis and S. aureus have been identified as lipoproteins, and the 32-kDa S. epidermidis lipoprotein has been shown to be a component of a novel, iron-regulated ABC transporter which shows organizational and sequence homologies with previously described transporters in several gram-positive and gram-negative bacterial pathogens.
At present it is unclear which specific solute(s) is bound by the 32-kDa lipoprotein in S. epidermidis. However, the iron-regulated nature of this ABC transporter provides further support for our previous suggestion that this staphylococcal cytoplasmic membrane lipoprotein may be involved in iron transport. This suggestion is supported by recent work by Dintilhac et al. (10, 11), who examined the DNA sequence of the S. epidermidis ABC transporter reported here and found structural similarities with a new family of solute binding proteins (cluster 9) that is distinct from the eight other families previously described by Tam and Saier (39). Members of cluster 9 share the property of proven or hypothetical binding of metallic cations, including Fe, Mn, and Zn. Our own recent unpublished data suggest that the S. epidermidis operon may also be regulated by Mn2+ availability, raising the possibility that the 32-kDa S. epidermidis lipoprotein may be able to bind and transport a variety of metal ions. We are currently attempting to generate S. epidermidis mutants to address this possibility.
The spatial distribution of the 32-kDa lipoprotein within the cell wall of S. epidermidis as shown by immunoelectron microscopy is consistent with those of other solute binding lipoproteins in gram-positive bacteria. These proteins are anchored to the cytoplasmic membrane by a lipid-modified N terminus, with the solute-binding component projecting into or in some cases through the cell wall where they are exposed on the bacterial surface. However, the observation that SitC is located primarily within the cell wall of S. epidermidis initially appears to be at variance with results from our earlier cell fractionation studies which showed that this protein was associated with the staphylococcal cytoplasmic membrane (36, 41). This apparent anomaly can be resolved if, following removal of the cell wall by lysostaphin treatment, SitC remains tethered to the cytoplasmic membrane by its lipid tail. The lipoprotein would then be pelleted with the cytoplasmic membrane fraction of the lysed staphylococcal cells.
This distribution would potentially allow the 32-kDa lipoprotein to function in both siderophore-dependent and -independent iron acquisition pathways. The lipoprotein could act as a ferric siderophore receptor either exposed on the cell surface or within the porous matrix of the cell wall, into which low-molecular-mass molecules such as siderophores readily penetrate. The involvement of lipoproteins in ferrihydroxamate siderophore iron uptake has been previously reported for Bacillus subtilis (35), and more recent studies with Corynebacterium diphtheriae IRP1 suggest that this iron-regulated lipoprotein may also function as a siderophore receptor (34). Alternatively, the 32-kDa S. epidermidis lipoprotein may be involved in shuttling iron from receptor-bound transferrin or lactoferrin across the cell wall to the cytoplasmic membrane prior to uptake into the cell. Experiments to identify the solute specificity of the 32-kDa lipoprotein in S. epidermidis are under way.
Our immunoblotting data obtained by using a monoclonal antibody specific for the 32-kDa lipoprotein are consistent with our earlier studies which showed that this antigen was conserved among S. epidermidis and S. aureus isolates but was generally not expressed by other staphylococcal species (36, 41). The other staphylococci do, however, produce one or more major lipoproteins in the 30- to 40-kDa size range as shown by SDS-PAGE analysis of Triton X-114 extracts (7), and we speculate that these lipoproteins may also be components of ABC transporters in these species. Based on these data, it also seems likely that the ∼36-kDa and 35-kDa iron-regulated membrane lipoproteins produced by S. epidermidis and S. aureus, respectively, may also be components of other ABC transporters which may also have a role in iron acquisition. We are currently attempting to clone the genes encoding these lipoproteins to confirm this possibility.
Despite the strong antigenic and potential functional homology between the 32-kDa lipoproteins of S. epidermidis and S. aureus, Southern blot and PCR analyses surprisingly failed to detect any homology between the genes encoding these proteins at the DNA level under the experimental conditions used. A previous study has indicated that even within the highly conserved staphylococcal gene encoding HSP60, species-specific variations exist which are sufficient to allow species differentiation by hybridization techniques (16). It is therefore conceivable that such species-specific variation within the staphylococcal ABC transporter gene sequences may also exist and may account for our findings.
Interestingly, the 32-kDa S. epidermidis lipoprotein also shows significant sequence homology with a number of proteins known to be adhesins in other gram-positive bacteria. In these other organisms, the lipoproteins are clearly surface exposed and available for interaction with appropriate tissue or other surface receptors (20, 37). The limited exposure of the 32-kDa antigen on the surface of S. epidermidis as assessed by electron microscopy suggests, however, that it is unlikely that this lipoprotein plays a significant role in the adhesion of S. epidermidis. However, the observation that in both S. epidermidis and S. aureus, significant quantities of membrane lipoproteins are released into culture supernatants under iron-restricted growth conditions in vitro and in vivo (7) may indicate an extracellular function for these lipoproteins. The mechanism of release of the lipoproteins from the staphylococcal surface is presently unclear, but our Triton X-114 phase partitioning and [3H]palmitate labelling studies indicate that the released lipoproteins remain acylated. Similar findings have been reported for extracellular lipoproteins from Streptococcus mutans (37). A possible toxic role for the staphylococcal lipoproteins is supported by recent studies which indicate that membrane proteins of L forms of S. aureus may stimulate cytokine release from mammalian cells in in vitro assays and may be lethal when administered to d-galactosamine-treated mice (1). The relationship between the lipoproteins described in the present studies and the toxic membrane proteins of 30 and 36 kDa isolated by Akashi et al. (1) is presently unclear but merits further investigation, since other bacterial lipoproteins have previously been shown to induce cytokine release from mammalian cells (15).
In addition to their potential toxic activity, the extracellular lipoproteins also induce a significant humoral antibody response in rats with chamber implants inoculated with staphylococci (27), and variable levels of antibodies to these lipoproteins are detectable in human sera from individuals with staphylococcal infection (36). This observation raises the serodiagnostic potential of these lipoproteins for some types of staphylococcal disease.
ACKNOWLEDGMENT
This work was supported by Programme Grant G9219778 from the Medical Research Council to P.W.
REFERENCES
- 1.Akashi A, Ono S, Kuwano K, Arai S. Proteins of 30 and 36 kilodaltons, membrane constituents of the Staphylococcus aureus L form, induce production of tumor necrosis factor alpha and activate human immunodeficiency virus type 1 long terminal repeat. Infect Immun. 1996;64:3267–3272. doi: 10.1128/iai.64.8.3267-3272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The amino acid sequence of Staphylococcus aureus penicillinase. Biochem J. 1975;151:197–218. doi: 10.1042/bj1510197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbuthnott J P, Arbuthnott E, Arbuthnott A D J, Pike W J, Cockayne A. Investigation of microbial growth in vivo: evaluation of a novel in vivo chamber implant system. FEMS Microbiol Lett. 1992;100:75–80. doi: 10.1111/j.1574-6968.1992.tb14022.x. [DOI] [PubMed] [Google Scholar]
- 4.Attwood T K, Beck M E, Bleasby A J, Degtyyarenko K, Michie A D, Parry-Smith D J. Novel developments with the PRINTS protein fingerprint database. Nucleic Acids Res. 1997;25:212–216. doi: 10.1093/nar/25.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1994. [Google Scholar]
- 6.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 7.Cockayne, A., S. Tandon, and P. Williams. Unpublished data.
- 8.Derbyshire J B. Further immunological studies in experimental staphylococcal mastitis. J Comp Pathol. 1961;71:146–158. doi: 10.1016/s0368-1742(61)80019-2. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dintilhac A, Claverys J P. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148:119–131. doi: 10.1016/S0923-2508(97)87643-7. [DOI] [PubMed] [Google Scholar]
- 11.Dintilhac A, Alloing G, Granadel C, Claverys J-P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 12.Domanski T L, Bayles K W. Analysis of Staphylococcus aureus genes encoding penicillin-binding protein 4 and an ABC-type transporter. Gene. 1995;167:111–113. doi: 10.1016/0378-1119(96)82965-9. [DOI] [PubMed] [Google Scholar]
- 13.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenno J C, Shaikh A, Spatafora G, Fives-Taylor P. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol Microbiol. 1995;15:849–863. doi: 10.1111/j.1365-2958.1995.tb02355.x. [DOI] [PubMed] [Google Scholar]
- 15.Ganeshkumar N, Arora N, Kolenbrander P E. Saliva-binding protein (SsaB) from Streptococcus sanguis 12 is a lipoprotein. J Bacteriol. 1993;175:572–574. doi: 10.1128/jb.175.2.572-574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh S H, Potter S, Wood J O, Hemmingsen S M, Reynolds R P, Chow A W. HSP60 gene sequences as universal targets for microbial species identification—study with coagulase-negative staphylococci. J Clin Microbiol. 1996;34:818–823. doi: 10.1128/jcm.34.4.818-823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 18.Henderson B, Poole S, Wilson M. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol Rev. 1996;60:316–341. doi: 10.1128/mr.60.2.316-341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann K, Stoffel W. Tmbase—a database of membrane spanning protein segments. Biol Chem. 1993;347:166. [Google Scholar]
- 20.Jenkinson H F. Adherence, coaggregation, and hydrophobicity of Streptococcus gordonii associated with expression of cell surface lipoproteins. Infect Immun. 1992;60:1225–1228. doi: 10.1128/iai.60.3.1225-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkinson H F. Adherence and accumulation of oral streptococci. Trends Microbiol. 1994;2:209–212. doi: 10.1016/0966-842x(94)90114-k. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander P E, Andersen R N, Ganeshkumar N. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene scaA, an ATP-binding cassette. Infect Immun. 1994;62:4469–4480. doi: 10.1128/iai.62.10.4469-4480.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay J A, Riley T V. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect Immun. 1994;62:2309–2314. doi: 10.1128/iai.62.6.2309-2314.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe A M, Lambert P A, Smith A W. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer C, Bierbaum G, Heidrich C, Reis M, Suling J, Iglesisas-Wind M, Kempter C, Molitor E, Sahl H. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis for a role of PepC in thioether formation. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- 27.Modun B, Williams P, Pike W J, Cockayne A, Arbuthnott J P, Finch R, Denyer S P. Cell envelope proteins of Staphylococcus epidermidis grown in vivo in a peritoneal chamber implant. Infect Immun. 1992;60:2551–2553. doi: 10.1128/iai.60.6.2551-2553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modun B, Kendall D, Williams P. Staphylococci express a receptor for human transferrin: identification of a 42-kilodalton cell wall transferrin-binding protein. Infect Immun. 1994;62:3850–3858. doi: 10.1128/iai.62.9.3850-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neikirk Andersen R, Ganeshkumar N, Kolenbrander P E. Cloning of the Streptococcus gordonii PK488 gene, encoding an adhesin which mediates coaggregation with Actinomyces naeslundii PK606. Infect Immun. 1993;61:981–987. doi: 10.1128/iai.61.3.981-987.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen J B K, Lampen J O. Membrane bound penicillinases in Gram positive bacteria. J Biol Chem. 1982;257:4490–4495. [PubMed] [Google Scholar]
- 31.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross J I, Eady E A, Cove J H, Cunliffe W J, Wooton J C. Inducible erythromycin resistance in staphylococcus is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990;4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 33.Saraste M, Sibbald P R. The P-loop—a common motif in ATP-binding and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt M P, Talley B G, Holmes R K. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:5364–5367. doi: 10.1128/iai.65.12.5364-5367.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider R, Hantke K. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol Microbiol. 1993;8:111–121. doi: 10.1111/j.1365-2958.1993.tb01208.x. [DOI] [PubMed] [Google Scholar]
- 36.Smith D G E, Wilcox M H, Williams P, Finch R G, Denyer S P. Characterization of the cell envelope proteins of Staphylococcus epidermidis cultured in human peritoneal dialysate. Infect Immun. 1991;59:617–624. doi: 10.1128/iai.59.2.617-624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutcliffe I C, Tao L, Ferretti J J, Russell R R B. MsmE, a lipoprotein involved in sugar transport in Streptococcus mutans. J Bacteriol. 1993;175:1853–1855. doi: 10.1128/jb.175.6.1853-1855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam R, Saier M H J. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viscount H B, Munro C L, Burnette-Curley D, Peterson D L, Macrina F L. Immunization with FimA protects against Streptococcus parasanguis endocarditis in rats. Infect Immun. 1997;65:994–1002. doi: 10.1128/iai.65.3.994-1002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcox M H, Williams P, Smith D G E, Finch R G, Denyer S P. Variation in the expression of cell envelope proteins of coagulase-negative staphylococci cultured under iron-restricted conditions in human peritoneal dialysate. J Gen Microbiol. 1991;137:2561–2570. doi: 10.1099/00221287-137-11-2561. [DOI] [PubMed] [Google Scholar]
- 42.Wray W, Bonlikas T, Wray V P, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]