Abstract
A number of gene silencing phenomena that inactivate genes at the post-transcriptional level have been identified. Due to its potential for studying gene function, post-transcriptional gene silencing (PTGS) has become an intense area of research. In this review we describe the different means of inducing PTGS and discuss the possible biological roles of these artificially induced phenomena. We also discuss other features of PTGS such as the mechanism of mRNA degradation, the nature of the silencing signal and the mechanism of PTGS inhibition by viral proteins.
Introduction
The introduction of transgenes or double-stranded RNA (dsRNA) into a variety of hosts can trigger post-transcriptional silencing of all homologous host genes and/or transgenes. Transgene-induced post-transcriptional gene silencing (PTGS) has been detected in plants (van der Krol et al., 1990) where it is called co-suppression, and in fungi where it is called quelling (Cogoni and Macino, 2000). In animals, PTGS can be induced by dsRNA in a process called RNA interference (RNAi) (Fire, 1999). In plants, PTGS can also be induced by viruses expressing host genes in a process called virus-induced gene silencing (VIGS) (Baulcombe, 1996). Viruses themselves can be the targets of the PTGS machinery (Baulcombe, 1996).
The principal common feature of all the PTGS phenomena is that they lead to a specific decrease in the level of the mRNA of both the homologous host gene and the introduced transgenes. In fact, most of the PTGS phenomena were identified by the characteristic loss of function of the endogenous gene. In plants, for example, introduction of a chalcone synthase transgene or a dihydroflavonol-4-reductase transgene produces plants with reduced/lack of floral pigmentation (Napoli et al., 1990). Neurospora crassa transformed with an al-1 transgene produces conidia lacking the characteristic orange color as a result of inactivation of the al-1 gene (Cogoni et al., 1996). In Caenorhabditis elegans, the function of a number of genes has been inferred from the phenotype of worms injected with dsRNA complementary to the gene of interest (Fraser et al., 2000). The subsequent decrease in mRNA levels is not due to a specific decrease in transcription (de Carvalho Niebel et al., 1995) but rather to its degradation, as silencing has been correlated with an increase in RNA degradation intermediates (Metzlaff et al., 1997; Holtorf et al., 1999). There is evidence that non-sense mediated mRNA decay (NMD), a mechanism that detects and degrades transcripts with a premature signal for termination of translation (Ruiz-Echevarria et al., 1996), is involved in the PTGS-related degradation of the mRNA, as some C. elegans genes are required for both RNAi and NMD (Domeier et al., 2000). The two mechanisms, however, are not completely overlapping. NMD, for instance, is dependent on translation of the mRNA, while the decrease in mRNA observed in the different PTGS phenomena is not prevented by inhibitors of translation (Holtorf et al., 1999). Also, the mRNA degradation associated with NMD begins with de-capping followed by 5′ to 3′ exonuclease degradation (Ruiz-Echevarria et al., 1996), while the degradation associated with PTGS appears to begin with endonucleotidic cleavage (Elbashir et al., 2001b).
RNA degradation
Insights into the mechanism of PTGS-related RNA degradation have been obtained in vitro. Cell-free extracts from Drosophila embryos were shown to process dsRNA into fragments of 21–23 nucleotides (nt) (Zamore et al., 2000). Similarly, small (25 nt) sense and anti-sense transgenic RNA molecules have been detected in plants undergoing PTGS (Hamilton et al., 1999), suggesting that these small RNAs are involved in silencing in vivo. Consistent with this, a gene called Dicer whose product can produce small RNA (25 nt) molecules from dsRNA was recently shown to participate in RNAi in Drosophila (Bernstein et al., 2001). Furthermore, synthetic 21–23 nt RNAs are sufficient to induce RNAi in vitro (Elbashir et al., 2001a) and in vivo (Elbashir et al., 2001b). In the in vitro system, mRNA homologous to the 21–23 nt RNAs was degraded and appeared as a ladder of bands of 21–23 bases, suggesting that the cleavage was directed by the 21–23 nt RNAs (Zamore et al., 2000). The co-purification of 21–23 nt RNAs with the RNA-induced silencing complex (RISC)—the target mRNA degrading activity—from extracts of Drosophila cells supports this notion (Hammond et al., 2000).
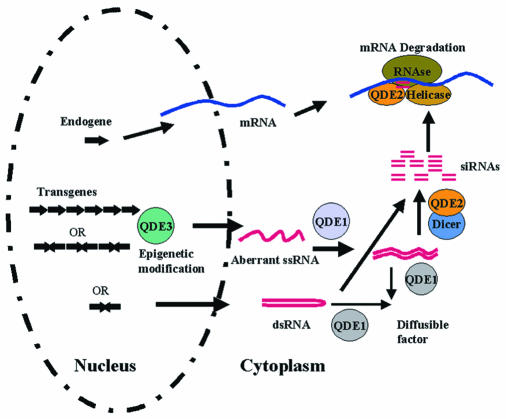
These data are in accord with a model of PTGS whereby dsRNA is processed by a RNase III type enzyme (Dicer) into small dsRNA molecules called small interfering RNAs (siRNAs) (Bass, 2000). siRNAs would direct a nuclease to the target mRNA, but degradation of the mRNA by the small RNA guided nuclease most likely requires other enzymatic activities. For instance, an RNA helicase could be required to denature the double-stranded small RNAs so as to allow pairing between the anti-sense siRNA and the mRNA. Potential candidates for this function include homologues of SDE3, an RNA helicase-like protein that is required for PTGS in plants (Dalmay et al., 2001) and Mut6, a putative DEAH-box RNA helicase that is required for PTGS in Chlamydomonas (Wu-Scharf et al., 2000). Adaptor proteins may also be required to transfer the siRNAs from Dicer to RISC, the activity that actually degrades the target mRNA, as these activities are biochemically separable (Bernstein et al., 2001). The products of the argonaute gene family could carry out this function (Figure 1). Members of this gene family have no obvious enzymatic activity but are required for PTGS in worms (RDE-1), fungi (QDE2) and plants (AGO1) (Fagard et al., 2000a). Consistent with an adaptor function, the products of these genes contain a putative protein–protein interaction motif called PAZ, which is also found in Dicer (Bernstein et al., 2001). Recently, it was reported that one of the Drosophila argonaute family members (AGO2) co-purifies with RISC and also co-immunoprecipitates with Dicer (Hammond et al., 2001), further supporting an adaptor function for this gene family.
Fig. 1. Model for post-transcriptional gene silencing in Neurospora. dsRNA can be produced either directly from inverted repeats or indirectly whereby an RdRP (QDE1) makes a complementary strand of an aberrant single-stranded (ss)RNA produced from the transgene. The aberrant RNA is a result of an epigenetic modification of the transgenic locus driven by the repetitive nature of the transgene and mediated by QDE3. Before Dicer processes the dsRNA into the siRNAs, the dsRNA is used as a template by RdRP (QDE1) to synthesize a diffusible factor. The siRNAs are subsequently transferred by QDE2 from Dicer to the mRNA-degrading enzyme. See text for additional details.
Transgenes and the synthesis of aberrant RNA
What is the nature of the silencing signal? It has long been postulated that the silencing signal is an ‘aberrant RNA’ molecule derived from the transgene, the dsRNA or a virus. This model has led to the search for a particular aberrant RNA, but in contrast to dsRNA-induced PTGS where the aberrant RNA is most likely the dsRNA itself, it is still unknown how the aberrant RNA(s) is (are) generated in transgene-induced PTGS. In fact, truncated RNAs, longer than full-length RNAs, anti-sense RNAs, double-stranded RNAs and chemically modified RNAs have all been found associated with PTGS (Meins, 2000).
Many models have been proposed to explain how transgenes can produce these abnormally processed RNAs (Baulcombe, 1996; Wassenegger and Pelissier, 1998; Meins, 2000) and more than one of them is likely to be correct. Abnormal RNAs such as dsRNAs, for instance, are most probably the result of read-through transcription of transgenes inserted as inverted repeats (IR) (Figure 1). This type of transcript has been detected in plants containing an IR of an anti-sense chalcone synthase transgene (Stam et al., 2000). Abnormal RNA such as truncated RNAs, on the other hand, could be produced as a result of DNA–DNA pairing (Baulcombe and English, 1996) and/or as a result of epigenetic changes at the transgenic loci. Although it would seem unlikely, some evidence suggests that a ‘nuclear step’ may be involved in some cases of PTGS. For instance, not only transcriptional gene silencing, but also PTGS, was released in plants when a silent transgenic plant was crossed with a plant defective in the SWI2/SNF2 chromatin remodeling component (Morel et al., 2000). Further support for a nuclear step comes from the observation that PTGS inhibition in plants by the viral protein Cmv2b requires nuclear localization (Lucy et al., 2000), and that a putative RecQ DNA helicase is required for quelling in Neurospora (Cogoni and Macino, 2000). Unless chromatin remodeling components and DNA helicase have a cytoplasmic function, these observations point to a nuclear step in PTGS.
The common denominator
Since different forms of nucleic acid (dsRNA, viruses and transgenes) can trigger similar silencing phenomena, and transgenes can produce many types of aberrant transcripts, many models of PTGS invoke a unifying silencing signal. It has been postulated that the different silencing phenomena converge at the dsRNA stage, as both transgenes and viruses could potentially produce dsRNA. Viruses may produce dsRNA as replication intermediates (Waterhouse et al., 2001) while transgenes, as described above, can produce dsRNA when inserted as inverted repeats. Viruses and transgenes that do not directly produce dsRNA could still produce dsRNA due to the activity of a RNA-dependent RNA polymerase (RdRP). It is generally envisioned that the aberrant RNAs produced from transgenes or viruses are used as templates by the RdRP to make dsRNA. In this context it is interesting to note that the QDE1 gene in N. crassa (Cogoni and Macino, 1999), the SDE1 gene in Arabidopsis (Dalmay et al., 2000) and the C. elegans EGO1 gene (Smardon et al., 2000) are all required for PTGS and are all homologous to a tomato gene encoding a protein with RdRP activity. The fact that an RdRP is also required for dsRNA-induced PTGS in C. elegans (Smardon et al., 2000) suggests that this enzyme has functions other than the synthesis of dsRNA from an aberrant RNA. Possibly, the RdRP could also be involved in the amplification step of silencing by synthesizing a diffusible silencing factor (see below). Although dsRNA might be the convergence point of multiple PTGS phenomena, it can only be as an intermediate rather than the actual silencing agent, as dsRNA is not readily detected in most cases of PTGS. In fact, dsRNA injected into flies is rapidly converted into small 22 or 23 nt RNAs (Yang et al., 2000). The executers of silencing are most likely these small 22 or 23 nt RNAs as they have been found associated with silencing induced by transgenes in plants (Hamilton et al., 1999), by dsRNA in flies (Yang et al., 2000) and by viruses in plants (Hamilton et al., 1999), and perhaps even by transgenes in fungi (C. Catalanotto, G. Azzalin, G. Macino and C. Cogoni, unpublished results). In fact, as mentioned previously, siRNAs are sufficient to induce RNAi in cultured cells (Elbashir et al., 2001a).
The systemic silencing signal
An interesting aspect of PTGS is the existence of a sequence-specific diffusible factor. The presence of a diffusible silencing factor was first proposed to explain the non-clonally spatial patterns of silencing in plants (Meins, 2000), as well as the spread of the silent state from a region of initiation to the rest of the plant (Voinnet et al., 1998). The existence and specificity of this diffusible signal was established by grafting experiments between silent transgenic plants and non-silenced transgenic plants, as silencing was transmitted from the silenced stock to the non-silenced scion (Palauqui et al., 1997). Quelling in fungi also involves a dominant diffusible factor, as heterokaryons containing a quelled and a non-quelled nucleus exhibit a quelled phenotype (Cogoni et al., 1996). In C. elegans, RNAi can be triggered in the whole worm by either injecting dsRNA into the body cavity or by ingestion of bacteria expressing dsRNA (Timmons et al., 2001). As with quelling, this factor is dominant because in crosses between an animal affected by RNAi and a wild-type animal, the offspring are affected (Grishok et al., 2000). The nature of the diffusible factor remains unknown and it is unlikely to be the siRNAs since recent data showed that these small RNAs are eliminated without affecting the spread of silencing (Mallory et al., 2001). This diffusible factor, which must be nucleic acid based in order to maintain the specificity, could be synthesized from dsRNA by the RdRP and is probably responsible for the amplification of silencing (Figure 1). This would explain how a few molecules of dsRNA are sufficient to trigger RNAi (Fire, 1999) and why an RdRP would be required for dsRNA-induced PTGS.
Inhibitors of PTGS
Once it had been demonstrated that plant viruses are targets of PTGS, the discovery that viruses encode proteins that inhibit PTGS (Voinnet et al., 2000) came as no surprise, for this seems to be a common evasive strategy of many viruses (Van Dyke, 1994). These proteins have been placed into three groups based on the step of PTGS they inhibit (Li and Ding, 2001). One group inhibits PTGS in all tissues of the plant, reversing PTGS to areas where it has been established. Suppression of PTGS by this group of proteins, which include HC-Pro from potyvirus, is associated with inhibition of the accumulation of the PTGS-associated 22 or 23 nt RNAs, but has no effect on the production of the diffusible silencing signal (Mallory et al., 2001). A cellular protein that interacted with HC-Pro in a two hybrid screen has also been shown to suppress PTGS (Anandalakshmi et al., 2000), but its effect on the accumulation of the siRNA has not been examined. The second group of PTGS inhibitors, which includes the Cmv2b protein of CMV, blocks the spread of PTGS to newly emerging tissue but has no effect on tissue where PTGS has been established (Brigneti et al., 1998). Nuclear localization of Cmv2b has been shown to be critical for the suppressor activity, suggesting that this protein blocks a ‘nuclear step’ of PTGS (Lucy et al., 2000). The third group thus far includes only the p25 protein of Potato Virus X, which was originally shown not to repress silencing (Brigneti et al., 1998). However, recent grafting experiments have shown that silenced stocks expressing p25 are unable to induce silencing of a scion irrespective of whether silencing has been induced by a transgene construct or a replicating virus (Voinnet et al., 2000). These data indicate that p25 prevents the spread of silencing, possibly by blocking the synthesis of the diffusible silencing signal.
What are the targets of the viral suppressors? We propose that in the case of p25 the target is the RdRP. If p25 were to inhibit the ability of RdRP to make dsRNA from an aberrant RNA and to synthesize the diffusible factor, it would explain why ectopic expression of p25 inhibits local PTGS induced by transgene but not by replicating virus (Voinnet et al., 2000), while still inhibiting the spread of PTGS induced by either replicating virus or transgene. The replicating virus would not require an RdRP to induce local PTGS because it either could produce dsRNA intermediates itself or it could encode its own RdRP. The transgene would generate an aberrant RNA that must be converted into dsRNA and therefore would require the activity of the RdRP (Figure 1). It is assumed in this model that the diffusible factor is synthesized by the host RdRP but not by the viral RdRP and that p25 is able to discriminate between the viral and cellular enzyme.
Biological role of PTGS
Although in the cases described above PTGS is triggered by artificial stimuli, the biological role of PTGS seems to be to prevent viral infection in plants and the mobilization of transposons in C. elegans. This conclusion has come from the observation that in C. elegans transposons are more active in RNAi defective mutants (Ketting et al., 1999; Tabara et al., 1999). Likewise, mutants defective in PTGS in plants are more susceptible to viral infection (Mourrain et al., 2000). In addition, viruses encode a number of factors that suppress PTGS (Voinnet et al., 2000), further supporting the notion that PTGS is a defense mechanism against viruses. PTGS/RNAi defective mutants such as EGO1 (Smardon et al., 2000), AGO1 (Fagard et al., 2000a) and Dicer (Knight and Bass, 2001) exhibit developmental defects, suggesting that either PTGS plays an essential role in development or that these genes have PTGS-independent developmental functions. The recent report that Dicer regulates the maturation of small temporal RNAs (stRNA) in both Drosophila (Hutvagner et al., 2001) and C. elegans (Grishok et al., 2001) supports the latter. stRNAs regulate developmental timing by binding to complementary sequences in the 3′ untranslated region of heterochronic genes and blocking their translation (Reinhart et al., 2000). stRNA are derived from longer precursor that are capable of forming double-stranded regions (Grishok et al., 2001). Dicer cleaves the longer precursor to generate stRNAs in much the same way that it cleaves dsRNA to generate siRNAs, except that stRNAs, unlike siRNAs, are single stranded (Hutvagner et al., 2001).
Perspectives
Rapid progress has been made in understanding the mechanism of PTGS. The recent report that RNAi can even function in cultured cells (Elbashir et al., 2001a) will increase the interest in PTGS and therefore this area of research will continue to grow. In the near future, we should see reports answering many key questions such as: how do transgenes generate the silencing signal? What is the nature of the diffusible silencing factor? What are the components of the mRNA degrading machinery? What are the targets of the viral inhibitors of PTGS? In addition, many of the genes identified by genetic analysis have not yet been characterized biochemically and we may get answers to further questions from this type of analysis. For example, we may learn whether or not the products of the QDE1 gene and its homologs have RNA-dependent RNA polymerase activity. If so, what are their substrates?
Acknowledgments
Acknowledgements
A.C. thanks Lisa Franchi, Emma Forrest and especially Carlo Cogoni for helpful discussion and comments on the manuscript. Work in the authors’ laboratory is supported by grants of The European Community (#QLK3-CT-2000-00078) and the Instituto Pasteur Fondazione Cenci Bolognetti.
References
- Anandalakshmi R., Marathe, R., Ge, X., Herr, J.M.,Jr, Mau, C., Mallory, A., Pruss, G., Bowman, L. and Vance, V.B. (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science, 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (1996) RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol., 32, 79–88. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. and English, J.J. (1996) Ectopic pairing of homologous DNA and post-transcriptional gene silencing in transgenic plants. Curr. Opin. Biotechnol., 7, 173–180. [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M. and Hannon, G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J., 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cogoni C., Irelan, J.T., Schumacher, M., Schmidhauser, T., Selker, E.U. and Macino, G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Cogoni C. and Macino, G. (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino, G. (2000) Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev., 10, 638–643. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton, A., Rudd, S., Angell, S. and Baulcombe, D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Horsefield, R., Braunstein, T.H. and Baulcombe, D.C. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis.EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho Niebel F., Frendo, P., Van Montagu, M. and Cornelissen, M. (1995) Post-transcriptional cosuppression of β-1, 3-glucanase genes does not affect accumulation of transgene nuclear mRNA. Plant Cell, 7, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeier M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L. and Mango, S.E. (2000) A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science, 289, 1928–1931. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. and Tuschl, T. (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W. and Tuschl, T. (2001b) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Boutet, S., Morel, J.B., Bellini, C. and Vaucheret, H. (2000a) AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M. and Vaucheret, H. (2000b) Systemic silencing signal(s). Plant Mol. Biol., 43, 285–293. [DOI] [PubMed] [Google Scholar]
- Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. and Ahringer, J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Grishok A., Tabara, H. and Mello, C.C. (2000) Genetic requirements for inheritance of RNAi in C. elegans. Science, 287, 2494–2497. [DOI] [PubMed] [Google Scholar]
- Grishok A. et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. (1999) A novel species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R. and Hannon, G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D. and Hannon, G. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Holtorf H., Schob, H., Kunz, C., Waldvogel, R. and Meins, F., Jr (1999) Stochastic and nonstochastic post-transcriptional silencing of chitinase and β-1, 3-glucanase genes involves increased RNA turnover-possible role for ribosome-independent RNA degradation. Plant Cell, 11, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T. and Zamore, P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp, T.H., van Luenen, H.G. and Plasterk, R.H. (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homologue of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans.Science, 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X. and Ding, S.W. (2001) Viral suppressors of RNA silencing. Curr. Opin. Biotechnol., 12, 150–154. [DOI] [PubMed] [Google Scholar]
- Lucy A.P., Guo, H.S., Li, W.X. and Ding, S.W. (2000) Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J., 19, 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C. et al. (2001) HC-Pro Suppression of Transgene Silencing Eliminates the Small RNAs but Not Transgene Methylation or the Mobile Signal. Plant Cell, 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins F. Jr (2000) RNA degradation and models for post-transcriptional gene-silencing. Plant Mol. Biol., 43, 261–273. [DOI] [PubMed] [Google Scholar]
- Metzlaff M., O’Dell, M., Cluster, P.D. and Flavell, R.B. (1997) RNA-mediated RNA degradation and chalcone synthase A silencing in petunia. Cell, 88, 845–854. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Mourrain, P., Beclin, C. and Vaucheret, H. (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol., 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Napoli C., Lemieux, C. and Jorgensen, R. (1990) Introduction of a chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui J.C., Elmayan, T., Pollien, J.M. and Vaucheret, H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R. and Ruvkun, G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Czaplinski, K. and Peltz, S.W. (1996) Making sense of nonsense in yeast. Trends Biochem. Sci., 21, 433–438. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N. and Maine, E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Stam M., de Bruin, R., van Blokland, R., van der Hoorn, R.A., Mol, J.N. and Kooter, J.M. (2000) Distinct features of post-transcriptional gene silencing by antisense transgenes in single copy and inverted T-DNA repeat loci. Plant J., 21, 27–42. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. and Mello, C.C. (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans.Cell, 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court, D.L. and Fire, A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans.Gene, 263, 103–112. [DOI] [PubMed] [Google Scholar]
- van der Krol A.R., Mur, L.A., Beld, M., Mol, J.N. and Stuitje, A.R. (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T.A. (1994) Analysis of viral-host protein interactions and tumorigenesis in transgenic mice. Semin. Cancer Biol., 5, 47–60. [PubMed] [Google Scholar]
- Voinnet O., Vain, P., Angell, S. and Baulcombe, D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer, C. and Baulcombe, D.C. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Wassenegger M. and Pelissier, T. (1998) A model for RNA-mediated gene silencing in higher plants. Plant Mol. Biol., 37, 349–362. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang, M. and Lough, T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf D., Jeong, B., Zhang, C. and Cerutti, H. (2000) Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science, 290, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Yang D., Lu, H. and Erickson, J.W. (2000) Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–2000. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A. and Bartel, D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]