Abstract
Recombination intermediates containing four-way (Holliday) junctions are generated during DNA repair and replication in many systems, including yeast mitochondrial DNA (mtDNA). In contrast, convincing evidence for recombination in mammalian mtDNA is lacking. We have used two-dimensional agarose-gel electrophoresis to analyse non-linear forms of mtDNA in human heart muscle. Replication intermediates from both the coupled and strand-asynchronous mtDNA replication pathways were detected. An additional class of non-linear molecules, with the electrophoretic properties of four-way junctions, was also prominent. These molecules were insensitive to topoisomerase I or RNase H, but were diminished by branch migration or RuvC treatment. Junctional molecules were detected in all regions of the mitochondrial genome, were found in myocardial DNA from young and old adults, but were present at lower levels in skeletal muscle and placenta. We suggest that they could represent intermediates of mtDNA repair, given their prevalence in the oxyradical-rich environment of heart muscle mitochondria.
INTRODUCTION
Recombination intermediates containing four-way (Holliday) junctions are generated during DNA repair and replication in many systems, including yeast mitochondrial DNA (mtDNA). They can be visualized by two-dimensional neutral–neutral agarose-gel electrophoresis (2DNAGE; Brewer and Fangman, 1988) as a so-called X-spike, located at double unit-length for the fragment being analysed (Friedman and Brewer, 1995). A recent study of replicating DNA from the slime mold Physarum polycephalum (Bénard et al., 2001) demonstrated that junctional molecules are formed post-replicatively between sister chromatids, and are probably involved in post-replicational DNA repair.
X-spikes are abundant in DNA undergoing amplification (Heck and Spradling, 1990). They can be enhanced by synchronization of DNA replication, e.g. in yeast rDNA (Brewer and Fangman, 1988), by loss of junction-resolving activities, such as Cce1p in yeast mitochondria (Lockshon et al., 1995), or by mutations that promote recombination, such as in the RMR3 helicase of yeast (Ivessa et al., 2000). Conversely, they are absent in mutants defective in Holliday junction formation, such as rad52 (Zou and Rothstein, 1997).
The existence of mtDNA recombination in mammals is controversial (Eyre-Walker, 2000), although it clearly occurs in some metazoans, e.g. mussels (Ladoukakis and Zouros, 2001). Uniparental inheritance precludes sexual recombination, but molecular recombination may still play a role in mtDNA maintenance. Rearranged mtDNAs associated with disease appear to be aberrant recombination products. They are also detected at a low level as sublimons in tissues of healthy individuals (Kajander et al., 2000). Intramolecular recombination has been inferred to occur in long-term culture to resolve partial duplications to wild-type molecules plus deletions (Tang et al., 2000).
Repair of some types of DNA damage, such as inter-strand cross-links, requires homologous recombination in bacteria (Sinden and Cole, 1978) and yeast nuclei (Abe et al., 1994), and such lesions are also repaired in mammalian mitochondria (LeDoux et al., 1992). Overexpression of mitochondrially targeted Escherichia coli RecA in human cells enhances mtDNA repair after bleomycin treatment (Paul et al., 2001), suggesting that it involves a natural recombination pathway. The relatively high prevalence of mtDNA sublimons in highly oxidative tissues, such as heart and skeletal muscle, compared with more quiescent tissues such as skin, supports the idea that mtDNA recombination events may be linked to repair, since these tissues generate the greatest concentration of DNA damaging, reactive oxygen species (ROS). Homologous recombination activity can be detected in mammalian mitochondrial extracts (Thyagarajan et al., 1996), and is sensitive to inhibition by RecA antibodies. Furthermore, the mitochondrial transcription factor mt-TFA can bind Holliday junctions in vitro (Ohno et al., 2000), and its yeast orthologue Abf2p stabilizes such junctions in vivo (MacAlpine et al., 1998).
Because of the high levels of sublimons observed previously in human heart muscle, we set out to use 2DNAGE to investigate the presence of mtDNA recombination intermediates in DNA samples from this tissue. Previous 2DNAGE analyses of mammalian mtDNA (Holt et al., 2000) revealed two types of mtDNA replication intermediate (RI), distinguishable by sensitivity to single strand-specific nucleases (SSNs), such as nuclease S1. SSN-sensitive RIs, as predicted by the orthodox, strand-asynchronous model of mtDNA replication (Clayton, 1982), predominate in cultured cells maintaining constant mtDNA copy number. SSN-resistant RIs, indicating conventional, strand-coupled DNA synthesis, predominate in cells recovering from copy number depletion. Both replication modes are evident in solid tissues. Analysis of heart muscle mtDNA in the present study now reveals the presence, in addition to these RIs, of prevalent recombination intermediates.
RESULTS
2DNAGE reveals prominent junctional species in human heart mtDNA
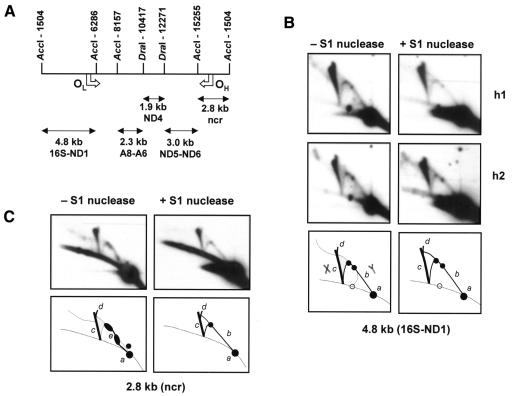
Human heart DNA was found to contain both classes of mtDNA RI described previously in human cells and tissues (Holt et al., 2000), with standard Y-arcs resistant to SSN treatment found in all fragments of the genome tested (Figures 1 and 2). From the OH-containing fragment, the major SSN-sensitive RIs formed a diffuse sigmoidal arc, with migration properties suggestive of extensive duplex regions. The SSN-resistant species from this fragment were similar to those seen previously in human placenta or cells recovering from mtDNA depletion, although with a more prominent, complete Y-arc. As in cells recovering from mtDNA depletion, the major SSN-sensitive species from the OL-containing fragment formed a diffuse arc below the track of the standard Y-arc (Figure 1), ending near to a prominent spot on the linear diagonal, which is probably a partial digestion product. An additional diffuse arc joined this position to the standard Y-arc.
Fig. 1. 2DNAGE analysis of origin regions of human heart muscle mtDNA. (A) Map of human mtDNA, arbitrarily linearized at the nucleotide pair (np) 1504 AccI site (Anderson et al., 1981), showing restriction fragments used as probes plus positions of orthodox mode replication origins (OH, OL). Gene names use conventional abbreviations. ncr, non-coding region. (B) AccI-digested heart muscle DNAs h1: 22-year-old male (individual s1 in Kajander et al., 2000), and h2: 80-year-old male (individual s2), with or without SSN (S1 nuclease) treatment, probed for the 4.8 kb OL-containing fragment. (C) Sample h1 probed for the 2.8 kb OH-containing AccI fragment. Sample h2 gave an identical result. Diagrammatic illustrations of the gels: dotted line, diagonal of linear molecules; open circle, partial digestion product; spot a, unit-length linear fragment; arc b, Y-form replication intermediates, as depicted in the bottom left-hand panel of (B); arc c, X-form junctional molecules, as depicted in the bottom left-hand panel of (B); arc d, bifurcation of the X-form arc, possibly representing molecules containing more than 1 junction; arc e, diffuse sigmoidal arc seen in the OH-containing fragment before S1 nuclease treatment. Other features, not discussed in the text, are unlabelled.
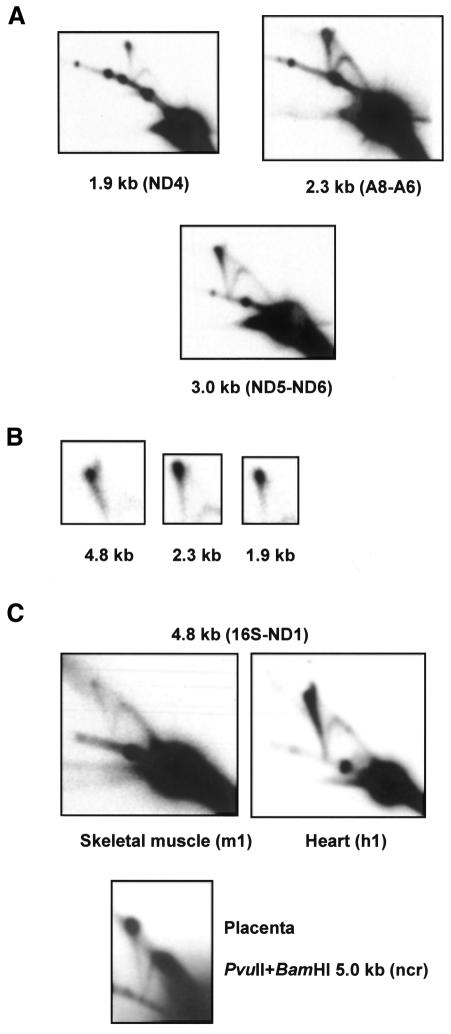
Fig. 2. 2DNAGE analysis of human mtDNA. (A) Heart muscle DNA h1, AccI+DraI digested and S1 treated, probed for non-origin fragments as per Figure 1A. (B) Comparable short exposures of the apex of the X-spike from different fragments, showing different relative signals compared with the apex of the Y-arc (bottom right of each panel), plus apical bifurcation. (C) AccI-digested DNAs (S1 untreated) from skeletal muscle (m1) and heart (h1) of same individual, probed for the 4.8 kb OL-containing fragment, plus PvuII+BamHI-digested placental mtDNA (S1 treated) probed for the 5.0 kb OH-containing fragment.
In addition, all probes detected a prominent class of SSN-resistant, non-linear molecules forming an X-spike (Figures 1 and 2). The intensity of the X-spike relative to that of the standard Y-arc appeared to be approximately in proportion to fragment size, but in each case the X-spike was the most prominent non-linear species detected. In addition, it appeared to be bifurcated near to its apex, the minor arm extending upwards, initially towards the direction of less retardation in the first dimension. The relative intensity of the minor compared with the major arm seemed again to be dependent on fragment size (Figure 2B), being greatest in the largest (4.8 kb) fragment probed, which also contains the light-strand origin for orthodox mode replication (OL). The X-spike was similar in all heart DNA samples analysed: those from young and aged individuals (h1, h2, ages 22 and 80, respectively) gave essentially identical patterns in parallel gel-blots (Figures 1 and 2).
Junctional molecules in mtDNA vary in abundance between tissues
To confirm that the prominent X-spike seen in human heart mtDNA was not an artefact of preparation due, for example, to the time interval between death and tissue extraction, we analysed in parallel a sample of skeletal muscle DNA taken from one of the same individuals. This gave a much fainter X-spike whose intensity was comparable with that of the standard Y-arc (Figure 2C). A similar X-spike was seen also in human placenta, most clearly in OH-containing fragments (Figure 2C; see also figures 4 and 7 of Holt et al., 2000).
Junctional mtDNA molecules have the properties of recombination intermediates
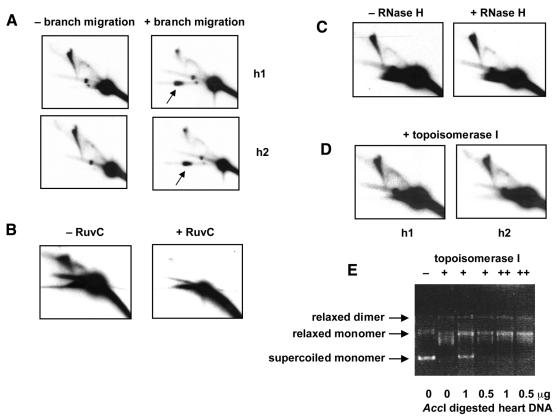
In principle, junctional molecules forming an X-spike on 2DNAGE might be joined by hemicatenation (where a strand of one duplex is wound around a strand of another), by four-way (Holliday) junctions, or covalently (e.g. by cross-linking). Junctions might also comprise RNA–DNA (as opposed to DNA–DNA) hybrids. To distinguish these various possibilities, we carried out a series of diagnostic treatments of heart mtDNA. In-gel branch migration (Johnson and Symington, 1993), carried out after first-dimension electrophoresis, resolved most DNA in the X-spike to material co-migrating in the second dimension with the monomeric fragment (Figure 3A). Only the apex region was retained. Both arms of the bifurcated region were diminished by branch migration. In contrast, RIs were essentially unaffected by this treatment, the single exception being stalled molecules migrating as a discrete spot on the standard Y-arc, which were preferentially removed (Figure 3A).
Fig. 3. Resolution of junctional molecules. 2DNAGE of AccI-digested human heart DNA samples (not S1-digested), probed for the 4.8 kb OL-containing fragment, after various treatments. (A) Samples h1 and h2 with or without treatment of the first-dimension gel slab to promote branch migration. Note the large increase in material from the X-spike now migrating in the second dimension with the mobility of the unit fragment (arrow). (B) Sample h2 (2 µg) incubated for 20 h in agarose gel plug with or without RuvC. Note the increase in heterogeneous double-stranded DNA digestion products running below the unit fragment, after RuvC treatment. (C) Sample h1 (2 µg) incubated for 1 h with or without 1 U RNase H. (D) Samples h1 and h2 treated with topoisomerase I prior to electrophoresis. (E) Validation of topoisomerase assay. Supercoiled plasmid DNA (500 ng) incubated either with (lane 1) or without (lane 2) 3 U topoisomerase I, or with 15 U of the enzyme in the presence of AccI-digested heart DNA as shown. Lanes marked ++ were incubated with an additional 6 U of the enzyme for a further 2 h, after overnight reactions. The selected conditions used for the main experiment completely remove supercoiling of the test plasmid, and hence should also separate any catenated molecules.
Treatment with the Holliday-junction resolvase RuvC in agarose gel plugs also greatly diminished the X-spike (Figure 3B). Although the treatment is less specific, removing also the Y-arcs of RIs as reported previously by Bénard et al. (2001), the junctional molecules were clearly degraded preferentially, compared with the unit length fragment. Heat denaturation also completely abolished the X-spike (data not shown).
In contrast, hemicatenanes, but not Holliday junctions or cross-linked molecules, should be resolved by decatenation with topoisomerase I. Topoisomerase I treatment under conditions that completely relaxed supercoiled DNA (Figure 3E), hence would be expected to remove any catenated structures, did not affect the X-spike in any detectable way (Figure 3D). RNase H treatment also had no effect (Figure 3C). All tests were therefore consistent with the X-spike consisting of DNA molecules joined by four-way recombination junctions.
DISCUSSION
Human heart mtDNA includes a prominent class of non-linear molecules, with the properties of recombination junctions. These migrate as an X-spike on 2DNAGE, are resolved under conditions that promote branch migration or resolution of four-way junctions, and are refractory to treatment with topoisomerase I, SSN or RNase H. The X-spike closely resembles those which accumulate in yeast mtDNA when resolution of four-way junctions is blocked by deletion of the CCE1 gene (Lockshon et al., 1995). This is the first clear demonstration of recombination intermediates in mammalian mtDNA.
Their abundance varies between tissues, being high in heart, intermediate in skeletal muscle or placenta, but low or absent in cultured cells (Holt et al., 2000). In the heart, their abundance is related, at least crudely, to the size of restriction fragment under analysis, implying that the distribution of junctions is random across the genome. Bifurcation of the X-spike into a major and a minor arm is also more pronounced in larger fragments. One possibility is that the minor arm consists of molecules joined by two Holliday junctions, creating a structure that is less extended, and hence migrates more conventionally in the first dimension.
In previous studies of RIs using 2DNAGE it was suggested that junctional molecules might arise as an artefact during DNA extraction, due to branch migration following annealing of nascent strands (Dijkwel et al., 1991). However, this study and an earlier one on Physarum (Bénard et al., 2001) show that standard Y-form RIs are essentially unaffected by prolonged incubation under conditions that promote branch migration. The only RIs affected were those that had stalled in vivo. As elsewhere (Michel, 2000), these may comprise ‘crowsfoot’ intermediates from which replication can be reinitiated by recombination. Crucially, the abundance of junctional molecules was very different in heart and skeletal muscle mtDNA extracted in parallel from tissues of the same individual, therefore is extremely unlikely to be a preparative artefact. In contrast, different heart DNA samples showed the same high level of junctional mtDNA molecules.
Junction formation could be a natural step in the initiation of DNA replication. However, this would be predicted to generate even more complex, branched species, that would migrate to the left of a conventional X-spike, being in total more than twice the unit fragment length. Although we cannot rule this out entirely, the presence of junctional molecules seems to be unrelated to the mtDNA replication mode. Thus, there is a striking similarity between the mtDNA RIs of heart and those of cultured cells recovering from mtDNA depletion (compare Figure 1B with figures 5a and b of Holt et al., 2000). In both cases strand-coupled RIs predominate, and the minority of SSN-sensitive RIs appear very similar, although quite different from those seen in cultured cells maintaining a constant mtDNA copy number. Nevertheless, the abundance of the X-spike is enormously different between heart and cultured cells undergoing copy number recovery.
The earlier finding of illegitimate recombination products (sublimons) at a low level in heart mtDNA prompted us to consider whether their accumulation might be related to the presence of recombination intermediates. We therefore compared heart DNA samples from two individuals having very different sublimon levels (<0.1 versus 92 copies per cell of the prevalent 3.75 kb sublimon class; Kajander et al., 2000). These contained almost indistinguishable amounts of junctional mtDNA molecules. In contrast, skeletal muscle DNA from one of the same individuals, with substantial sublimon levels (eight copies per cell), had much lower amounts of junctional mtDNA. Thus, there appears to be no correlation between the amounts of sublimons and of junctional molecules in a given DNA sample. Nevertheless, the slow, age-related accumulation of sublimons in heart muscle (O.A. Kajander, P.J. Karhunen and H.T. Jacobs, manuscript in preparation) might still be a consequence of the presence of an active homologous recombination system in this tissue, promoting occasional, aberrant recombination events.
In Physarum, kinetic analysis of synchronised cells showed that junctional molecules in nuclear DNA arise post-replicatively and are resolved in a subsequent step (Bénard et al., 2001), suggesting that they are involved in a programmed, post-replicative process. Two possibilities were proposed, both linked to DNA repair, that may apply also to mtDNA. The first is that recombination junctions form in response to DNA lesions that cause incomplete nascent strand synthesis, and are an intrinsic step in the repair of such lesions. The second is that by forming a physical link between daughter molecules they ensure their close proximity, to allow time for DNA repair if needed. Subsequent branch migration would be impeded by heterology, which may then recruit other proteins to carry out repair of the lesions thus detected.
A similar involvement of junctional molecules in post-replicative DNA repair in mitochondria is highly plausible. They are abundant in heart muscle, the tissue with the highest metabolic throughput of mitochondrial oxidative phosphorylation (OXPHOS), and also the highest production of DNA-damaging ROS. Loss of mitochondrial ROS-detoxifying enzymes such as MnSOD results in early neonatal death in the mouse, associated with dilated cardiomyopathy (Li et al., 1995). Heart mitochondria contain much higher levels of the major mitochondrial endonuclease (endonuclease G) than other tissues (Houmiel et al., 1991). Its tissue distribution reflects OXPHOS capacity rather than mtDNA content, suggesting a role in recombinational repair of oxidative damage (Gerschenson et al., 1994). It is a relatively non-specific enzyme with a preference for short GC tracts, hence is a good candidate for initiating frequent strand exchanges as part of a generalized mechanism for promoting post-replicative repair.
Although active repair of oxidatively damaged DNA is an attractive explanation for the abundant mtDNA recombination intermediates in heart, a comprehensive tissue survey and a direct experimental test will now be needed to verify this. Moreover, we cannot exclude the radically different interpretation that they accumulate in heart because for some reason they cannot be resolved in this tissue.
METHODS
DNA extraction and digestion. Autopsy-derived DNA from human tissues (Kajander et al., 2000) or human placental mtDNA (Holt et al., 2000) were digested with restriction endonucleases or RNase H under conditions recommended by the manufacturers (New England Biolabs or Promega). S1 nuclease (MBI Fermentas) treatment used 1 U of enzyme in the supplied reaction buffer for 1 min at 37°C. Topoisomerase I (Gibco-BRL) treatment used conditions under which 0.5 µg of supercoiled plasmid DNA could be relaxed. One microgram of AccI-digested DNA was incubated overnight with 15 U of the enzyme at 37°C, and then for an additional 2 h with a further 6 U of the enzyme, in the manufacturer’s recommended reaction buffer. RuvC treatment was carried out in agarose gel plugs in 25 mM Tris–HCl pH 8.0, 1 mM dithiothreitol, 0.1 mg/ml bovine serum albumen, 6% glycerol, 10 mM MgCl2, for 20 h at 37°C, using 18 µg of the enzyme (a kind gift from Professor R.G. Lloyd) on 2–3 µg of DNA.
2DNAGE and Southern hybridization. One to five micrograms of DNA were used per lane for 2DNAGE (Brewer and Fangman, 1988; Friedman and Brewer, 1995). After the first dimension, 1.2 V/cm for 24 h at 4°C in a 0.4% agarose gel in TBE buffer without ethidium bromide (EB), the gel was equilibrated with TBE containing 300 ng/ml EB. Excised lanes were rotated through 90°. Cooled, 1% agarose gel containing 300 ng/ml EB was cast around them. After the second dimension (6 V/cm for 4 h at 4°C in TBE containing 300 ng/ml EB with buffer recirculation), Southern blotting was carried out by standard capillary transfer. To promote branch migration of possible Holliday junction structures, gel slabs from the first-dimension electrophoresis were incubated for 4 h at 65°C in 10 mM Tris–HCl, 0.1 M NaCl, 0.1 mM EDTA pH 8.0. Control gel slabs were incubated for 4 h at room temperature. For probes, the following human mtDNA fragments were PCR amplified as described previously (Kajander et al., 2000), using 20mer primer oligonucleotides: non-coding region (np 35–646), 16S/ND1 (np 3012–3692), A8-A6 (np 8460–9107), ND4 (np 10 951–11 651) and ND5-ND6 (np 13 569–14 338). Fragment identity was confirmed by direct sequencing. Probes were labelled by random priming using Oligolabelling kit (Amersham Pharmacia Biotech) and [α-32P]dCTP (Amersham Pharmacia Biotech; 3000 Ci/mmol). Hybridization and wash conditions were as used previously (Kajander et al., 2000). The signal was detected by autoradiography for between 2 and 36 h. Filters were stripped and reprobed as described previously (Kajander et al., 2000).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Hans Spelbrink and David Sherratt for useful discussions, and Bob Lloyd and Stuart Ingleston for supplying RuvC. The work was supported by the Academy of Finland, UK Medical Research Council, Tampere University Hospital Medical Research Fund, Yrjö Jahnsson Foundation, the Finnish Foundation for Alcohol Research and the Pirkanmaa Regional Funds of the Finnish Cultural Foundation.
REFERENCES
- Abe H., Wada, M., Kohno, K. and Kuwano, M. (1994) Altered drug sensitivities to anticancer agents in radiation-sensitive DNA repair deficient yeast mutants. Anticancer Res., 14, 1807–1810. [PubMed] [Google Scholar]
- Anderson S. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- Bénard M., Maric, C. and Pierron, G. (2001) DNA replication-dependent formation of joint DNA molecules in Physarum polycephalum. Mol. Cell, 7, 971–980. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell, 55, 637–643. [DOI] [PubMed] [Google Scholar]
- Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.A., Vaughn, J.P. and Hamlin, J.L. (1991) Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol. Cell. Biol., 11, 3850–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. (2000) Do mitochondria recombine in humans? Phil. Trans. R. Soc. Lond. B, 355, 1573–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer, B.J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol., 262, 613–627. [DOI] [PubMed] [Google Scholar]
- Gerschenson M., Low, R.L. and Loehr, J. (1994) Levels of the mitochondrial endonuclease during rat cardiac development implicate a role for the enzyme in repair of oxidative damage in mitochondrial DNA. J. Mol. Cell. Cardiol., 26, 31–40. [DOI] [PubMed] [Google Scholar]
- Heck M.M. and Spradling, A.C. (1990) Multiple replication origins are used during Drosophila chorion gene amplification. J. Cell Biol., 110, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt I.J., Lorimer, H.E. and Jacobs, H.T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell, 100, 515–524. [DOI] [PubMed] [Google Scholar]
- Houmiel K.L., Gerschenson, M. and Low, R.L. (1991) Mitochondrial endonuclease activity in the rat varies markedly among tissues in relation to the rate of tissue metabolism. Biochim. Biophys. Acta, 1079, 197–202. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou, J.Q. and Zakian, V.A. (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell, 100, 479–489. [DOI] [PubMed] [Google Scholar]
- Johnson R.D. and Symington, L.S. (1993) Crossed-stranded DNA structures for investigating the molecular dynamics of the Holliday junction. J. Mol. Biol., 229, 812–820. [DOI] [PubMed] [Google Scholar]
- Kajander O.A., Rovio, A.T., Majamaa, K., Poulton, J., Spelbrink, J.N., Holt, I.J., Karhunen, P.J. and Jacobs, H.T. (2000) Human mtDNA sublimons resemble rearranged mitochondrial genomes found in pathological states. Hum. Mol. Genet., 9, 2821–2835. [DOI] [PubMed] [Google Scholar]
- Ladoukakis E.D. and Zouros, E. (2001) Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol., 18, 1168–1175. [DOI] [PubMed] [Google Scholar]
- LeDoux S.P., Wilson, G.L., Beecham, E.J., Stevnsner, T., Wassermann, K. and Bohr, V.A. (1992) Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis, 13, 1967–1973. [DOI] [PubMed] [Google Scholar]
- Li Y.B. et al. (1995) Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature Genet., 11, 376–381. [DOI] [PubMed] [Google Scholar]
- Lockshon D., Zweifel, S.G., Freeman-Cook, L.L., Lorimer, H.E., Brewer, B.J. and Fangman, W.L. (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell, 81, 947–955. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Perlman, P.S. and Butow, R.A. (1998) The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl Acad. Sci. USA, 95, 6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Ohno T., Umeda, S., Hamasaki, N. and Kang, D. (2000) Binding of human mitochondrial transcription factor A, an HMG box protein, to a four-way DNA junction. Biochem. Biophys. Res. Commun., 271, 492–498. [DOI] [PubMed] [Google Scholar]
- Paul R., Dalibart, R., Lemoine, S. and Lestienne, P. (2001) Expression of E. coli RecA targeted to mitochondria of human cells. Mutat. Res., 486, 11–19. [DOI] [PubMed] [Google Scholar]
- Sinden R.R. and Cole, R.S. (1978) Repair of cross-linked DNA and survival of Escherichia coli treated with psoralen and light: effects of mutations influencing genetic recombination and DNA metabolism. J. Bacteriol., 136, 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Manfredi, G., Hirano, M. and Schon, E.A. (2000) Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol. Biol. Cell, 11, 2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B., Padua, R.A. and Campbell, C. (1996) Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem., 271, 27536–27543. [DOI] [PubMed] [Google Scholar]
- Zou H. and Rothstein, R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]