Abstract
Chloroplasts import post-translationally most of their constituent polypeptides via two distinct translocon units located in the outer and inner envelope. The protein import channel of the translocon of the outer envelope of chloroplasts, Toc75, is the most abundant protein in that membrane. We identify a novel Toc75 homologous protein, atToc75-V, a prominent protein that is clearly localized in the chloroplastic outer envelope. Phylogenetic analysis indicates that Toc75-V is more closely related to its prokaryotic ancestors than to Toc75 from plants. The presence of a second translocation channel suggests that alternative, previously unrecognized import routes into chloroplasts might exist.
INTRODUCTION
Chloroplasts have to import post-translationally most of their protein constituents from the cytosol. This process is facilitated by two supramolecular machines located in the chloroplastic outer and inner envelope, respectively (Keegstra and Cline, 1999; Küchler and Soll, 2001). Both the translocon at the outer envelope of chloroplasts (Toc complex) and the translocon at the inner envelope of chloroplasts (Tic complex) (Schnell et al., 1997) act jointly during the translocation reaction (Schnell and Blobel, 1993; Alefsen et al., 1994). Due to the endosymbiotic origin of chloroplasts (Margulis, 1970), several of the identified Tic and Toc subunits, such as Toc75, Tic55 and Tic20, are of prokaryotic origin (Heins and Soll, 1998; Reumann and Keegstra, 1999). Other subunits, such as the nucleotide dependent receptor polypeptides Toc159 and Toc34 or Tic110, the preprotein translocation channel of the inner envelope, show no clear prokaryotic origin and might have been added to the translocon during the conversion of an endosymbiont to an organelle.
Toc75 from pea is the most prominent chloroplastic outer envelope protein. It was identified early as a putative Toc component (Waegemann and Soll, 1991; Perry and Keegstra, 1994; Schnell et al., 1994) that can form an aqueous ion channel in vitro (Hinnah et al., 1997). Therefore, Toc75 most likely represents the preprotein translocation pore (Hinnah et al., 1997). In Arabidopsis, three genes coding for Toc75 homologues, namely atToc75-I, atToc75-III and atToc75-IV, can be identified (Jackson-Constan and Keegstra, 2001). Only one of these genes, atToc75-III (located on chromosome III), is represented by an expressed sequence tag (EST), whereas atToc75-I and atToc75-IV (located on chromosomes I and IV, respectively) are not, indicating that the latter are not expressed or are only expressed in special organs or under specific developmental conditions (Jackson-Constan and Keegstra, 2001). No gene insertion mutants of atToc75-III have been described to date, which suggests that Toc75 function is essential for chloroplast biogenesis. Here, we describe a prominent chloroplastic outer envelope protein from pea that represents a clear homologue to Toc75 from pea and Arabidopsis and to the prokaryotic predecessor (Bölter et al., 1998; Reumann et al., 1999) in both photosynthetic and non-photosynthetic bacteria.
RESULTS AND DISCUSSION
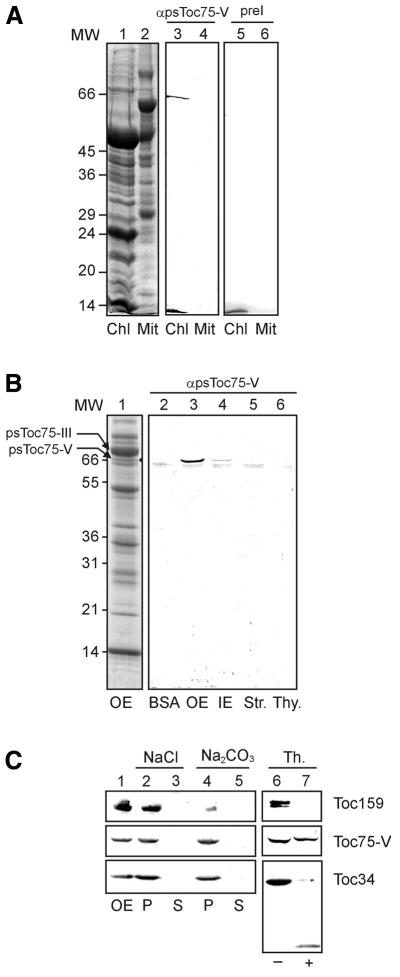
Before raising an antiserum in rabbits to a specific chloroplast protein, we routinely tested the preimmune sera for non-specific reactions. Several preimmune sera recognized a polypeptide in the outer envelope membrane of pea chloroplasts, with an apparent molecular weight on SDS–PAGE of 66 kDa (data not shown). In this area, only one prominent protein that migrates between Toc75 and Toc64 could be detected (Figure 1B). The SDS–PAGE purified protein was used to raise a specific antiserum in a rabbit whose preimmune serum did not show any cross-reactions with purified chloroplasts (Figure 1A). The serum against the 66 kDa polypeptide specifically recognized one protein in purified pea chloroplasts but not in pea leaf mitochondria. The 66 kDa protein is clearly localized in the purifed outer envelope membrane fraction. No immune reactive protein could be detected in thylakoids or in the stroma, whereas a minor cross-reaction was observed in the inner envelope membrane (Figure 1B). These results indicate that the 66 kDa protein is a genuine chloroplast protein that might share antigenic epitopes with proteins from bacteria. The rabbits used to raise antisera might be infected by bacteria and thus developed antibodies to polypeptides of the pathogen.
Fig. 1. Localization of a novel chloroplast protein. (A) Purified chloroplasts (Chl) or mitochondria (Mit; equivalent to 100 µg of protein) from pea leaves were separated by SDS–PAGE and either stained by Coomassie Blue (lanes 1 and 2) or immunoblotted and incubated with preimmune (lanes 3 and 4) or immune (lane 5 and 6) serum against Toc75-V. Numbers on the left indicate molecular weight (MW) markers (in kDa). (B) Different chloroplast subfractions, 15 µg of protein of each outer envelope (OE), inner envelope (IE), stroma (Str.) or thylakoids (Thy.) were used for immunoblotting. As a control, 1 µg of BSA was used to detect non-specific background. Lane 1 shows a Coomassie-stained SDS–PAGE of purified outer envelope membrane (300 µg). (C) Outer envelope membranes were either treated (+) or not treated (–) with the protease thermolysin, and the presence of Toc75-V, Toc159 and Toc34 was subsequently analysed by immunoblotting (lanes 6 and 7). Lanes 1–5 show purified outer envelopes (lane 1) extracted with either 1 M NaCl or 0.1 M Na2CO3 and separated into a soluble (S) and insoluble (P) fractions. The presence of Toc75-V, Toc159 and Toc34 was determined by immunoblotting.
In order to characterize the 66 kDa protein further, the purified outer envelope membranes were either treated with the protease thermolysin to determine whether the polypeptide contains surface exposed epitopes or extracted with 1 M NaCl or 0.1 M Na2CO3. The results (Figure 1C) demonstrate that the 66 kDa protein behaves as an integral membrane protein that is not released from the membrane by pH 11. It is further deeply imbedded into the membrane, which renders it inaccessible to protease. Other outer envelope proteins such as Toc159 and Toc34 also behave as integral membrane proteins but are susceptible to the protease thermolysin (Figure 1C).
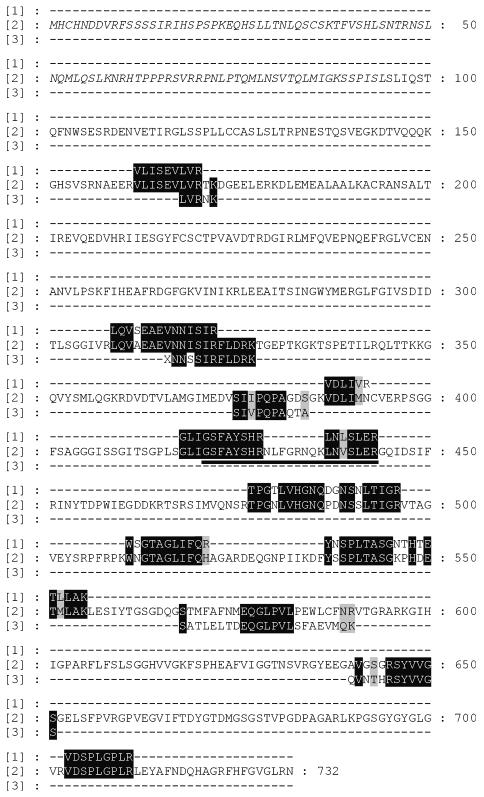
In order to identify the 66 kDa outer envelope polypeptide from pea, it was cleaved by endoproteinase Glu C or trypsin, and peptides were sequenced by Edman degradation (Figure 2, lane 3). Peptides obtained after trypsin treatment were also analysed by mass spectroscopy (Figure 2, lane 1). Two peptides obtained by these different approaches overlapped, demonstrating that the same protein was sequenced (Figure 2). When the sequences were used to search the MIPS database, they could be aligned to two EST clones from Arabidopsis AF 360205 and AY 040053. All peptides obtained by either Edman degradation or mass spectrometry showed sequence similarity to the EST clones (Figure 2). Both AF 360205 and AY 040053 locate to the same gene on chromosome V in Arabidopsis, at5g19620. The polypeptide deduced from the EST sequences has a length of 732 ammio acids, a calculated molecular weight of 79 936 Da and a pI value of 8.41. Chloro P predicts a chloroplast targeting sequence of 93 ammio acids (Figure 2), which would leave a calculated molecular weight for the mature form of 69 000 Da, close to the estimated size from SDS–PAGE. The pI of the mature form is 6.37. This is typical for a chloroplast precursor protein, which in general contains an excess of positively charged amino acids in the presequence (von Heijne et al., 1989; Jackson-Constan and Keegstra, 2001). Our protein sequencing data show that the polypeptide deduced from the genomic clone at5g19620 contains an omission of 23 amino acids in the central region of the sequence, due to a computational error in the predicted splice site. Two peptides, GLIGSFAYSHR and LNLSLER, are located in this region (Figure 2).
Fig. 2. Peptide fragments and deduced amino acid alignment with atToc75-V. Purified outer envelope membranes from pea chloroplasts were separated by SDS–PAGE, and the protein band present at ∼66 kDa (indicated as psToc75-V in Figure 1B) was used for amino acid determination. Lane 1 shows peptides deduced by mass spectrometry. Lane 3 shows peptide sequences determined by Edman degradation. Lane 2 shows the amino acid sequence of atToc75-V deduced from the EST clones AF 360205, AY 040053 and at5g19620 that had the highest homology to the peptides determined from pea. The predicted presequence is written in italics. The peptide that is missing in at5g19620 due to a computational splice error is underlined.
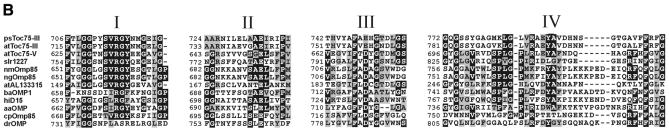
Alignment of the EST clones with the database yielded strong homologies to the protein import channel from chloroplasts Toc75 (Figure 3A). The identity to atToc75-III, psToc75 and the cyanobacterial synToc75 is 22, 22 and 31%, respectively. Together with the corresponding calculated E values of 6 × 10–16 (at), 4 × 10–20 (ps) and 2 × 10–68 (syn), respectively, these data indicate that the 66 kDa protein from pea represents a bona fide homologue to Toc75. We propose to name the protein atToc75-V or psToc75-V, depending on the parent organism (Schnell et al., 1997).
Fig. 3. atToc75-V shows distinct homologies to plant and prokaryotic usher proteins. (A) An alignment that compares atToc75-V with synToc75 (slr1227) and atToc75-III (T6H20230). Identical amino acid residues are boxed in black, and homologue exchanges are shaded in grey. (B) Conserved C-terminal domains in Toc75 proteins from plants and bacterial outer membrane proteins. ps, Pisum sativum L36858; at, Arabidopsis thaliana; syn, Synechoystis slr1227; nmOmp85, Neisseria meningitis outer membrane protein 85, AAF 40639; ng, Neisseria gonorrhoe, AAC 17600; ba, Brucella abortus, AAA 96788; hi, Haemophilus influenza, F 64102; aa, Aquifex aeolicus, AE 000733; cp, Chlamydia pneumoniae, AAD 18449; dr, Deinococcus radiodurans, AE 001898.
atToc75-V shows a higher identity to the cyanobacterial synToc75 than to the ‘direct’ Arabidopsis homologue atToc75-III. Therefore, we asked whether atToc75-V could be related to other polypeptides from non-photosynthetic bacteria. Indeed, atToc75-V shows a much stronger similarity to a class of bacterial transport proteins than to atToc75-III, as indicated by very low E values. Closely related proteins can be found, for example, in Neisseria meningites, Neisseria gonorrhoe, Brucella abortus, Haemophilus influenza and Aquifex aeolicus (Figure 3B). These usher proteins most likely facilitate the export of hemolysins. At the same time, they represent prominent surface antigens, which provoke the immune response upon infection (Thomas et al., 1990; Flack et al., 1995). We conclude that atToc75-V represents the most ancestral or earliest form of a Toc75-like channel, which was further modified during endosymbiosis.
The Toc75 channels and their bacterial homologues are most highly conserved in the C-terminal region. Here, four domains that allow these proteins to cluster into one family can be observed (Figure 3B). Domain I (Reumann et al., 1999) contains a block of conserved amino acids that could also be responsible for the immune response in different sera of rabbits (see above). Domains II, III and IV, which have not been recognized before, are largely defined by conserved glycine and proline residues as well as the aromatic amino acids phenylalanine and tyrosine. According to a topology model of psToc75, these four domains, which stretch over a region of ∼100 amino acids, could be inserted in the membrane (Svesknikova et al., 2000) and could be crucial for channel function. It has been proposed (Reumann and Keegstra, 1999) that the Toc75-like channel derived from an ancient prokaryotic channel of smaller size and evolved by partial gene duplication in the N-terminal region of the protein. It is important to point out that, in the genome of Arabidopsis thaliana (AL 133315), there exists an open reading frame of 328 amino acids that shows the typical signature found in domains I–IV (Figure 3B). Neither location nor function of this smaller protein is known at present, but it should be very interesting to determine its transport capacity.
Speculation
We believe that atToc75-V represents the most ancestral form of a Toc75-like channel in chloroplasts. It could represent a link between the prokaryotic usher proteins from which a eukaryotic organellar transport channel developed during endosymbiosis. The fairly abundant presence of atToc75-V (∼5–10% of atToc75-III; compare Figure 1) indicates that it has retained special functional properties that are required for the export or import of a subset of macromolecules, such as polypeptides. However, Toc75-V is not detected in isolated oligomeric Toc complexes, which also contain Toc34 and Toc159, indicating that it functions independently of these receptor polypeptides or together with components that have not been described yet (Figure 4). Alternatively, atToc75-V might be involved in the translocation of highly hydrophobic α-helical transmembrane solute transporters of the inner envelope membrane, the route of which is not fully understood. On the other hand, atToc75-V could function in the export of biomolecules from chloroplasts like its prokaryotic ancestor in bacteria. So far, Arabidopsis tDNA insertion mutants are not available to answer these questions.
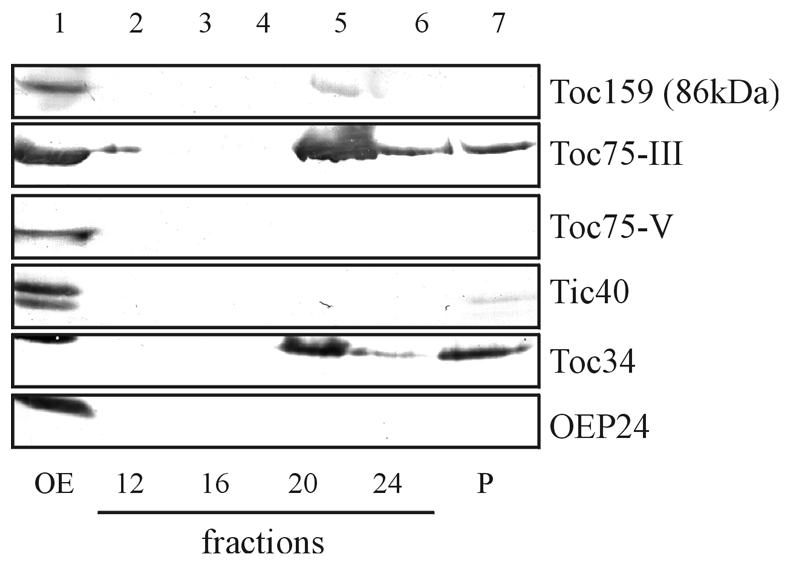
Fig. 4. Toc75-V is not present in the isolated Toc complex. Purified outer envelope membranes from pea chloroplasts were solubilized in 1.5% decylmaltoside for 10 min at ambient temperature and fractionated at 200 000 g on a linear sucrose density gradient (25–70%, w/v). Lane 1 shows an immunoblot of total solubilized membranes using antisera against Toc159 (the presence of the preteolytic fragment of 86 kDa is shown), Toc75-III, Toc75-V, Tic40, Toc34 and the outer envelope protein OEP24. Lanes 2–7 show immunoblots of different fractions of the sucrose density gradient using the same antisera as in lane 1. The Toc complex consisting of the subunits Toc159, Toc75-III and Toc34 is concentrated in fraction 20, whereas Toc75-V did not enter the gradient.
METHODS
Plant growth and chloroplast fractionation.
Pea plants (Pisum sativum var. golf ) were grown in a greenhouse for 10–12 days before harvest. Chloroplasts were isolated and purified by silica sol gradients (Mourioux and Douce, 1981). Chloroplasts were incubated in hypertonic buffer and lysed by 50 strokes in a Dounce homogenizer (Keegstra and Youssif, 1986). Outer and inner envelope membranes were separated from purified chloroplasts by sucrose density centrifugation (Keegstra and Youssif, 1986). Envelope membranes were stored at –80°C until further use.
In-gel digestion.
In-gel digestion was performed as described previously (Rosenfeld et al., 1992; Jeno et al., 1995; Shevchenko et al., 1996) at room temperature under shaking. The protein band of interest was excised from the SDS polyacrylamide gel (Laemmli, 1970) with a sterile razor blade, cut into small pieces, transferred into 1.5 ml Eppendorf tubes and washed with 100–150 µl of water, then with 50% (v/v) acetonitril and finally with 50% (v/v) acetonitril and 0.1 M NH4HCO3. Gel pieces were shrunk with acetonitril and dried down in a vacuum centrifuge. For reduction and alkylation, gel particles were swollen in 10 mM DTT and 0.1 M NH4HCO3 for 45 min at 56°C, washed twice with acetonitril and incubated for 30 min in the dark in 55 mM iodoacetamid and 0.1 M NH4HCO3. Washing of gel pieces was continued with 0.1 M NH4HCO3 and acetonitril until Coomassie was removed and colourless gel particles were dried down in a vacuum concentrator. They were rehydrated by ice-cold trypsin (Roche, Germany, sequencing grade) digestion buffer [for 120 µl: 15 µl of trypsin (25 µg per 250 µl of 1 mM HCl), 50 µl of H2O, 50 µl of 0.1 M NH4HCO3, 5 µl of 1% (w/v) CaCl2] and incubated for 45 min on ice. The remaining supernatant was removed, replaced by the same buffer without trypsin and put at 37°C with a little shaking overnight. For the extraction of peptides, gel pieces were covered with a sufficient volume of 25 mM NH4HCO3, incubated for 15 min at 37°C and pelleted. Gel pieces were incubated in acetonitril for another 15 min at 37°C and the supernatant was collected. Gels were then incubated in 5% (v/v) formic acid and treated with acetonitril, and peptides were extracted as described above. The two extracts were combined and dried down in a vacuum concentrator.
For amino acid sequencing by Edman degradation, peptides were generated according to Cleveland et al. (1977) using either trypsin or endopeptidase Glu C as protease. Peptides were separated by SDS–PAGE, blotted onto PVDF membranes (Towbin et al., 1979) and used for sequencing.
Desalting and concentrating of samples.
The dried protein digest was first dissolved in 2 µl of 40% (v/v) formic acid and 8 µl of water. Prior to the analysis in the mass spectrometer, peptides were purified in a Millipore ZipTip (Millipore, Bedford, MA). The procedure was performed according to the manufacturer’s instructions, except that the tip was prewetted by 100% methanol and equilibrated by washing in 5% (v/v) formic acid. After binding of peptides and washing with 5% (v/v) formic acid, peptides were eluted in 60% (v/v) methanol and 5% (v/v) formic acid and injected in NanoES spray capillaries (Protana, Odense, Denmark).
Mass spectrometry.
Peptide analysis was carried out on a Micromass Q-TOF-1.5 hybrid mass spectrometer (Micromass, Altrincham, UK) according to the manufacturer′s instructions. The measurements were perfomed at capillary voltages between 650 and 900 V and a cone voltage of 45 V. Spectra were usually recorded from 300 to 1500 m/z. For fragmentation of peptides in the MS/MS mode, the collision energy was raised from 4 to 18 or up to 42, depending on the individual peptide. Scanning time was usually 1 min, and argon was used as collision gas.
Sequencing, database search and alignments.
MS/MS spectra were sequenced manually, or the PepSeq program (Micromass) was used. For the identification of the protein, the EMBL (http://www.narrador.embl-heidelberg.de/GroupPages/pagelink/peptidesearchpage.html), MIPS (http://mips.gsf.de/proj/thal/db/search/search_frame.html) and SWISS-PROT (http://www.expasy. ch/sprot/) databases were searched by similarity or BLAST searches. Search parameters were set as recommended by the database programs. Alignments were carried out with Clustal X.
Immunological methods.
Outer envelope polypeptides were separated by SDS–PAGE (Laemmli, 1970) and stained as above. The polypeptide bands in question were excised from the gel, ground in liquid N2 and used to inject a rabbit. Antisera were used in immunoblots (Towbin et al., 1979) using anti-rabbit alkaline phosphatase and nitroblue tretrazolium and bromo chlorophenol indophenol as dye. The antiserum was named αpsToc75-V and used at 1:1000 dilution.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by the Deutsche Forschungsgemeinschaft, SFB TR 1 and the Fonds der Chemischen Industrie (to J.S. and L.E.).
REFERENCES
- Alefsen H., Waegemann, K. and Soll, J. (1994) Analysis of the chloroplast protein import machinery. J. Plant Physiol., 144, 339–334. [Google Scholar]
- Bölter B., Soll, J., Schulz, A., Hinnah, S. and Wagner, R. (1998) Origin of a chloroplast protein importer. Proc. Natl Acad. Sci. USA, 95, 15831–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Fischer, S.G., Kirschner, M.W. and Laemmli, U.K. (1977) Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem., 252, 1102–1106. [PubMed] [Google Scholar]
- Flack F.S., Loosmore, S., Chong, P. and Thomas, W.R. (1995) The sequencing of the 80-kDa D 15 protective surface antigen of Haemophilus influenzae. Gene, 156, 97–99. [DOI] [PubMed] [Google Scholar]
- Heins L. and Soll, J. (1998) Mixing the prokaryotic and the eukaryotic? Curr. Biol., 8, 215–217. [DOI] [PubMed] [Google Scholar]
- Hinnah S.C., Hill, K., Wagner, R., Schilcher, T. and Soll, J. (1997) Reconstitution of a chloroplast protein import channel. EMBO J., 16, 7351–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D. and Keegstra, K. (2001) Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol., 125, 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeno P., Mini, T., Hintermann, E. and Horst, M. (1995) Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem., 224, 451–455. [DOI] [PubMed] [Google Scholar]
- Keegstra K. and Cline, K. (1999) Protein import and routing systems of chloroplasts. Plant Cell, 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K. and Youssif, A.E. (1986) Isolation and characterisation of chloroplast envelope membranes. Ann. Rev. Plant Physiol., 40, 471–501. [Google Scholar]
- Küchler M. and Soll, J. (2001) From nuclear genes to chloroplast localized proteins. Plant Sci., 161, 379–389. [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T 4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Margulis L. (1970) Origin of Eukaryotic Cells. Yale University Press, New Haven, CT.
- Mourioux G. and Douce, R. (1981) Slow passive diffusion of ortho phosphate between isolated chloroplasts and suspending medium. Plant Physiol., 67, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E. and Keegstra, K. (1994) Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell, 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. and Keegstra, K. (1999) The endosymbiotic origin of the protein import machinery of chloroplastic envelope membranes. Trends Plant Sci., 4, 302–307. [DOI] [PubMed] [Google Scholar]
- Reumann S., Davila-Aponte, J. and Keegstra, K. (1999) The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc. Natl Acad. Sci. USA, 96, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J. Capdevielle, J., Guillemot, J.C. and Ferrara, P. (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203, 173–179. [DOI] [PubMed] [Google Scholar]
- Schnell D.J. and Blobel, G., (1993) Identification of intermediates in the pathway of protein import into chloroplasts and their localization to envelope contact sites. J. Cell Biol., 120, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler, F. and Blobel, G. (1994) Isolation of components of the chloroplast protein import machinery. Science, 266, 1007–1012. [DOI] [PubMed] [Google Scholar]
- Schnell D.J., Blobel, G., Keegstra, K., Kessler, F., Ko, K. and Soll, J. (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol., 7, 303–304. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm, M., Vorm, O. and Mann, M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Svesknikova N., Grimm, R., Soll, J. and Schleiff, E. (2000) Topology studies of the chloroplast protein import channel. Biol. Chem., 381, 687–693. [DOI] [PubMed] [Google Scholar]
- Thomas W.R., Callow, M.G., Dilworth, R.J. and Audesho, A.A. (1990) Expression in Escherichia coli of a high-molecular-weight protective surface antigen found in nontypeable and type b Haemophilus influenzae. Infect. Immun., 58, 1909–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin A., Staehlin, T. and Gordon, J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitro cellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn, J. and Herrmann, R.G. (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem., 180, 535–545. [DOI] [PubMed] [Google Scholar]
- Waegemann K. and Soll, J. (1991) Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J., 1, 149–158. [Google Scholar]