Abstract
We isolated and characterized a new nuclease (NurA) exhibiting both single-stranded endonuclease activity and 5′–3′ exonuclease activity on single-stranded and double-stranded DNA from the hyperthermophilic archaeon Sulfolobus acidocaldarius. Nuclease homologs are detected in all thermophilic archaea and, in most species, the nurA gene is organized in an operon-like structure with rad50 and mre11 archaeal homologs. This nuclease might thus act in concert with Rad50 and Mre11 proteins in archaeal recombination/repair. To our knowledge, this is the first report of a 5′–3′ nuclease potentially associated with Rad50 and Mre11-like proteins that may lead to the processing of double-stranded breaks in 3′ single-stranded tails.
INTRODUCTION
One of the most severe DNA lesions produced by damaging agents are double-stranded breaks (DSBs) that are also induced by disintegration of DNA replication forks. In Bacteria, DSBs are repaired by homologous recombination, whereas, in Eukarya, they are repaired either by homologous or non-homologous recombination (Kowalczykowski et al., 1994; Pâques and Haber, 1999). In Archaea, mechanisms involved in DSB repair have not been clearly identified. Nevertheless, the study of such processes in hyperthermophilic archaea is of a special interest, since these microorganisms are continuously exposed to DNA damaging temperatures and are among the most radioresistant organisms, indicating that DSB repair might be very efficient (Kopylov et al., 1993; DiRuggiero et al., 1997; Gerard et al., 2001). Despite the lack of genetic tools, proteins such as recombinases and Holliday’s junction resolvases have been characterized, showing that homologous recombination also occurs in hyperthermophilic archaea (for a review, see Seitz et al., 2001).
In all organisms studied so far, homologous recombination requires the processing of DSBs in a form that can be utilized by recombinases. These proteins (bacterial RecA, eukaryal Rad51 and Rad51 paralogs and archaeal RadA and RadB) are among the most phylogenetically conserved recombination/repair proteins in the three Domains of life (Aravind et al., 1999; Seitz et al., 2001), and they all require 3′ single-stranded (ss) tails for their loading and the subsequent strand invasion of a homologous DNA duplex.
In Bacteria, the major pathway of DSB processing is quite well understood. This process is performed by the RecBCD complex harboring helicase, endonuclease and both 3′–5′ and 5′–3′ exonuclease activities. This complex acts in conjunction with cis-acting chi sites on the genome that modulate the relative efficiency of the two exonuclease activities, leading to the production of 3′ overhangs (Anderson and Kowalczykowski, 1997). Besides this pathway, other bacterial systems are involved in DSB processing, such as proteins from the RecFOR pathway, and the SbcC–SbcD complex. However, the role of the SbcC and SbcD proteins in such processes is not totally understood (Cromie and Leach, 2001).
In Eukarya, which lack both RecBCD and RecFOR homologs, the processing of DSBs involves the Rad50 and Mre11 proteins, which are homologs to bacterial SbcC and SbcD proteins, respectively (Sharples and Leach, 1995). These proteins act in association with a third partner (for a review, see Haber, 1998): Xrs2 in yeast, and Nbs1 in human (Xrs2 and Nbs1 do not share obvious sequence similarities but could be functional analogs). However, biochemical characterization of the complex does not explain how it performs DSB processing: the Rad50–Mre11 complex exhibits both ss endonuclease and 3′–5′ double-stranded (ds) exonuclease activities, like their bacterial counterparts (Furuse et al., 1998; Trujillo et al., 1998; Usui et al., 1998); the addition of Nbs1 to the human Rad50–Mre11 complex induces partial unwinding of duplex DNA that can account for the production of 3′ tails by endonuclease cleavage, but Nbs1 also induces an ATP dependent switch in endonuclease specificity that leads to cleavage of 3′ protruding tails. Other partners, such as a helicase and/or a 5′–3′ nuclease, should thus be involved in order to produce the 3′ tails required for recombinase loading and DNA strand invasion (Paull and Gellert, 1999).
In Archaea, comparative genomic analyses failed to detect any archaeal RecBCD or RecFOR homologs. In contrast, homologs of Rad50/SbcC and Mre11/SbcD proteins (but not Xrs2 or Nbs1) have been identified in all archaeal genomes (Aravind et al., 1999; Seitz et al., 2001). These proteins have been called Rad50 and Mre11 as in Eukarya and might play a role in recombination/repair in Archaea. The characterization of these proteins from the hyperthermophilic archaeon Pyrococcus furiosus showed that they form a complex exhibiting the same activities as their eukaryal and bacterial counterparts (Hopfner et al., 2000a).
In the present report, we show that rad50 and mre11 homologs of most thermophilic archaea are organized in an operon-like structure with a third gene encoding a protein conserved in all thermophilic species. Characterization of the protein from Sulfolobus acidocaldarius shows that it corresponds to both an ss endonuclease and a 5′–3′ exonuclease able to act on ss and ds substrates. Multiple alignment of all archaeal homologs shows three conserved motifs defining a new nuclease family that we propose to call NurA. This nuclease might be involved, in association with Rad50–Mre11 complex, in the resection of DSBs in hyperthermophilic archaea.
RESULTS
mre11 and rad50 genes are linked to a conserved p39 (nurA) gene in most thermophilic archaeal species
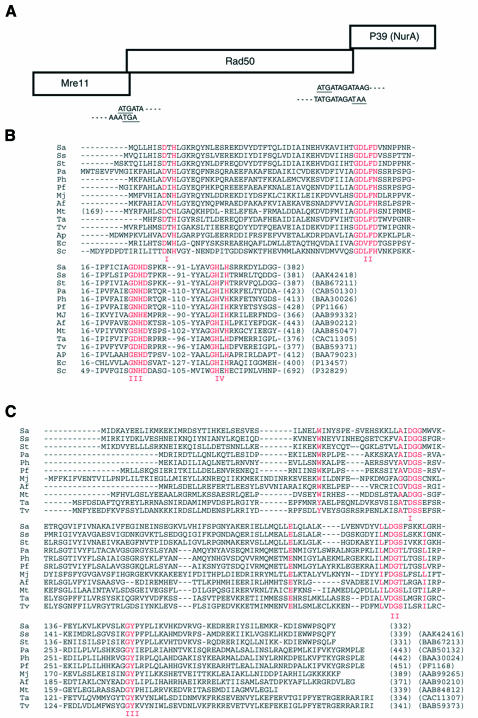
In a previous study, we isolated a gene encoding a putative Rad50 homolog from S. acidocaldarius (Elie et al., 1997). Sequencing of the adjacent regions showed that this gene is organized in an operon-like structure with two other genes, the three genes overlapping each other (Figure 1A). The upstream gene (1149 nt) encodes a predicted 44 kDa protein that exhibits the four phosphodiesterase motifs characteristic of Mre11 proteins (Figure 1B). The downstream gene (999 nt) encodes a predicted 39 kDa protein that does not show obvious sequence similarities with proteins in the databases. However, visual inspection of each alignment obtained with PSI-BLAST program allowed the detection of one homolog in the three thermophilic archaea, Methanococcus jannaschii, Methanobacterium thermoautotrophicum and Aeropyrum pernix, with E values of 7 × 10–15, 1 × 10–10 and 1 × 10–5, respectively. These values are significant, since a cut-off of E < 5 × 10–3 is employed for inclusion of sequences in the position-specific weight matrices (PSI-BLAST NCBI). By sequential use of these three open reading frames (ORFs) as independent queries, we found one homolog in each of the other thermophilic archaeal genomes (E values from 4 × 10–4 to 1 × 10–39). Finally, one homolog was found in the two Sulfolobales genomes recently sequenced (37% identities with the S. acidocaldarius P39 amino-acid sequence). Multiple alignment of all archaeal sequences performed by hand revealed three conserved motifs (Figure 1C). Two of them, localized in the N-terminal part of the sequences, contain the same triplet DGS (motif I, WXnAXDGS; and motif II, EXnhhhhDGS; h indicating a hydrophobic residue), whereas the third one, GY, is found near the C-terminal end. These motifs have not been reported yet, and their functional significance is unknown. Genomic context analysis revealed that, as in S. acidocaldarius, mre11, rad50 and p39 (nurA) genes are organized in the same order in an operon-like structure in 9 of the 11 sequenced thermophilic archaeal genomes from both the Euryarchaea and Crenarchaea phyla (M. jannaschii and M. thermoautotrophicum are the two exceptions).
Fig. 1. (A) Operon-like organization of mre11, rad50 and p39 (nurA) genes found in most hyperthermophilic archaea. Overlapping sequences with underlined start and stop codons are from S. acidocaldarius. Mre11, Rad50 and P39 sequences from S. acidocaldarius reported in this manuscript have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers CAD26844, CAD26845 and CAD26846, respectively. (B) Multiple alignment of Mre11/SbcD sequences showing the four phosphoesterases motifs. Species abreviations: Sa, Sulfolobus acidocaldarius; Ss, S. solfataricus; St, S. tokodaii; Pa, Pyrococcus abyssi; Ph, P. horikoshii OT3; Pf, P. furiosus; Mj, Methanococcus jannaschii; Af, Archeoglobus fulgidus; Mt, Methanobacterium thermoautotrophicum; Ta, Thermoplasma acidophilum; Tv, T. volcanium; Ap, Aeropyrum pernix; Ec, Escherichia coli; Sc, Saccharomyces cerevisiae. A 169 amino-acid N-terminal extension has been deleted from the Mt ORF. The total number of amino acids and accession numbers are given in parentheses at the end of each sequence. Accession numbers for the Pf sequence were provided by the Utah Genome Center. (C) Multiple alignment of P39 (NurA) sequences from thermophilic archaea. The Ap sequence has been omitted, the alignment showing too many gaps, possibly caused by sequencing errors. Only conserved or equivalent amino acids present in all sequences are shown in red.
The P39 protein or ‘NurA’ exhibits ss endonuclease and 5′–3′ ss and ds exonuclease activities
The S. acidocaldarius p39 gene was cloned into pET30 vector to produce the protein with a His6-leader peptide that could be entirely removed by protease cleavage. The recombinant protein was purified to near homogeneity by affinity chromatography on Ni2+-NTA agarose and ion-exchange chromatography on Source 30S and was finally recovered in the non-absorbed fraction of a second Ni2+-NTA agarose column after enterokinase digestion (Figure 2). The protein migrates as a 36 kDa polypeptide following SDS–PAGE, which is in good agreement with the predicted molecular mass. As sequence analyses did not reveal any indication about the protein function, we first tested a putative DNA-binding activity by gel-shift assay and observed the total disappearance of the DNA substrate. Nuclease activity was thus tested with different ss and ds DNA at 70°C. As shown in Figure 3, circular ss DNA is completely degraded after a 5 min incubation, whereas supercoiled DNA is slowly converted to the nicked form and to few linear forms after further incubations. No activity was observed in the presence of Mg2+ or in the absence of Mn2+ (not shown). These results show that P39 exhibits a Mn2+ dependent ss endonuclease activity on closed circular DNA. The activity detected on supercoiled DNA might be explained by the cleavage of ss regions induced by local melting of the substrate at 70°C or by secondary structures. P39 activity was also tested on end-labeled ss and ds linear DNA (only one labeled strand). Analysis of the reaction products on denaturing polyacrylamide gels (Figure 4) shows that, on 5′ end-labeled DNA, only one labeled product corresponding to a small oligonucleotide of about five nucleotides in length is obtained without any trace of intermediate products (95% of the initial radioactivity was recovered in this product even after longer incubation and with a higher protein–DNA ratio; data not shown). In contrast, incubation of P39 with 3′ end-labeled DNA generates intermediate products the length of which slowly decreased upon incubation times (relative amounts of reaction products have been quantified with phosphoimager). The same pattern was obtained on ss and ds linear DNA with a lower degradation efficiency for ds DNA. These results show that P39 protein is associated with an exonuclease activity that degrades ss and ds linear DNA from the 5′ ends to the 3′ ends releasing small oligonucleotides. This exonuclease activity also required Mn2+ (data not shown).
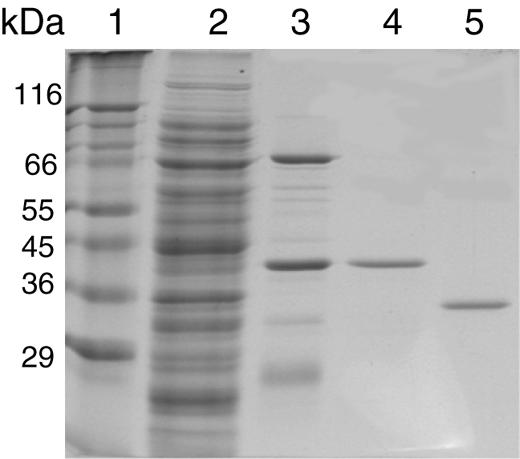
Fig. 2. Purification of recombinant P39 protein. Purification steps were analyzed by SDS–PAGE and Coomassie Blue staining. Lane 1, molecular weight standards; lane 2, soluble fraction of overproducing extracts; lane 3, Ni2+-NTA agarose pool; lane 4, Source 30S pool; lane 5, Ni2+-NTA agarose wash after enterokinase digestion.
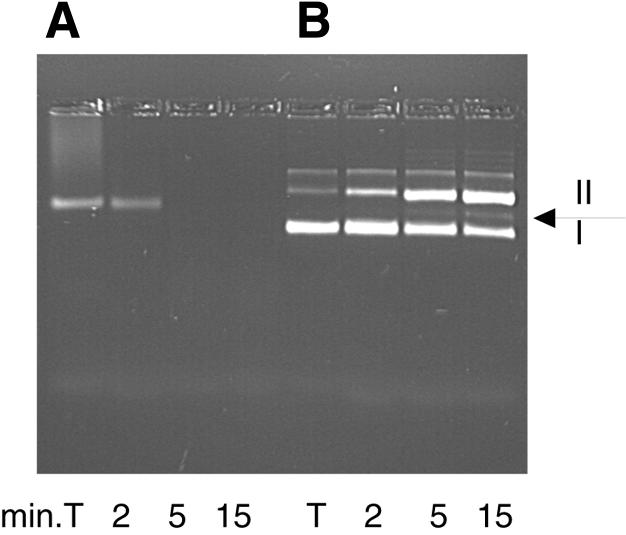
Fig. 3. P39 nuclease activity on ΦX174 circular ss DNA (A) and supercoiled pBR322 DNA (B). I, supercoiled DNA; II, nicked DNA. Linear DNA is indicated by the arrow. In lane T, DNA was incubated without P39 for 15 min.
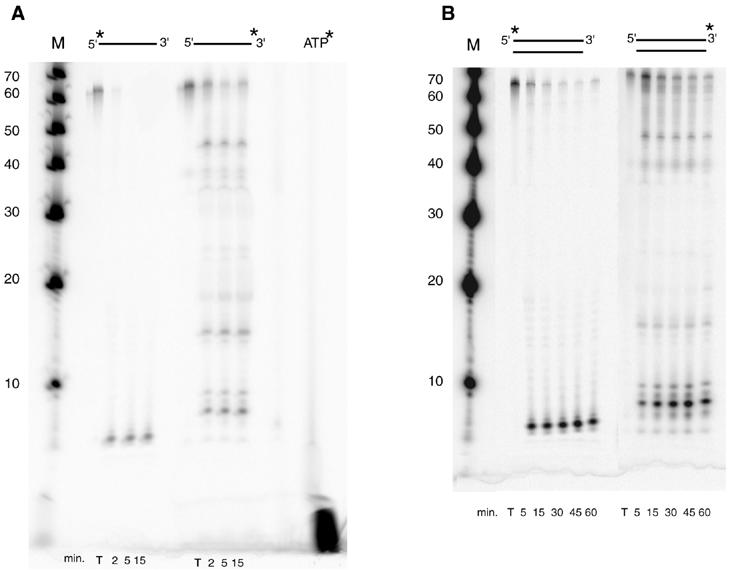
Fig. 4. Time-course of P39 activity on ss (A) and ds (B) end-labeled linear DNA. The asterisk indicates the position of labeling. M, 3′ end-labeled 10 bases DNA ladder; T, DNA incubated without P39 for 15 min (A) or 60 min (B).
All activities associated with P39 protein are recovered at 70°C, which corresponds to the growth temperature of S. acidocaldarius.
DISCUSSION
Besides RecA/Rad51/RadA recombinases, Rad50/SbcC and Mre11/SbcD proteins are the only universal DNA recombination/repair proteins. In Bacteria, these proteins are at least involved in two pathways: recombination processes and elimination of palindromic sequences during DNA replication (Connelly and Leach, 1996). In Eukarya, Rad50–Mre11 proteins constitute, with the Xrs2/Nbs1 protein, the core element of many different DNA metabolism pathways, i.e. homologous and non-homologous recombination, telomeres maintenance, cell’s response checkpoint to DSBs and formation of meiotic DSBs (for a review, see Haber, 1998). These proteins should also play an important role in DNA metabolism pathway(s) in Archaea.
In this study, we report the identification of a new p39 gene organized in an operon-like structure with the rad50 and mre11 homologs from the hyperthermophilic archaeon S. acidocaldarius. Characterization of the P39 protein shows that it exhibits both a ss endonuclease activity on closed circular DNA and an exonuclease activity on linear ss and ds DNA, as reported for yeast Mre11. P39 protein degrades linear DNA from the 5′ ends to the 3′ ends, the opposite direction of Mre11 exonuclease, with a preference for ss DNA substrate. In each case, reaction products are small oligonucleotides, as reported for the exonuclease activities associated with Escherichia coli RecBCD complex. All activities are manganese dependent, as in the case of nucleases activities associated with Mre11/SbcD proteins from Bacteria, Eukarya and Archaea.
Moreover, we show that this nuclease is phylogenetically conserved in most thermophilic archaea. The archaeal homologs only exhibit low-level amino-acid sequence identity but display three conserved motifs that differ from nuclease motifs described previously (Aravind et al., 1999; Makarova et al., 2002). Nevertheless, these motifs contain invariant residues that have been shown to participate in catalysis in several endonucleases and exonucleases families (Beese and Steitz, 1991; Aravind et al., 1999). In particular, the carboxylate residues may serve as ligands to divalent metal ions that catalyze the phosphoryl transfer reaction, and both a glycine and a tyrosine residue have been shown to be involved in 5′–3′ exonuclease activity of E. coli DNA polymerase I (Xu et al., 1997). At present, we have failed to detect obvious homologs of this nuclease in Bacteria and Eukarya by iterative BLAST analyses, and we thus propose to call this new nuclease ‘NurA’ (for ‘Nuclease repair of Archaea’).
Finally, we show that the rad50, mre11 and nurA genomic context is conserved in most thermophilic archaea. These data strongly suggest that the three proteins are involved in the same molecular pathway, since organization of the three genes has been preserved from extensive genes rearrangements that occur in prokaryotic genomes (Huynen and Bork, 1998). Considering the 5′–3′ direction of the exonuclease activity of NurA, an attractive hypothesis is that, in thermophilic archaea, Rad50, Mre11 and NurA proteins act together in the processing of DSBs at the initiation step of homologous recombination. In that case, the exonuclease activities associated with the three proteins (Mre11 3′–5′ activity and NurA 5′–3′ activity) should be regulated in order to produce 3′ overhangs. This regulation could be performed either by protein–protein interactions, by ATP, since it has been shown that the 3′–5′ exonuclease activity of P. furiosus Rad50-Mre11 is ATP dependent (Hopfner et al., 2000b), and/or by other factors. Characterization of the concerted action of the three proteins from S. acidocaldarius and a search for potential additional partners are presently in progress in our laboratory.
Possibly, archaeal NurA has bacterial and eukaryal homologs that do not exhibit enough sequence similarity to be detected in classical BLAST searches. Our identification of nuclease motifs typical for NurA should help in the detection of such remote homologs.
METHODS
Sequences and computer analyses. Cloning and sequencing of S. acidocaldarius operon were performed as described previously (Elie et al., 1997). Comparative sequences analyses were performed by PSI-BLAST program (Altschul and Koonin, 1998) and multiple alignment by CLUSTAL W program (Thompson et al., 1994) or by hand. Genomic context analyses were performed by the GENOMAPPER program developed by Y. Zivanovic in our laboratory.
Overproduction and purification of P39 protein. The p39 gene was amplified by PCR using the S. acidocaldarius λgt11 clone described previously (Elie et al., 1997) and inserted in pET30 Ek/LIC vector (Novagen). As E. coli and S. acidocaldarius present differences in codon usage, E. coli BL21(DE3)pLysS cells were cotransformed with pET30-p39 and a plasmid bearing E. coli tRNAArg(AGA, AGG) and tRNAIle(AUA) genes. Inductions were performed overnight at room temperature after a cold shock of 1 h at 4°C. Cells were resuspended in buffer A [20 mM HEPES pH 7.5, 1 M NaCl, 0.03% (v/v) Tween-20, 1 mM PMSF, 5 mM β-mercaptoethanol, 1 µg/ml pepstatine and leupeptine] and disrupted by sonication, and the soluble fraction was loaded onto a 1 ml Ni2+-NTA agarose column (Qiagen). After a 10 vol. wash in buffer A and a 5 vol. wash with 20 mM imidazole in buffer B (buffer A containing 50mM NaCl), proteins were eluted with 200 mM imidazole. Imidazole was removed with a PD-10 column (Bio-Rad) in buffer B and the fraction was loaded onto a 1 ml Source 30S column (Pharmacia). After a 10 vol. wash in buffer B, a 30 ml linear salt gradient from 50 mM to 1 M NaCl was applied. P39 was eluted at 430 mM NaCl. Fractions were dialyzed against buffer B and loaded onto a 0.5 ml Ni2+-NTA agarose column, and enterokinase (Novagen) digestion was performed on the column overnight at 16°C. P39 protein was recovered by a simple wash with buffer B.
Nuclease activities. Purified P39 protein (0.2 pmol) was incubated with 0.1 pmol of DNA in 20 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MnCl2, 0.5 mM DTT and 100 µg/ml BSA at 70°C. Reactions were stopped by the addition of 1% SDS and 1 mg/ml Proteinase K with a further incubation at 50°C for 30 min, and DNA was extracted by phenol–chloroform. Reaction products were either analyzed by electrophoresis on 0.7 % agarose gel and ethidium bromide staining (circular DNA) or on 10% polyacrylamide/7 M urea gels which were scanned by phosphoimager (linear DNA). Linear ss DNA corresponds to the 65mer oligonucleotide, GCTATCATGGAATCTCGTTTATCAGACTGGAATTCAAGCGCGAGCTCGAATAAGAGCTACTGTGG, either 3′ end-labeled with [α-32P]ddATP (3000 Ci/mmol, Amersham) and terminal deoxynucleotidyl transferase (Promega) or 5′ end-labeled with [γ-33P]ATP (2500 Ci/mmol, Amersham) and T4 polynucleotide kinase (Promega). Linear dsDNA corresponds to the same 3′ or 5′ end-labeled oligonucleotide annealed to the complementary oligonucleotide.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. Bühler and F. Mastsunaga for helpful discussions and critical reading of the manuscript. We also thank Y. Zivanovic and B. Labedan for their help in computer analyses and N. Joly for his contribution in p39 expression. This work was supported by Electricité de France and the Association pour la Recherche sur le Cancer (ARC). F.C. was supported by a fellowship from the ARC.
REFERENCES
- Altschul S.F. and Koonin, E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- Anderson D.G. and Kowalczykowski, S.C. (1997) The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev., 11, 571–581. [DOI] [PubMed] [Google Scholar]
- Aravind L., Walker, D.R. and Koonin, E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese L.S. and Steitz, T.A. (1991) Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.C. and Leach, D.R. (1996) The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells, 1, 285–291. [DOI] [PubMed] [Google Scholar]
- Cromie G.A. and Leach, D.R. (2001) Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol. Microbiol., 41, 873–883. [DOI] [PubMed] [Google Scholar]
- DiRuggiero J., Santangelo, N., Nackerdien, Z., Ravel, J. and Robb, F.T. (1997) Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol., 179, 4643–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elie C., Baucher, M.F., Fondrat, C. and Forterre, P. (1997) A protein related to eucaryal and bacterial DNA-motor proteins in the hyperthermophilic archaeon Sulfolobus acidocaldarius. J. Mol. Evol., 45, 107–114. [DOI] [PubMed] [Google Scholar]
- Furuse M., Nagase, Y., Tsubouchi, H., Murakami-Murofushi, K., Shibata, T. and Ohta, K. (1998) Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J., 17, 6412–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard E., Jolivet, E., Prieur, D. and Forterre, P. (2001) DNA protection mechanisms are not involved in the radioresistance of the hyperthermophilic archaea Pyrococcus abyssi and P. furiosus. Mol. Genet. Genomics, 266, 72–78. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1998) The many interfaces of Mre11. Cell, 95, 583–586. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher, A., Shin, D., Fairley, C., Tainer, J.A. and Carney, J.P. (2000a) Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol., 182, 6036–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher, A., Shin, D.S., Craig, L., Arthur, L.M., Carney, J.P. and Tainer, J.A. (2000b) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Huynen M.A. and Bork, P. (1998) Measuring genome evolution. Proc. Natl Acad. Sci. USA, 95, 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylov V.M., Bonch-Osmolovskaya, E.A., Svetlichnyi, V.A., Miroshnichenko, M.L. and Skobkin, V.S. (1993) γ-irradiation resistance and UV-sensivity of extremely thermophilic archaebacteria and eubacteria. Mikrobiologiya, 62, 90–95. [Google Scholar]
- Kowalczykowski S.C., Dixon, D.A., Eggleston, A.K., Lauder, S.D. and Rehrauer, W.M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind, L., Grishin, N.V., Rogozin, I.B. and Koonin, E.V. (2002) A DNA repair system specific for thermophilic Archaea and Bacteria predicted by genomic context analysis. Nucleic Acids Res., 30, 482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F. and Haber, J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T.T. and Gellert, M. (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev., 13, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz E.M., Haseltine, C.A. and Kowalczykowski, S.C. (2001) DNA recombination and repair in the archaea. Adv. Appl. Microbiol., 50, 101–169. [DOI] [PubMed] [Google Scholar]
- Sharples G.J. and Leach, D.R. (1995) Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol., 17, 1215–1217. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo K.M., Yuan, S.S., Lee, E.Y. and Sung, P. (1998) Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem., 273, 21447–21450. [DOI] [PubMed] [Google Scholar]
- Usui T., Ohta, T., Oshiumi, H., Tomizawa, J., Ogawa, H. and Ogawa, T. (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell, 95, 705–716. [DOI] [PubMed] [Google Scholar]
- Xu Y., Derbyshire, V., Ng, K., Sun, X.C., Grindley, N.D. and Joyce, C.M. (1997) Biochemical and mutational studies of the 5′-3′ exonuclease of DNA polymerase I of Escherichia coli. J. Mol. Biol., 268, 284–302. [DOI] [PubMed] [Google Scholar]