Abstract
tRNA function is based on unique structures that enable mRNA decoding using anticodon trinucleotides. These structures interact with specific aminoacyl-tRNA synthetases and ribosomes using 3D shape and sequence signatures. Beyond translation, tRNAs serve as versatile signaling molecules interacting with other RNAs and proteins. Through evolutionary processes, tRNA fragmentation emerges as not merely random degradation but an act of recreation, generating specific shorter molecules called tRNA-derived small RNAs (tsRNAs). These tsRNAs exploit their linear sequences and newly arranged 3D structures for unexpected biological functions, epitomizing the tRNA “renovatio” (from Latin, meaning renewal, renovation, and rebirth). Emerging methods to uncover full tRNA/tsRNA sequences and modifications, combined with techniques to study RNA structures and to integrate AI-powered predictions, will enable comprehensive investigations of tRNA fragmentation products and new interaction potentials in relation to their biological functions. We anticipate that these directions will herald a new era for understanding biological complexity and advancing pharmaceutical engineering.
eTOC blurb
tRNA-derived small RNAs (tsRNAs) are increasingly recognized as critical regulators of cellular physiology. Kuhle et al. review how tsRNA functionalities are intricately linked to their 3D structures and their dynamic modulation by modifications and binding partners, highlighting the promise of new experimental and AI-based approaches to tackle their biological complexity.
Introduction:
The transfer RNA (tRNA) stood at the center of an entire research era dedicated to the elucidation of the molecular mechanisms underlying cellular information processing. First discovered in 1958 and identified as the small adaptor molecule that Francis Crick had predicted in his ‘adaptor hypothesis’ in 1954, tRNAs became a central focus of biochemists’ and molecular biologists’ efforts to unravel the mysteries of biological complexity1–3. Contemporaneously, aminoacyl-tRNA synthetases (aaRSs) were identified as the enzymes that catalyze the ATP-dependent matching of each amino acid with its cognate tRNA4,5, thereby establishing the rules of the universal genetic code. Another milestone was reached when, in 1965, the complete chemical structure of yeast tRNAAla was elucidated6, followed, about eight years later, by the 3D X-ray crystal structure of tRNAPhe from yeast7,8. These studies revealed an intricate, highly condensed secondary and tertiary structure, which finally provided a framework to understand the moleclular basis of genetic code expression and protein biosynthesis. It served as a model to study the recognition principles in protein-RNA interactions, the role of modified nucleotides and metal ion coordination in RNA folding and function, and it lent credibility to the idea that, during the early phases of cellular evolution, catalytically active RNAs could have predated protein-based enzyme catalysis9. The complex structure-function relationship of tRNA biology established that for RNAs, as for proteins, function follows form.
Recent years have seen renewed interest in tRNA biology, driven by studies into the unexpected functions of tRNAs in shaping cellular responses to environmental cues. These new tRNA functions range from adaptive protein synthesis to signaling in diverse regulatory networks. In particular, the discovery of tRNA-derived small RNA (tsRNA), and the striking complexity of their biological functions, epitomize the tRNA ‘renovatio’ [Latin, meaning renewal, renovation, and rebirth] as regulatory small RNAs.
In this review, we focus on the overall complexity of the emerging human tRNA biology and, with a few illustrative examples, highlight its basis and physiological importance. A particular emphasis is thereby put on the structures of tsRNAs to understand their biological functions, a concenpt that was first shaped more than 50 years ago by the study of tRNAs.
The initial discoveries that tRNA fragmentation generates functional small RNAs, known as tRNA-derived small RNAs (tsRNAs), came as a surprise10. However, it has since been proven to be an evolutionarily conserved process and is now recognized as an ancient method of small RNA biogenesis, regulated by various ribonucleases (RNases) and tRNA modifications, and highly responsive to dynamic cellular conditions11. This revelation has stimulated a new direction in tRNA biology and aligns well with the idea that nature is efficient, conservative, and tends to repurpose available molecular components with functional importance. In this case, the regulated tRNA fragmentation and selective retention of tsRNAs exemplify an adaptive and interconnected phenomenon resulting from evolutionary refinement. We emphasize the crucial point that tRNA fragmentation is not merely a process of degradation but one of creation, whereby numerous tRNA fragments (tsRNAs) are generated, and then deployed for a particular biological function.
The highly regulated process of tsRNA biogenesis and functionality also highlights the importance of continuing to explore the diverse functions of previously neglected small non-coding RNAs (sncRNAs)12. In fact, this exploration stimulated studies of other sncRNA types derived from the fragmentation of various RNA precursors, including rRNAs, snoRNAs, snRNAs, Y RNAs, and vault RNAs, among others12−14. Our understanding of their biogenesis, functions, and interactions continues to evolve as we explore the endless frontier of the RNA world.
Although the topic of tsRNA has been reviewed elsewhere11,15, here we aim to provide a fresh perspective by focusing on tRNA-derived structures – the cornerstone of their functionality. This focus is driven by our updated understanding of tRNA’s role in many unexpected functions that depend on unique structures, and their dynamic regulation. Importantly, many tsRNAs exert their functions not only through linear sequences for base-pairing under the well-defined RNAi doctrine, but also by employing diverse secondary and tertiary structures that generate various binding potentials.
Delving into these new functions starts with a deeper understanding of RNA modifications and subcellular environments that control the folding dynamics and interactions of tRNAs and tsRNAs. In pursuit of these objectives, we discuss state-of-the-art methods for analyzing tRNA/tsRNA sequences and their modifications, methods for studying tsRNA 3D structures, and the state of emerging AI-based predictions. Finally, we envision how these efforts, centered on tRNA structures and fragmentation, bring new excitement for understanding both biological complexity and for how to apply this understanding to new classes of therapeutic medicines.
1. The intricate world of tRNA structures
a. RNA structure defines its function - tRNA as a paradigm
In 1966, Francis Crick noted that “it almost appears as if tRNA were Nature’s attempt to make an RNA molecule play the role of a protein”2. Like for proteins, the functional tRNA fold can be decomposed into a primary, secondary, and tertiary structure16. The primary structure is usually comprised of 76–90 nucleotides. It adopts a characteristic cloverleaf-like secondary structure, in which the four arms are designated as acceptor stem, dihydrouridine (D) stem-loop, anticodon stem-loop, and TψC (T) stem-loop. The terminal D-loop and T-loop then join to form an intricate network of tertiary interactions, which arranges the individual structural subdomains into an L-shaped overall fold that defines tRNAs from all three domains of life. Notably, while Watson-Crick pairs of the typical A-type RNA helix define the tRNA secondary structure, the tertiary fold relies almost exclusively on interactions between non-Watson-Crick pairs. This highlights the importance of non-WC pairs to expand the diversity of possible interactions in functional RNA folding17,18.
tRNA function is exquisitely sensitive to this conserved L-shaped architecture16,19. On the one hand, it forms the uniform platform for interactions that are common to all tRNAs, from interactions with processing enzymes that sample the shapes and overall dimensions of tRNA subdomains, to their precise fit into the ribosome. On the other hand, tRNAs also present unique, highly differentiated surfaces that enable a diversity of specific interactions that are idiosyncratic to each tRNA19,20. A familiar example is the anticodon-codon interaction between tRNA and mRNA. Another example is the way in which aminoacyl-tRNA synthetases (aaRSs) pick out their cognate tRNAs, and simultaneously reject all others, to establish the rules of genetic code expression.
b. tRNA structure in extra-translational functions
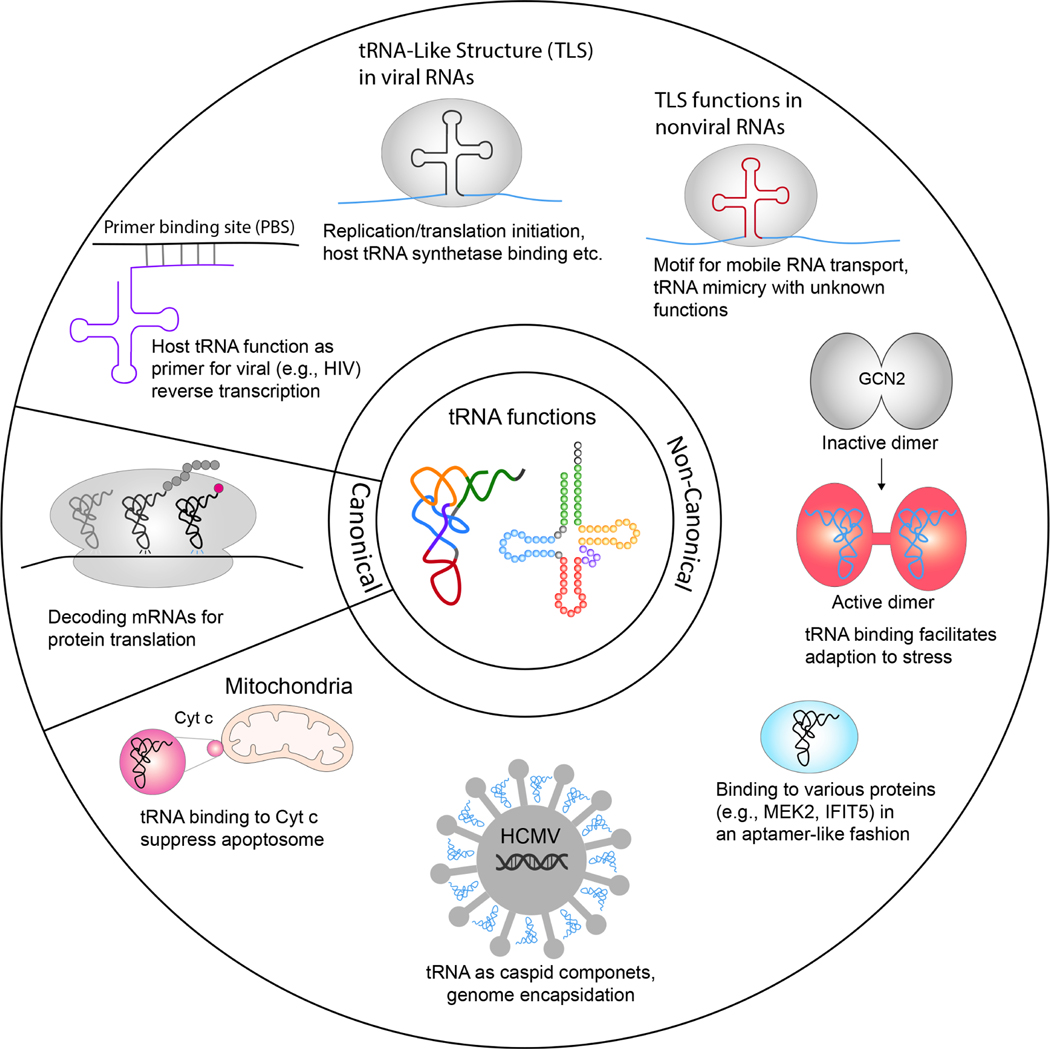
tRNAs were long considered to be largely confined to their role as adaptors in translation. Recent years have seen increasing evidence for critical functions of tRNAs outside of the core translation process10 (Figure 1). In one of the earliest documented examples, tRNAs were demonstrated to act as key metabolic sensors in the integrated stress response to nutrient deprivation21. In this example, their deacylated form binds and activates Gcn2. Other examples include tRNA binding to cytosolic Cytochrome c, leading to the suppression of apoptosome formation22, to the interferon-induced tetratricopeptide repeat protein IFIT5 and schlafen 11, to promote cellular antiviral defense mechanisms23,24, and to the mitogen-activated protein kinase 2 (MEK2) in pancreatic cancer cells, possibly affecting its role in cell cycle progression and proliferation25. Interestingly, the correct identification of MEK2, along with various other human proteins, as a tRNA-binding protein was based on a machine learning algorithm that specifically searched for a protein’s binding potential to the unique L-shaped tRNA structure26. Similarly, a recent study revealed how the capsid-bound tegument protein pp150 on human cytomegalovirus (HCMV) uses a shape-complementary positively charged patch to bind host tRNAs27, possibly to provide stability to the viral capsid and facilitate viral genome encapsulation.
Figure 1. Canonical and non-canonical tRNA functionality.
Illustration of canonical tRNA function in decoding the genetic code of mRNAs and deliver cognate amino acids to the ribosome for protein synthesis. Shown also is the expanding non-canonical tRNA functions in the cell. These non-canonical functions leverage the structural features of tRNA to interact with specific proteins, thereby regulating cellular physiology and mediating critical viral-host interactions across various biological processes.
Viruses also use tRNA-like structures (TLS) to hijack pre-existing tRNA functional networks in the host cell. For example, the 3ʹ-ends of various RNA viruses use structural mimics of entire tRNAs or its subdomains as recognition signal for replication initiation28. Similarly, translation initiation on many viruses is promoted by structures in the 3’-UTR, as well as by internal ribosomal entry sites (IRES) that mimic tRNAs, or their anticodon domains, to facilitate ribosome recruitment and reading frame selection29,30. More recent evidence shows how the brome mosaic virus uses a conformationally dynamic TLS to bind the host tyrosyl-tRNA synthetase (TyrRS), a binding process that differs greatly from how tRNA typically binds to the same enzyme. This highlights how dynamic structural changes of viral TLS can manipulate host machinery through non-standard interactions31. Accumulating evidence also suggests that TLS are not confined to viruses, but occur frequently in non-viral mRNAs as well as in non-coding RNAs from bacteria, yeast, plants, and humans32–39. It is obvious from these examples that the use of the tRNA shape as a regulatory, aptamer-like element is far more widespread than was appreciated up until only recently40. Moreover, they highlight the potential for TLS-containing RNAs to interfere with central cellular processes, a potential that may be exploited extensively by tRNA-derived small RNA.
c. RNA modifications regulate conformational plasticity and information content.
The unique overall fold of tRNA has a high degree of structural plasticity16,41. While its ‘ground state’ is the overall L-shaped fold, the tRNA body has the capacity to undergo local structural changes or global twisting and bending to adapt for interactions. Post-transcriptional modifications modulate a tRNA’s stability and structural dynamics42,43. With human nuclear-encoded tRNAs containing on average 11–13 modifications per molecule44,45, they have the highest density of post-transcriptional modifications among all RNAs. These modifications range from simple methylations to highly sophisticated hypermodifications that greatly increase the information content and functional diversity of tRNAs in vivo42,46.
Modifications as simple as methylations can have diverse and position-dependent effects on the tRNA structure43. For instance, nucleobase methylation on the Watson-Crick edge (e.g., m1A, m1G, and m3C) interferes with canonical Watson-Crick H-bonding, while at the same time promoting alternative, non-canonical interactions. On the other hand, methylation of nucleobases outside of the Watson-Crick edge, such as 5-methylcytosine (m5C), does not interfere with Watson-Crick base-pairing but increases the stability of stacking interactions47. Similarly, pseudouridylation (ψ) increases the stability of stem regions, whereas dihydrouridine (D) destabilizes the stacking interactions required for an A-type RNA helix so that loop formation is favored42,43.
The strategic deposition or removal of modifications in the tRNA structure thus can increase rigidity in one region, while increasing flexibility in another. Recent studies have begun to unravel the dynamic interplay between factors that encode (writers) or remove (erasers) tRNA modifications in response to varying cellular conditions, environmental stresses, and their biological consequences. These functional effects range from stress-induced tRNA degradation and adaptive translation to the site-specific endonucleolytic cleavage of tRNAs into tsRNAs with distinct biological funcitons48,49.
2. tRNA fragmentation: a rebirth of function
a. Brief survey of current state
Early studies of tsRNAs originated from the observation that tRNA fragmentation increases under various stress conditions in both unicellular organisms and mammalian cells50–56. These findings not only led to the discovery of tsRNAs’ role in regulating cellular responses to stress conditions but also identified the enzymes responsible for cleaving tRNAs into tsRNAs53–55,57,58. Further research based on traditional small RNA sequencing revealed that tsRNAs can be significantly more enriched than other well-studied small RNAs (e.g., miRNAs, piRNAs) even under physiological conditions, such as in mature sperm59. In this context, sperm tsRNAs serve a role in carrying hereditary information, by contributing to what we call the ‘sperm RNA code’60. This code plays a critical role in mediating the paternal epigenetic inheritance of environmental stressors61–68. Additionally, tsRNAs have been detected in the sera of a wide range of species under physiological conditions69,70. They are highly responsive to infections and to various pathological conditions, thus making them promising candidates for novel biomarkers of diseases, including cancer and neurological disorders71–74.
Recent advancements in small RNA-sequencing techniques that overcome modifications harbored by tsRNAs led to the detection of a greater abundance of tsRNAs across various tissues and cells75,76. Further studies have shown that RNA modifications alter the function of tsRNAs63,77–79. These discoveries put tsRNA research into the limelight, branching into several directions, including the regulation of biogenesis, disease associations, and the study of fundamental functional principles (Figure 2). Significantly, although some tsRNAs function in an RNAi-like manner58,80–84, a growing body of evidence suggests that the biological activities of many tsRNAs are based on their secondary and tertiary structures (Figure 3).
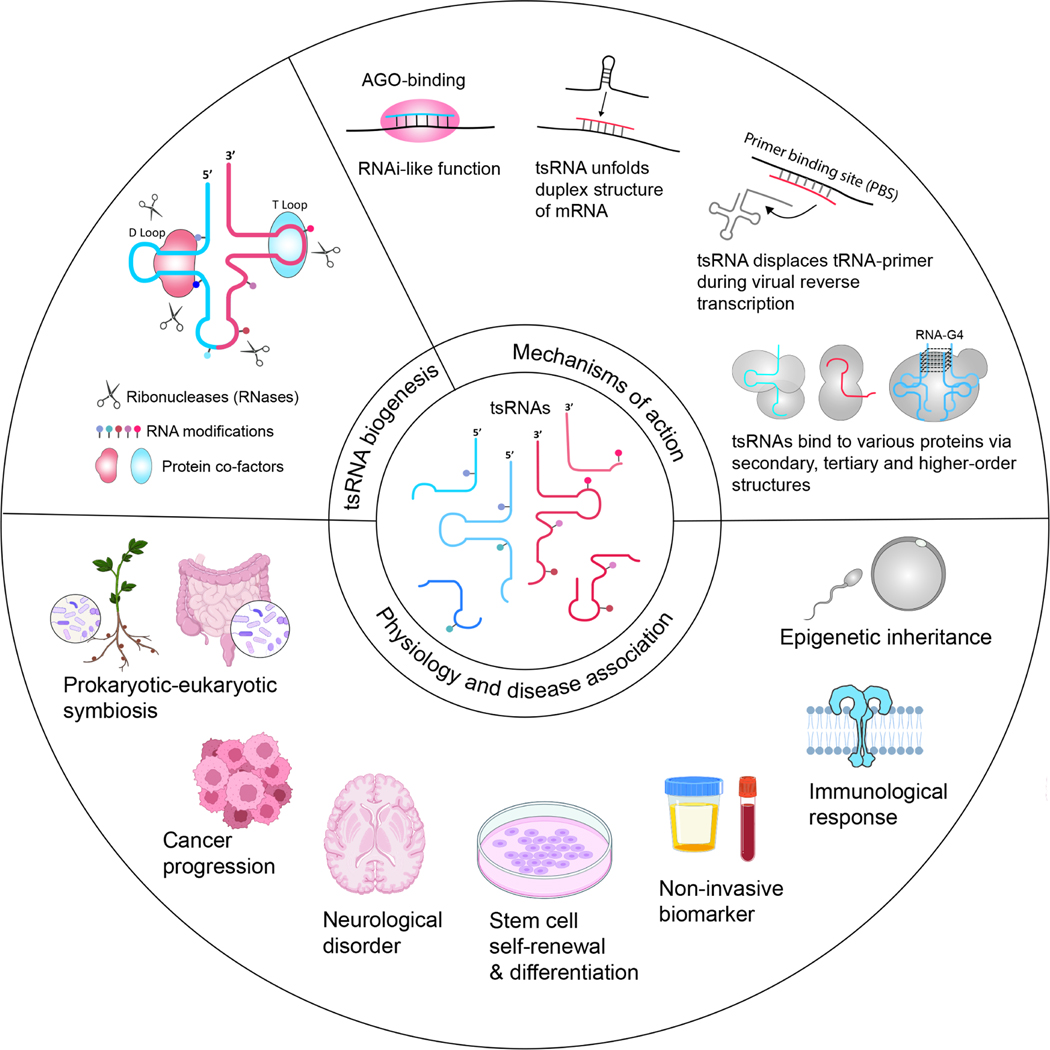
Figure 2. Overview of the expanding field of tsRNA research.
This illustration depicts the three main branches of current tsRNA research. These branches encompass the study of tsRNA biogenesis, which involves various tRNA modifications, RNases, and regulatory proteins. The molecular mechanisms of actions, and the roles of tsRNAs in both physiological functions and disease associations, is based on previous research and review articles10,11,15,82,214–218.
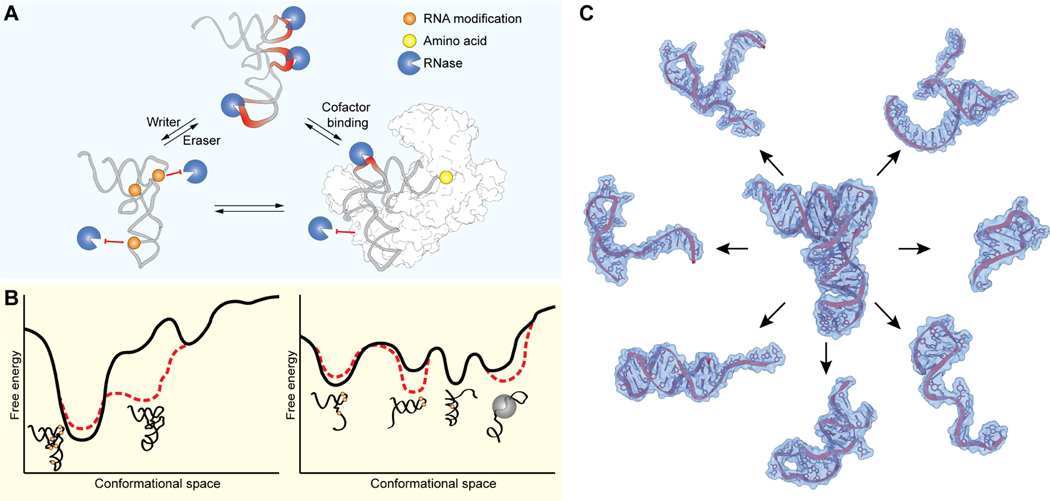
Figure 3. Structural plasticity in tRNAs and tsRNAs and their modulation by RNA modifications and cofactor binding.
(A) Illustration showing how post-transcriptional RNA modifications and tRNA-binding proteins act to regulate RNase-mediated tRNA cleavage and tsRNA formation. (B) Schematic presentation of the conformational free-energy landscapes of tRNAs (left) and tsRNAs (right) and their modulation in response to changes e.g., in modification state or ligand binding. (C) Structural view of predicted tsRNAs derived from a single tRNAAla precursor.
b. RNA modifications control tsRNA formation
A central question regards the mechanisms underlying the regulation and specificity of tRNA fragmentation, which may be referred to as the “tRNA fragmentation code.” This code encompasses: the susceptibility of certain tRNAs to fragmentation due to their structures and modifications, the determination of cleavage sites by ribonucleases, and the selective retention of specific fragments (e.g., 3’ vs. 5’ halves) following enzymatic nicking. Accumulating evidence points to a deep connection between tRNA modifications and tsRNA formation. Individual modifications may have both stimulatory and inhibitory effects on tRNA fragmentation, and their impact depends on the specific tRNA and cleavage site. tRNA fragments mostly arise from mature tRNAs, with most tsRNAs generated through endonucleolytic cleavage in the loop regions, where the accessibility of the tRNA backbone for cleavage can regulate tsRNA formation. As an example, the endonuclease angiogenin (ANG) requires unstructured, single-stranded regions to catalyze hydrolysis of the phosphodiester backbone. Certain modifications, such as cytosine-C5 methylation (m5C) deposited by the writers DNMT2 and NSUN2, protect tRNAs from endonucleolytic cleavage by ANG and reduce 5’ tRNA half production (Figure 3). Conversely, deletion of DNMT2 and NSUN2 increases tRNA susceptibility to cleavage into tsRNAs63,85–89.
TRMT10A-mediated N1-methylguanine (m1G) modification on G9 of tRNAGln stabilizes tRNAGln and prevents tsRNA production90. On the other hand, TRMT10A deficiency-induced G9 hypomethylation leads to apoptosis in β-cells due to the accumulation of 5’ tsRNAGln. Likewise, increased levels of N1-methyladenine (m1A) and 3-methylcytidine (m3C) in tRNAs in response to reduced levels of the tRNA demethylases ALKBH1 and ALKBH3 prevent ANG-mediated tRNA cleavage and the formation of specific tsRNA subsets91,92.
Further fragmentation regulation is provided by modifications in the anticodon wobble position 34 of tRNAs. For instance, 2′-O-methylation of C34 in human elongator tRNAMet prevents stress-induced cleavage by angiogenin93. Simlarly, queuosine (Q) modification, incorporated by the QTRT1/QTRT2 complex into position 34 of certain tRNAs, was also reported to protect tRNAs against angiogenin-mediated cleavage94.
While most reported cases point to a protective effect of modifications against tRNA fragmentation, some studies suggest that modifications can promote tsRNA biogenesis. PUS7-catalyzed pseudouridylation (Ψ) at position U8 of tRNAs increases levels of several types of TOG-containing (i.e., a terminal oligo-guanine motif) short 5’ tsRNAs in stem cells, suggesting that Ψ8 may promote cleavage of these tRNAs78. In another example, the m22G26 modification in tRNALeu(CAA) is protective against fragmentation at the anticodon-loop, but promotes cleavage at the V-loop95. Finally, the 5-methoxycarbonylmethyl-2-thiouridine (mcm5S2) modification at the anticodon wobble position of yeast tRNAs promotes site-specific cleavage into tsRNAs by the Kluyveromyces lactis γ-toxin96,97.
c. tRNA-binding proteins act as cofactors in tsRNA formation:
Post-transcriptional modifications and tRNA structural dynamics affect interactions with tRNA-binding proteins (tRBPs). Interestingly, several recent studies suggest that the tRNA aminoacylation state (determined by the activity of aminoacyl-tRNA synthetases) may serve as a cue for tsRNA formation. First evidence came from a study of sex-hormone-dependent cancer cells, which express a highly specific subset of ANG-dependent tsRNAs from tRNALys and tRNAHis, with the release of aminoacylated 3’ tsRNAs98. Upregulation of the corresponding 5’ tsRNAs promotes cancer cell proliferation through an unknown mechanism. A more recent study found that 22 nt 3’ tsRNAs from tRNALeu, tRNAAla, and tRNAGly are fully aminoacylated under normal growth conditions in HeLa cells99. Interestingly, the levels of the tRNALeu-derived tsRNA, which specifically enhances translation of a subset of mRNAs encoding ribosomal proteins (RPS) in human and mouse cells100,101, depend on the aminoacylation state and the presence of the leucyl-tRNA synthetase (LeuRS). Depletion of eEF1A, the elongation factor that delivers aminoacylated tRNAs to the ribosome, further increases the expression of these charged 3’ tsRNAs. Besides the functional implications of these results for a possible regulatory circuit between tRNA-charging levels, RPS translation, and ribosome biogenesis, they also raise the intriguing possibility that aaRSs and other tRBPs may serve as regulatory co-factors in tRNA fragmentation as a response to cellular cues (Figure 3A−C).
Further support for this notion comes from a study in the ciliate Tetrahymena, which showed that the Ago/Piwi protein Twi12 binds tRNAs, promotes their fragmentation, and specifically retains 18–22 nt long 3’ tsRNAs that stimulate the association of Twi12 with other proteins and import into the nucleus102. Moreover, Hasler et al. showed that the lupus autoantigen (La) protein functions as an RNA chaperone for pre-tRNAs to promote correct folding and processing into mature tRNA. Yet, some pre-tRNAs can adopt alternative pre-microRNA-like hairpin structures which are efficiently cleaved by Dicer to produce tsRNAs. Accordingly, reduced levels of La result in elevated levels of cellular tsRNAs103.
d. tsRNA structure – beyond the RNAi doctrine:
The default view of tsRNAs is that of unstructured small non-coding RNAs, in which linear sequence information mediates biological function by antisense matching to target sequences. To some degree, this view may be a vestige of the historical context when research was focused on RNAi-based post-transcriptional control of gene expression. Studies are now uncovering additional layers of biological information outside of the RNAi-doctrine, which is encoded in the structural properties of tsRNAs.
RNA has the capacity to fold into an array of complex structural states (Figure 3). Depending on its length, sequence, and external cellular conditions, a given RNA may populate an ensemble of alternative conformations with distinct compactness, stabilities, and flexibilities104,105. The dynamic equilibrium between these alternative states is dictated by the nature of the RNA’s conformational free-energy landscape, which can lie anywhere on a spectrum between a steep topography, populated by a few stable and highly favored conformations, and a flat landscape with many unstable structural states that readily interconvert into each other. Much of the functional complexity of RNAs is rooted in their ability to dynamically adapt this conformational landscape in response to specific cellular signals, which includes binding of other macromolecules or small molecule ligands, changes in temperature or ion concentrations, chemical modifications, and mutations104,106,107. The folding of mature tRNA under normal conditions is an example for a steep free-energy landscape (Figure 3B). For the majority of nucleotide positions in a tRNA the possibilities for alternative conformations and interactions are highly constrained to a few favored states16. The landscape changes dramatically in the event of tRNA cleavage, and the subsequent release of tsRNAs from constraints imposed by the tRNA fold. Depending on the cleavage site, modification state, and length, a tsRNA enters a new structure space (Figure 3C). The tsRNA conformation that mediates biological function may involve structural features inherited from the tRNA precursor, thereby giving rise to TLSs, or may adopt entirely unrelated conformational states.
In the following, we discuss recent findings that demonstrate the importance to understand tsRNAs as structured molecules, which mediate biological functions by binding to and thereby regulating other RNAs and proteins. Post-transcriptional modifications are a particular focus because they are increasingly recognized for their key role in modulating the composition of the cellular tsRNA pool and its response to cellular and environmental changes. It is important to note that additional factors such as small molecule ligands, ion concentrations, and temperature, among others, are likely to contribute to the structural and functional dynamics of cellular tsRNAs.
tsRNA binding regulates localization and activities of RBPs and other RNAs:
RNA-binding proteins (RBPs) typically are thought of as proteins that bind RNA to change the fate or function of the bound RNAs. However, recent evidence shows that the regulatory relationship between RNAs and RBPs goes both ways108. An example is the association of tsRNAs with the Twi12 in the ciliate Tetrahymena, where tsRNAs regulate nuclear RNA metabolism and cell growth109. As Couvillion et al.102 showed, cytosolic Twi12 binds tRNAs, leading to their on-protein cleavage into 5’ and 3’ tsRNAs. Of these, the 18–22 nt long 3’ tsRNAs are selectively retained to stimulate assembly of Twi12 into a larger RNP complex with exonuclease Xrn2 and Tan1 and its relocalization into the nucleus102. tsRNA-binding to Twi12 thus acts as a trigger to stimulate nuclear RNA metabolism and rRNA processing110. Interestingly, the Twi12-associated tsRNA pool correlates with the overall abundances of cellular tRNAs, and not with a specific or a few specific tRNAs. This lack of specificity in tRNA selection implies that selection is based on the tRNA shape rather than on specific sequence features.
In another example, Saikia et al.111 showed that tsRNAs are part of the cellular anti-apoptotic, pro-survival response to stress. In the intrinsic pathway that initiates apoptosis, Cytochrome c (Cyt c) is released from mitochondria into the cytosol and interacts with Apoptotic protease-activating factor 1 (Apaf-1) to form the apoptosome and activate caspase 9. In cells exposed to hyperosmotic stress, angiogenin-induced tRNA cleavage leads to the accumulation of 5’ and 3’ tsRNAs (tRNA halves) that selectively bind released Cyt c to form stable ribonucleoprotein complexes. The formation of these complexes leads to decreased apoptosome formation, inhibition of caspase 9 cleavage, and increased cell survival. Notably, tsRNAs bind Cyt c with higher affinity than full-length tRNAs111, which are also known to inhibit apoptosis via direct binding to Cyt c and suppression of apoptosome formation in healthy unstressed cells22. Thus, tRNAs and tsRNAs may form a two-tiered adaptive response to prevent cells from entering apoptotic states under normal and stressed conditions, respectively, with tsRNAs providing a swift first response to stress-induced Cyt c release before other anti-apoptotic factors, such as the Inhibitor of Apoptosis (IAPs) proteins, become available.
AaRSs are another prominent target for riboregulation by tsRNAs. Keam et al.112 showed that a 19 nt long 5’ tsRNA from tRNAGln associates with the human multisynthetase complex (MSC), a large aggregation of multiple aaRSs into a single complex. Paradoxically, although this tsRNA was first identified as a member of a subclass of short 5′ tsRNAs that repress global translation in a linear sequence-independent manner (meaning that no common sequence elements in the tsRNAs were required for this activity)113, Keam et al. reported that 5’ tsRNAGln binding to the MSC increases translation of ribosomal and poly(A)-binding proteins112. In another study, Mleczko et al. found that several 3′ tsRNAs and one 5′ tsRNA associate with aaRSs in yeast, resulting in translation inhibition by reducing tRNA-specific as well as global aminoacylation by aaRSs114.
tsRNAs have also emerged as one of the most diverse classes of so-called ribosome-associated non-coding RNAs (rancRNAs), small RNAs that directly bind the ribosome in a stress-dependent manner and mediate translation regulation115. Ribosome-associated tsRNAs have been identified in bacteria116, yeast114,117,118, plants119, and mammals113,120,121. One of the best studied examples are 20–44 nt long 5’ tsRNAs produced in the halophilic archaeon H. volcanii in response to environmental stress56. One specific 26-nt long 5’ tsRNAVal, is induced by alkaline stress and suppresses global translation by binding to the 16S rRNA of small ribosomal subunits near the mRNA binding channel. Interestingly, this appears to be achieved through two distinct mechanisms: First, by displacing mRNAs from the translation initiation complex and second, by inhibiting peptide bond formation during elongation56,122. Similarly, in various mammalian model systems, a constitutively expressed 35-nt long 5’ tsRNAPro associates with ribosomes and mediates global translation repression120. This tsRNA binds directly to the 18S rRNA of the small ribosomal subunit close to the subunit interface, presumably leading to ribosomal stalling and the accumulation of a specific low-molecularweight by-product that was identified as the released peptidyl-tRNA.
While most rancRNAs described so far inhibit translation, a recent study showed that tsRNAs may also stimulate ribosomal translation. Analyzing the small ncRNA interactome of ribosomes in the protozoan parasite Trypanosoma brucei, Fricker et al.123 identified a 3’ tsRNAThr that was particularly abundant under nutrient deprivation and in stationary phase, which associates with ribosomes and stimulates translation by facilitating mRNA loading during recovery from starvation conditions123.
Competitive tsRNAs binding to RNA-binding proteins regulates mRNA stability and translation and modulates cancer progression:
The Y-Box Binding Protein 1 (YBX1) is an abundant RBP involved in a variety of cellular pathways. One of its key functions is to bind and stabilize thousands of transcripts, resulting in a vast YBX1-dependent regulon with broad consequences for diverse cellular functions. Among these, YBX1 is one of the most overexpressed oncogenes in human cancers, where it stabilizes pro-oncogenic transcripts. Hypoxia induces a subpopulation of anticodon-stem-loop-containing tsRNAs derived from tRNAGlu, tRNAAsp, tRNAGly, and an intron-containing tRNATyr.124 These tsRNAs counteract the YBX1-stabilized tumor-promoting transcripts in breast cancer cells. Mechanistically, they sequester YBX1 away from oncogenic mRNAs, resulting in their destabilization and subsequent suppression of cancer metastasis. Interestingly, YBX1 is known to specifically recognize and stabilize m5C modified mRNA transcripts, in part by recruiting the RBP human antigen R (HUR, also known as ELAVL1)125,126. Each of the four tRNAs giving rise to YBX1-interacting tsRNAs contain one or more m5C deposition sites47. Whether m5C modifications in the anticodon-loop or other parts play a role in the tsRNA-mediated sequestration of YBX1 remains to be determined.
A similar mechanism of tsRNA-mediated riboregulation was recently revealed for the RBP Nucleolin (NCL)127. NCL is a multifunctional RNA-binding protein that controls early steps of ribosome biogenesis in the nucleolus but is also abundantly found in the cytosol where it binds transcripts to modulate cell proliferation, survival, and apoptosis. As Falconi et al.127 showed, cytoplasmic NCL forms a tight and highly specific complex with a 32 nt long tsRNA derived from the 3’ half of tRNAGlu(UUC), which thereby leads to the competitive displacement of NCL from other transcripts. Among the mRNAs more exposed is that encoding tumor suppressor p53, which leads to increased p53 expression and thus modulation of cancer cell growth. Interestingly, RNA-binding by NCL relies on a single-stranded recognition motif (U/G)CCCG(A/G) within a stem-loop structure. In the thermodynamically most stable fold predicted for the 3’ tsRNAGlu, this motif is buried within the double-stranded stem region inherited from the original T-stem-loop architecture. Efficient NCL binding may thus require alternative folding of the tsRNA, possibly correlated with changes in its modification state.
Notably, YBX1- and NCL-binding tsRNAs are downregulated in metastatic cancer cells127,128, suggesting that their dysregulation promotes aberrantly elevated translation of oncogenic transcripts. Like the mTOG-ψs discussed below78, these tsRNAs may thus be required to suppress aberrant protein synthesis programs.
Interestingly, an opposite effect on NCL activity was recently reported for a 5’ tsRNA derived from tRNACys, which is upregulated during breast cancer progression and required for breast cancer metastatic lung colonization128. Contrary to the effect by 3’ tsRNAGlu, the 5’ tsRNACys binds NCL and drives its oligomerization with pro-metastatic metabolic transcripts Mthfd1l and Pafah1b1. This oligomerization creates a higher-order ribonucleoprotein complex, which protects bound transcripts from degradation and increases their stability and expression. Identification of the binding site by CLIP and crosslinking-induced modification sites showed that NCL-binding depends on two G-rich motifs in the tsRNA, one of which corresponds to the 5’TOGs known to be involved in RG4 formation (see below) and the other to the loop region of the D-stem-loop. Whether the 5’ tsRNACys molecules associate into an RG4, and whether this plays a role in NCL oligomerization remains unclear at present, as does a potential role for RNA modifications.
tsRNAs form quaternary G-quadruplex structures to target initiation factors and induce stress granule formation.
Quaternary structure formation among tsRNAs has been suggested for their role in stress granule formation and global translation regulation. An important mechanism of general translation repression is the condensation of mRNAs, translation initiation factors, and ribosomal subunits into non-membrane-enclosed subcellular compartments called stress granules (SGs). While it is now well established how eIF2α phosphorylation in the context of the integrated stress response promotes SG formation, 5’ tsRNAs, produced by ANG-cleavage under stress, also play an important role in an alternative, eIF2α-independent pathway of SG formation53. The Anderson lab determined that select ~30 nt long 5’ tsRNAs from tRNAAla and tRNACys, which bear a conserved motif of four to five guanine residues at their 5’ end (5’TOGs), form intermolecular RNA G-quadruplexes (RG4) to competitively displace the translation initiation factor eIF4A/E/G (eIF4F) complex from m7G-capped mRNAs129,130. Mechanistically, the HEAT1 domain of eIF4G, the central scaffolding protein of the eIF4F complex, directly binds to RG4s, thereby inhibiting the scanning step of translation initiation and leading to the formation of stress granules in an eIF2α-independent manner131. The same 5’ tsRNAs were also found to associate with the cold shock domain of YBX1. Although YBX1 is dispensable for translation initiation factor displacement from capped mRNAs132, it facilitates SG assembly and may thereby contribute to the efficiency of global translational repression129,132–134.
Modification-induced functional switch in tsRNA-mediated translation control of embryonic stem cell maintenance and differentiation.
Recent experiements support a critical functional role of tsRNAs and their dynamic regulation by post-transcriptional modifications during cell differentiation and embryogenesis78,79. In one specific example, Guzzi et al. showed using human ESCs (hESCs) that the pseudouridine (ψ) synthase PUS7 directs the formation of ~18 nt long TOG-containing 5’ tsRNAs (termed mini- or mTOG-ψs) from tRNAAla, tRNACys, and tRNAVal by depositing a ψ at the U8 position (ψ8) of these tRNAs78,79. Like the longer (~30 nt) TOG-containing 5’ tsRNAs130,131, mTOG-ψs interfere with eIF4F complex assembly and lead to global translational repression. Underlying this effect is the association of mTOG-ψs with the RRM domains of polyadenylate-binding protein 1 (PABPC1), a central component of the 5ʹ cap-binding translation initiation complex, which prevents recruitment of the translational co-activator PABPC1-interacting protein 1 (PAIP1). This results in a strong translational repression of transcripts containing pyrimidine-enriched sequences (PES) at the 5′-UTR, which includes mRNAs encoding components of the protein synthesis machinery.
Notably, PUS7 is upregulated and leads to an enrichment of mTOG-ψs in embryonic stem cells, whereas PUS7 and mTOGs are rapidly downregulated during embryonic differentiation78. Thus, mTOGs appear to be part of a post-transcriptional regulatory circuit to control gene expression during stem cell commitment and development. Consistently, PUS7 depletion impairs tsRNA-mediated translation regulation and leads to aberrantly increased protein biosynthesis. Growth and differentiation defects in hESCs as well as defective germ layer specification are the result, which suggests that mTOG-ψs are required to selectively inhibit aberrant protein synthesis programs78,79. Notably, while the association of mTOGs with PABPC1 is enhanced by ψ8, mTOGs with unmodified U8 cannot displace eIF4F, but instead show a preference for YBX1. PUS7-mediated pseudouridylation may thus act as a structural ‘switch’ to rewire the mTOG-interactome landscape and ensure tight spatiotemporal control of gene expression in the process of stem cell commitment.
Further evidence for the ability of modifications to modulate the ‘information capacity’ of a tsRNA pool is provided by the role of the m5C writer DNMT2 during stem cell differentiation and early embryonic development. DNMT2-dependent m5C deposition in position C38 of various tRNA species is known to inhibit angiogenin-induced tsRNA generation86. Accordingly, loss of m5C38 deposition in DNMT2−/− knock-out mice leads to a significant increase in tsRNA levels in the bone marrow88. This increase is accompanied by a reduction of the hematopoietic stem and progenitor cell population and cell-autonomous defects in their differentiation. These results are consistent with various studies that suggest a broad role of DNMT2 in the control of tsRNA generation86,89, and the finding that loss of DNMT2 activity leads to differentiation and developmental defects135. In another line of investigation, the sperm-derived tsRNAs (and other small RNAs) derived from mice kept on a high-fat diet (HFD) contain significantly elevated levels of m5C and m2G base modifications, possibly as a result of HFD-induced upregulation of DNMT2 in the epididymis61,63. These effects could be part of the ‘sperm RNA code’ that transmits paternally acquired metabolic traits60,63. For example, transfection of an m5C38 modified vs. unmodified sperm-derived 39-nt 3’ tsRNAGly led to distinct transcriptomic responses in NIH/3T3 cells, possibly due to modification-induced structural changes in tsRNAs that contribute to distinct coding signatures in the ‘sperm RNA code’ and the transmission of epigenetic information. This is supported by the fact that the modification state of C38 induces a significant structural change in the 3’ tsRNAGly, with the m5C38-modified tsRNA exhibiting an increased susceptibility to RNase degradation63. Similar observations were made for heat-shock-induced tsRNAs in Drosophila, which were longer-lived and more abundant in DNMT2 knockout flies compared to tsRNAs from wild-type flies136.
Nicked tRNAs and tsRNA dimers confer increased stability and may serve as reservoirs of tsRNAs with diverse quaternary structures.
Following tRNA cleavage, the resulting tsRNAs are usually viewed as separate molecules with independent functions. Yet, two recent studies suggest that a large proportion of tsRNAs remains associated in stable nicked tRNA complexes between 5’ and 3’ fragments, both in human cells and in extracellular human biofluids137,138. Although no specific function was linked with nicked tRNAs, they were suggested to serve either as a stable reservoir for tsRNAs or to mediate independent functions that require a structurally intact, but translationally inactive, tRNA fold. Evidence for such a specific function comes from a recent study in the bacterium Salmonella enterica. Here, a nicked tRNA directly binds the transcriptional activator RtcR and thereby activates expression of the rtcBA RNA repair operon137. Interestingly, the interaction of nicked tRNA with RtcR relies on the presence of a 2’,3’ cyclic phosphate in the 5’ tsRNA from its endonucleolytic cleavage, supporting the notion that distinct terminal phosphate states, resulting from tRNA cleavage by distinct endonucleases, may be yet another important factor determining the structures and intermolecular interactions of tsRNAs139,140. Finally, in addition to the stable complexes of 5’ and 3’ tsRNAs in nicked tRNAs, Tosar et al.141 show that 5’ tsRNAs from tRNAGly can form homodimers as well as heterodimers with 5’ tsRNAGlu, resulting in tertiary and quaternary structures that are distinct from those expected in monomeric tsRNAs or nicked tRNAs.
Taken together, these results further highlight the vast complexity of tsRNAs at the three-dimensional level, which is based on their capacity to adopt diverse ensembles of specific secondary, tertiary and quaternary structures.
3. Emerging methods centered on tRNA and tsRNA sequences, modifications, and structures
The prediction of the 3D structure of tRNA in 1969 is considered as a milestone in the emergence of bioinformatics142. By integrating limited tRNA sequence information, secondary structure predictions, and experimental data, Levitt was able to derive a complex 3D model that provided valuable guidence for subsequent attempts to experimentally determine the tRNA tertiary structure by means of X-ray crystallography8,143,144. The early work by Levitt and others145,146 demonstrated the great potential that computational modeling holds to support experimental approaches to unravel the structural properties and biological functions of RNAs. At the same time, it also highlighted the need to integrate a wide range of high-quality, high-throughput experimental data to increase the accuracy and thus credibility of computational structure predictions to gain functional insights.
Despite it being 60 years since the first tRNA sequence was reported, and despite thousands of sequences published since then, obtaining accurate tRNA/tsRNA sequences and modification states is still a challenge. The error in the assignment of a single base pair, as occurred with E. coli tRNAAla, can severely impede interpretations and discoveries based on a single sequence147. In the most recent works, computational and AI-based predictions of structures of tsRNAs require complete accuracy of primary sequences and modifications. Thus, newer technologies for sequencing and emerging techniques to examine tsRNA structures, through combining experimental, computational, and AI-driven predictions, are providing a more systematic approach to identification of tsRNAs and their interactome inside the cell. These issues are discussed below.
a. Resolving primary sequences
The presence of RNA modifications and structures that block reverse transcription during cDNA production make tRNAs and tsRNAs difficult to sequence148,149. The problem is compounded by adaptor ligation challenges caused by unique RNA termini generated during tsRNA biogenesis98. Recent methodological advancements have aimed at overcoming these challenges, either through enzymatic conversions140,150 to remove blocking modifications (e.g., m1A, m1G, m3C)76, or by the use of highly processive reverse transcriptases (e.g., TGIRT, BoMoC, and MarathonRT)151–153, which can be advantageously employed to deduce the RNA’s modification state154. These methods thoroughly reviewed elsewhere12,155, have led to ever-increasing precision in deciphering the full repertoire of tRNAs and tsRNAs in different tissue and cell contexts. They are also being utilized as tools in translational research to identify distinct tRNA/tsRNA signatures associated with disease conditions153,156.
b. Multiplexed mapping of RNA modifications
The complex modifications deposited on tRNAs/tsRNAs are sources of biological information that is an integral part of what controls RNA stability, structure, and interaction potentials46,157. Databases (e.g., Modomics)44 catalog site-specific tRNA modifications gathered over decades from a variety of studies. Most of these studies focused on individual modifications and, therefore, offer a static perspective that fails to capture the dynamic regulatory processes of RNA modifications in a tissue or cell-specific context. Next-generation methods that directly sequence tRNA/tsRNA and simultaneously identify all modifications are needed to investigate dynamic regulation of tRNA/tsRNA modifications. Two classes of methods are currently applied to map multiple types of RNA modifications directly and quantitatively on tRNA/tsRNAs. These methods are based on advanced mass spectrometry (MS) and nanopore technology.
Because the input RNAs for MS are typically digested into smaller pieces or single nucleotides, most studies utilizing MS focus on quantifying RNA modifications rather than on site-specific mapping. A recent innovation uniformly degrades tRNA into a mass ladder, and thereby allows the direct “reading” of RNA sequence and modification information from the mass shift along the ladder158,159. This approach is conceptually reminiscent of Sanger DNA sequencing, which exploits a DNA ladder12. Although the method requires further development to achieve high throughput capabilities and the analysis of multiple tRNAs/tsRNAs from mixed RNA types, it can be readily implemented for de novo sequencing of enriched full length tRNAs/tsRNAs, enabling simultaneous mapping of multiple RNAs along with the tRNA sequence itself. This approach could be used to address immediate questions such as tRNA/tsRNA modification variations in different tissue and cells, as well as under normal and disease conditions.
Recently, nanopore-based direct RNA sequencing has used the remarkable structures of natural membrane ion channels to identify RNA pieces. This identification is based on changes in ionic currents as the RNAs pass through the pore160. Due to its unique features, nanopore technology has revolutionized direct DNA and RNA sequencing and holds significant potential for directly identifying associated RNA modifications that produce distinguishable ion currents. Contemporary studies employed nanopore-based approaches to sequence native tRNA populations, and to provide quantitative estimations of both tRNA abundances and modification dynamics161. Although current methods cannot yet identify modifications de novo and require existing tRNA sequence/modification information as a reference, this direction can be further enhanced through deep-learning algorithm training for high-throughput analysis of tRNA/tsRNA sequence and modifications. This training must be based on various standard RNA sequences with multiple modifications inserted at different positions, or it can use the information obtained from deciphered tRNA/tsRNA sequences with known modification maps. Of course, a large dataset is needed to ensure accuracy of the deep-learning analysis.
c. Analyzing tRNA and tsRNA structures with experimental, computational, and AI-based prediction
Traditional computational approaches:
Computational methods serve as a straightforward approach for gaining structural insights into RNA molecules. These high-throughput RNA structure prediction techniques primarily rely on sequence information, making them a popular choice for analyzing RNA structures. Conventional computational approaches for RNA structure prediction include minimum free energy (MFE) based methods (e.g., Mfold, RNAfold, UNAFold, RNAstructure, MC-Fold, and MC-Sym)162–166. These approaches assume that the most stable secondary structure of an RNA molecule minimizes free energy based on thermodynamic principles. MFE-based methods can be extended to further provide probabilistic information about alternative RNA structures by calculating base-pairing probabilities for each nucleotide position in the RNA sequence using partition functions from statistical thermodynamics. Thus, each tsRNA sequence can be used to generate multiple predicted secondary structures according to MFE ranking (Figure 4A). Other computational methods, such as stochastic context-free grammars (SCFGs)167,168, appear to offer a more rigorous probabilistic framework for RNA secondary structure predictions. While these computational methods have been successful in predicting some RNA secondary structures and are being improved through knowledge-based approaches, they inherently face limitations in predicting the dynamic nature of RNA folding. Because MFE-based algorithms predict secondary structures based on the lowest free energy, they tend to maximize the number of Watson-Crick pairs, which underestimates the importance of non-Watson-Crick interactions in RNA folding. Moreover, limitations arise from unknown subcellular environments and challenges posed by highly modified RNAs, such as those tsRNAs where modifications can alter base-pairing principles.
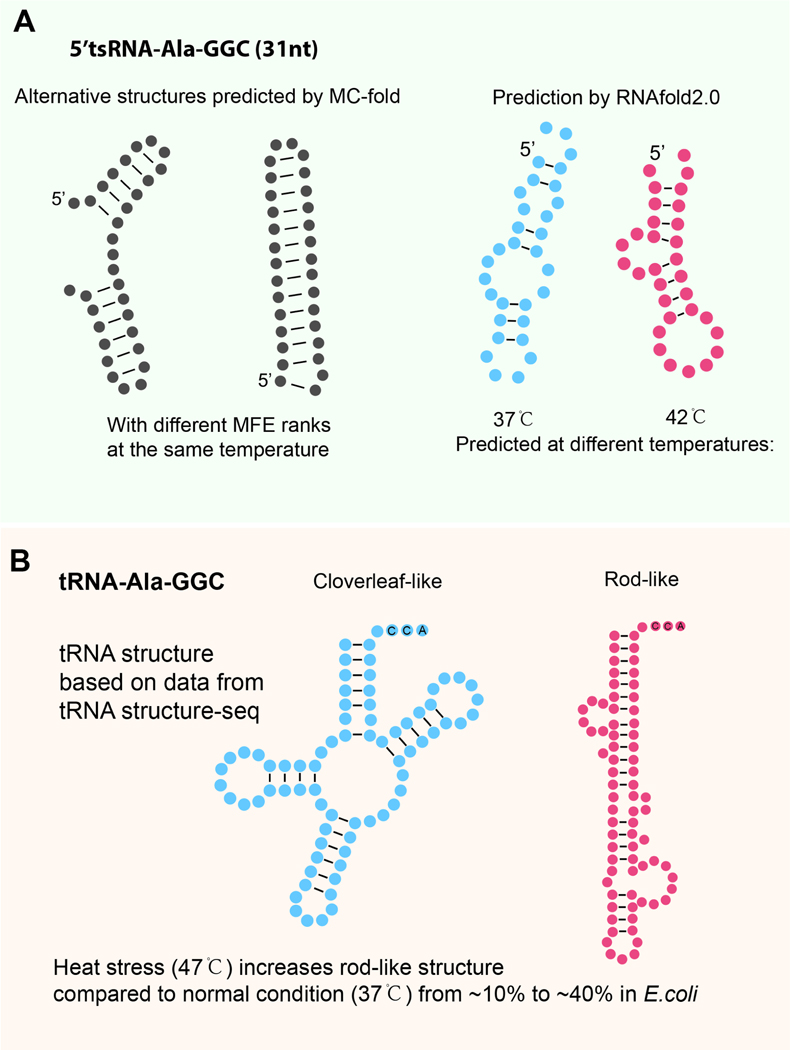
Figure 4. Interchangeable structures of tRNAs and tsRNAs depending on the environment and local context.
(A) Illustration showing predicted tsRNAAla secondary structure with differential MFE ranking (using MCfold165), or with different temperature settings (using RNAstructure166). These predicted structures do not consider the impact of RNA modifications and the changing local context, and therefore need further experimental validations. (B) Illustration showing that heat stress triggers conformational changes in bacterial tRNAAla, increasing the proportion of rod-like shapes while decreasing the prevalence of the classic cloverleaf tRNA structure. These structures are based on experimental data from tRNA structure-seq175.
Traditional experimental approaches:
Traditionally, high-resolution methods such as X-ray crystallography, NMR, and more recently, cryo-electron microscopy (cryo-EM) have been used to verify secondary and tertiary RNA structures. Although NMR and cryo-EM have the capacity to provide information on an RNA’s conformational ensembles and dynamics, they are typically limited to a few favored structural states that occupy deep valleys in a free-energy landscape. Except for NMR, the small size of tsRNA presents an additional challenge for these traditional structural methods. Other traditional methods used for RNA structure determination that may bypass these limitations include in-line or chemical probing169, as well as site-direct mutagenesis, which have been applied successfully to determine secondary structures in solution. However, obtaining a comprehensive view of the conformational ensemble and dynamic changes for the numerous tsRNA and tRNA sequences within a cell remains a challenge.
Novel experimental technologies with high-throughput analyses:
Beyond computational prediction, new experimental technologies combine chemical, biological, and bioinformatic approaches for the analysis of RNA structures and interactions. These advances, which have gained momentum over the past decade, can be categorized into two main branches. The first reveals secondary RNA structures by detecting RNA backbone flexibility based on chemical probing, such as SHAPE-seq, Structure-Seq, and derivative methods170–174. The essence of these methods uses chemical probes, such as SHAPE agents or dimethyl sulfate (DMS), to target nucleotides in accessible, single-stranded regions (i.e., those without base-pairing). These probes add chemical RNA modifications that interfere with reverse transcription (by stopping RT or introducing misincorporations). High-throughput sequencing is then used to detect and quantify these stops/misincorporations. From this, the secondary structural profile of the RNA can be inferred. Notably, the latest development in this technical branch, called “tRNA structure-seq”, was employed to determine the dynamic structural changes of tRNA under heat stress in vivo and to identify tRNA modification changes using misincorporation signatures175. Remarkably, this work showed that even tRNAs, once considered to have relatively rigid structures, undergo dramatic shape changes that include rod-like structures under heat stress (Figure 4B). This method could soon be used for studying tsRNA secondary structure. In particular, it could address the question of structural heterogeneity between similar tsRNA sequences with differing terminal lengths (e.g., 18 nt vs. 22 nt), as these length differences have been shown to result in very different modes of action176. Furthermore, this method could also help to directly investigate how the variations in RNA modifications among tsRNAs contribute to their structural diversity.
The second branch of technical development identifies RNA-RNA interactions from proximity chemical crosslinking (e.g., by Psoralen or its derivative AMT, or amotosalen). These methods, including PARIS, SPLASH, LIGR-seq, and PARIS2177−181 determine long-range interactions between different RNAs, as well as alternative RNA conformations within long RNAs (e.g., rRNA). This approach has numerous derivatives and improvements, such as improving RNA capturing efficiency. Recently, the method identified viral-host RNA interactions (e.g., Zika, SARS-CoV-2) to reveal virus infection strategies. However, chemical crosslinking cannot efficiently probe protein-protected regions, so that these methods generally identify more exposed areas. Because inter- and intra-RNA-RNA interactions are commonly stabilized and mediated by proteins, RIC-seq (RNA in situ conformation sequencing) was developed to address the limitations of direct RNA-RNA crosslinking182. RIC-seq captures protein-mediated RNA-RNA proximal interactions in situ and can yield information regarding RNA-RNA spatial interactions in cells. This branch of technology could be further adapted for tsRNA research, proving useful in identifying tsRNA structures and exploring the RNA interactome of tsRNAs within cells. For example, it could provide new insights into tsRNA interaction sites within ribosomes and other nuclear RNPs. Indeed, by reanalyzing a previous database from CLASH (cross-linking, ligation, and sequencing of hybrids) – an experimental approach used to identify RNA–RNA duplexes associated with Argonaute proteins in vivo183,184 – researchers have not only uncovered numerous tsRNA–mRNA hybrids but also found evidence of interactions between tsRNA and rRNA, tsRNA and snRNA, and with other small non-coding RNAs185–187. These findings strongly suggest a complex tsRNA interactome in both the cytoplasmic and nuclear compartments, which may lay the foundation for understanding their modes of action.
AI-based deep learning and challenges caused by RNA flexibility:
AI-based prediction of RNA structure primarily relies on machine learning algorithms, particularly deep learning techniques, to predict secondary and tertiary RNA structures, and RNA-protein binding potentials. AI algorithms require large high-quality training data sets, which includes experimentally validated 3D RNA structures and sequences, secondary structures based on data from chemical probing that provide information on structural flexibility, and base-pairing interactions188,189. While databases continue to grow, they are nonetheless limited at this time.
For example, the prediction of RNA 3D structures uses high-quality structural data deposited in the Protein Data Bank. The availability of this information is far more limited compared to sequencing-based data. And, of course, the deposited 3D structures represent a static view that does not capture the dynamic nature of RNAs. For 3D RNA structure prediction, the recently developed approach Atomic Rotationally Equivalent Scorer (ARES)190 uses a strategy that learns the geometric arrangements of each atom. This strategy is a promising direction, because it enables accurate structural model prediction based on limited known RNA conformations.
Despite the success of accurate protein structure prediction enabled by AlphaFold191 and RoseTTAFold192, predicting RNA structure is inherently more challenging. RNA is more flexible, and a single RNA sequence can adopt multiple secondary, tertiary, or higher-order structures, which are expanded upon by the possibilities for structures based on RNA-RNA interactions189. Further complexity arises because each conformation potentially serves a different function. For example, atomic force microscopy (AFM) revealed that the same RNA primary sequence can simultaneously exhibit completely different 3D structures under near-physiological solution conditions193. Most of these structures showed ligand binding abilities that were idiosyncratic to the structure and suggested therefore functional relevancy. This potential repertoire of structures for a single RNA poses significant challenges for AI-based predictions. These challenges are particularly obvious for tsRNAs and tRNAs that are in highly compartmentalized cells. In such locales, RNA structures are influenced by factors that engender distinct functions within different contexts (Figure 4A,B).
AI-based prediction demands context-specific data from high-quality experiments, from which the specific conditions for a particular RNA structure can be obtained and used as the basis for deep learning. Taking tsRNAs as an example, their secondary and tertiary structures would depend on specific conditions, such as normal vs. stress conditions in tissues and cells, and nuclear vs. cytoplasmic compartments, which are intricately linked with their RNA modification status and binding partners. Thus, the AI-training process must deploy deep learning of context-dependent information, such as the repertoire of surrounding RNA sequences and proteins, which in turn influence/stabilize each other’s structures and interactions, and thereby exert specific functions in each context.
4. Future perspectives
The study of tsRNAs serves as a striking example of biological complexity, where the fragmentation of tRNAs has given rise to a new world of structural and functional diversity10. This wholly unexpected development represents a clear ‘renovatio’, a rebirth of tRNA functionality. tsRNAs have been identified across all domains of life11, which in turn speaks to their critical role in cellular evolution. Notably, their emergence predates other more widely studied small RNAs, such as miRNAs. This is due to tsRNA biogenesis being controlled by ancient enzymes that predate the emergence of Dicer (only found in eukaryotes) in the prokaryotic world, and regulated by even more ancient mechanisms controlling RNA modifications, potentially derived from a ribozyme-dominated RNA world11.
In fact, the exploration of tRNA fragmentation principles and the function of tsRNAs is now being generalized to study a wider spectrum of small non-coding RNAs (sncRNAs) fragmentated from a range of ancient structured RNAs, including rRNAs, snRNAs, snoRNAs, Y RNAs, Vault RNAs, among others12. Some of these share similar biogenesis pathways, such as being cleaved by RNase A, T2, L families, with their fragmentation patterns and efficiencies also controlled by the RNA modifications they carry194. This strongly suggests that the degradation and fragmentation of longer structured RNAs into sncRNAs, followed by selective retention, may represent the earliest form of sncRNA biogenesis12.
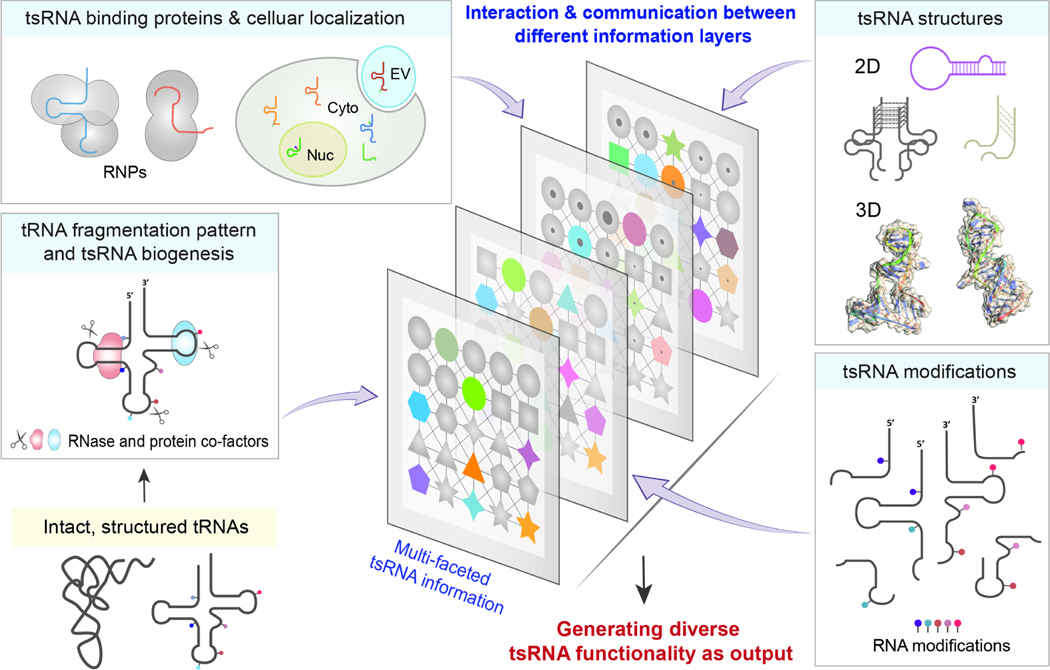
Furthermore, the exploration of the functions of tsRNAs and other sncRNAs has led to the revelation that there is a wider spectrum of functional principles of sncRNAs, extending beyond the RNAi doctrine. This is deeply rooted in the versatile structural signatures of RNAs, enabling a fundamental interaction potential with other molecules such as RNAs, proteins, and metabolites. This generates an astronomical number of combinations, resulting in the information capacity needed for the construction of the complex cells we observe today. The future integration of multi-layered information through advanced AI-based deep learning, such as considering aspects of tsRNA sequence, fragmentation pattern, site-specific RNA modifications, RNA structures, tsRNA binding proteins, and subcellular compartmentalization—will enable novel and deeper insights and the understanding of the diverse functionality of tsRNAs (Figure 5).
Figure. 5. Integrating multi-layers of tsRNA information enables diverse tsRNA functionality.
Multi-faceted data obtained from various technologies regarding tsRNA sequence/fragmentation pattern, site-specific RNA modifications, RNA structures, tsRNA binding proteins and subcellular compartmentalization would enable future high-dimensional analyses with the advancement of deep learning algorism, leading to better understand of the fundamental modes of tsRNA action, and thus their biological roles in specific contexts. RNase: ribonuclease; RNPs: ribonucleoprotein particles; EV: extracellular vesicle; Cyto: cytoplasma; Nuc: nucleus.
Finally, tsRNAs and their dysregulation are increasingly recognized as central players in a variety of human diseases, from neurological disorders to cancer195−200. Accordingly, tsRNAs are emerging as promising targets with significant therapeutic potential196,201–203, and a rapidly growing body of evidence suggests extracellular tsRNAs as diagnostic and prognostic biomarkers for liquid biopsies72,196,204,205. A deeper understanding of tsRNA biogenesis and function will therefore provide a necessary framework for future pharmaceutical engineering, particularly about RNA modifications and structural aspects of engineered sncRNAs. Approaches to therapeutically target specific RNAs require comprehensive knowledge of their structural dynamics and the functions associated with them206,207. This requirement will be aided by advances in the development of computational tools that predict RNA structures and their interaction potential in vivo190,208–210. This knowledge is also essential for the emerging field of tRNA therapy211. For instance, in the study of suppressor tRNA therapy, engineered suppressor tRNAs are designed to recognize and decode stop codons (termination codons) in mRNA during translation212,213. By doing so, they can treat diseases caused by nonsense mutations (including cystic fibrosis, Duchenne muscular dystrophy, and Dravet syndrome), where suppressor tRNAs restore full-length protein synthesis by bypassing premature stop codons. However, when suppressor tRNAs are delivered to target cells, tsRNAs with specific functions may arise and introduce side effects. Consequently, researchers should consider the delivery quantity, cleavage/retention patterns of the resulting tsRNAs, and their potential functions. Investigating this level of complexity will broaden our understanding of the principles governing the organization and operation of biological systems overall, ultimately enabling more informed treatment approaches for human diseases.
Acknowledgements
Research in P.S. laboratory was supported by donations from an anonymous source and the National Foundation for Cancer Research. Q.C. laboratory is in part supported by NIH (R01HD092431 and R01ES032024)
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crick FH (1958). On protein synthesis. Symp Soc Exp Biol 12, 138–163. [PubMed] [Google Scholar]
- 2.Crick FH (1966). The genetic code--yesterday, today, and tomorrow. Cold Spring Harb Symp Quant Biol 31, 1–9. [PubMed] [Google Scholar]
- 3.Hoagland MB, Stephenson ML, Scott JF, Hecht LI, and Zamecnik PC (1958). A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231, 241–257. [PubMed] [Google Scholar]
- 4.Hoagland MB, Keller EB, and Zamecnik PC (1956). Enzymatic carboxyl activation of amino acids. J Biol Chem 218, 345–358. [PubMed] [Google Scholar]
- 5.Schimmel PR, and Soll D. (1979). Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem 48, 601–648. [DOI] [PubMed] [Google Scholar]
- 6.Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, and Zamir A. (1965). Structure of a Ribonucleic Acid. Science 147, 1462–1465. 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 7.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, and Rich A. (1974). Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185, 435–440. 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 8.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, and Klug A. (1974). Structure of yeast phenylalanine tRNA at 3 A resolution. Nature 250, 546–551. 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 9.Higgs PG, and Lehman N. (2015). The RNA World: molecular cooperation at the origins of life. Nat Rev Genet 16, 7–17. 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- 10.Schimmel P. (2018). The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol 19, 45–58. 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, Zhang X, Shi J, Yan M, and Zhou T. (2021). Origins and evolving functionalities of tRNA-derived small RNAs. Trends in biochemical sciences 46, 790–804. 10.1016/j.tibs.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Zhou T, and Chen Q. (2022). Exploring the expanding universe of small RNAs. Nature cell biology 24, 415–423. 10.1038/s41556-022-00880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wajahat M, Bracken CP, and Orang A. (2021). Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int J Mol Sci 22. 10.3390/ijms221910193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert M, Benmoussa A, and Provost P. (2019). Small Non-Coding RNAs Derived From Eukaryotic Ribosomal RNA. Noncoding RNA 5. 10.3390/ncrna5010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su Z, Wilson B, Kumar P, and Dutta A. (2020). Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annual review of genetics. 10.1146/annurev-genet-022620-101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giegé R, Juhling F, Putz J, Stadler P, Sauter C, and Florentz C. (2012). Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA 3, 37–61. 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 17.Westhof E, and Fritsch V. (2000). RNA folding: beyond Watson-Crick pairs. Structure 8, R55–65. 10.1016/s0969-2126(00)00112-x. [DOI] [PubMed] [Google Scholar]
- 18.Leontis NB, and Westhof E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512. 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beuning PJ, and Musier-Forsyth K. (1999). Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 52, 1–28. . [DOI] [PubMed] [Google Scholar]
- 20.Giegé R, Sissler M, and Florentz C. (1998). Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26, 5017–5035. 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong J, Qiu H, Garcia-Barrio M, Anderson J, and Hinnebusch AG (2000). Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6, 269–279. [DOI] [PubMed] [Google Scholar]
- 22.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, and Yang X. (2010). tRNA binds to cytochrome c and inhibits caspase activation. Molecular cell 37, 668–678. 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katibah GE, Lee HJ, Huizar JP, Vogan JM, Alber T, and Collins K. (2013). tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Molecular cell 49, 743–750. 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, and David M. (2012). Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491, 125–128. 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Chow CR, Ebine K, Lee J, Rosner MR, Pan T, and Munshi HG (2016). Interaction of tRNA with MEK2 in pancreatic cancer cells. Sci Rep 6, 28260. 10.1038/srep28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisien M, Wang X, Perdrizet G 2nd, Lamphear C, Fierke CA, Maheshwari KC, Wilde MJ, Sosnick TR, and Pan T. (2013). Discovering RNA-protein interactome by using chemical context profiling of the RNA-protein interface. Cell Rep 3, 1703–1713. 10.1016/j.celrep.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YT, Strugatsky D, Liu W, and Zhou ZH (2021). Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150. Nature communications 12, 5513. 10.1038/s41467-021-25791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, and Kieft JS (2014). The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 511, 366–369. 10.1038/nature13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costantino DA, Pfingsten JS, Rambo RP, and Kieft JS (2008). tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol 15, 57–64. 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Au HH, Cornilescu G, Mouzakis KD, Ren Q, Burke JE, Lee S, Butcher SE, and Jan E. (2015). Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection. Proc Natl Acad Sci U S A 112, E6446–6455. 10.1073/pnas.1512088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonilla SL, Sherlock ME, MacFadden A, and Kieft JS (2021). A viral RNA hijacks host machinery using dynamic conformational changes of a tRNA-like structure. Science 374, 955–960. 10.1126/science.abe8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levi O, and Arava Y. (2019). mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol 17, e3000274. 10.1371/journal.pbio.3000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Chen X, Wolin SL, and Xiong Y. (2018). Structural Basis for tRNA Mimicry by a Bacterial Y RNA. Structure 26, 1635–1644 e1633. 10.1016/j.str.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz-Toledano R, and Gomez J. (2015). Messenger RNAs bearing tRNA-like features exemplified by interferon alfa 5 mRNA. Cell Mol Life Sci 72, 3747–3768. 10.1007/s00018-0151908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romby P, Brunel C, Caillet J, Springer M, Grunberg-Manago M, Westhof E, Ehresmann C, and Ehresmann B. (1992). Molecular mimicry in translational control of E. coli threonyl-tRNA synthetase gene. Competitive inhibition in tRNA aminoacylation and operator-repressor recognition switch using tRNA identity rules. Nucleic Acids Res 20, 5633–5640. 10.1093/nar/20.21.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frugier M, and Giege R. (2003). Yeast aspartyl-tRNA synthetase binds specifically its own mRNA. J Mol Biol 331, 375–383. 10.1016/s0022-2836(03)00767-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, Thieme CJ, Kollwig G, Apelt F, Yang L, Winter N, Andresen N, Walther D, and Kragler F. (2016). tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell 28, 1237–1249. 10.1105/tpc.15.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Huang J, Wu S, Zheng Q, Liu P, Feng H, Su X, Fu H, Xi Q, and Wang G. (2020). The tRNA-like small noncoding RNA mascRNA promotes global protein translation. EMBO Rep 21, e49684. 10.15252/embr.201949684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilusz JE, Whipple JM, Phizicky EM, and Sharp PA (2011). tRNAs marked with CCACCA are targeted for degradation. Science 334, 817–821. 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, Li X, and Wang G. (2022). tRNA-like structures and their functions. FEBS J 289, 5089–5099. 10.1111/febs.16070. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn CD (2016). RNA versatility governs tRNA function: Why tRNA flexibility is essential beyond the translation cycle. Bioessays 38, 465–473. 10.1002/bies.201500190. [DOI] [PubMed] [Google Scholar]
- 42.Lorenz C, Lunse CE, and Morl M. (2017). tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 7. 10.3390/biom7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motorin Y, and Helm M. (2010). tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944. 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 44.Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, Kurkowska M, Shirvanizadeh N, Destefanis E, Groza P, et al. (2022). MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic acids research 50, D231–D235. 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, and Agris PF (2011). The RNA Modification Database, RNAMDB: 2011 update. Nucleic acids research 39, D195–201. 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T. (2021). The expanding world of tRNA modifications and their disease relevance. Nature reviews. Molecular cell biology 22, 375–392. 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 47.Bohnsack KE, Hobartner C, and Bohnsack MT (2019). Eukaryotic 5-methylcytosine (m(5)C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes (Basel) 10. 10.3390/genes10020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirchner S, and Ignatova Z. (2015). Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nature reviews. Genetics 16, 98–112. 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 49.Kim HK, Yeom JH, and Kay MA (2020). Transfer RNA-Derived Small RNAs: Another Layer of Gene Regulation and Novel Targets for Disease Therapeutics. Mol Ther 28, 2340–2357. 10.1016/j.ymthe.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SR, and Collins K. (2005). Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. The Journal of biological chemistry 280, 42744–42749. 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 51.Thompson DM, Lu C, Green PJ, and Parker R. (2008). tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103. 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, and Qu LH (2008). Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic acids research 36, 6048–6055. 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamasaki S, Ivanov P, Hu GF, and Anderson P. (2009). Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185, 35–42. 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X. (2009). Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS letters 583, 437–442. 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 55.Thompson DM, and Parker R. (2009). The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185, 43–50. 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gebetsberger J, Zywicki M, Kunzi A, and Polacek N. (2012). tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 260909. 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YS, Shibata Y, Malhotra A, and Dutta A. (2009). A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes & development 23, 2639–2649. 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, and Kay MA (2010). Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695. 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, et al. (2012). A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22, 1609–1612. 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Shi J, Rassoulzadegan M, Tuorto F, and Chen Q. (2019). Sperm RNA code programmes the metabolic health of offspring. Nature reviews. Endocrinology 15, 489–498. 10.1038/s41574-019-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400. 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 62.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J, et al. (2018). Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 20, 535–540. 10.1038/s41556018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarker G, Sun W, Rosenkranz D, Pelczar P, Opitz L, Efthymiou V, Wolfrum C, and Peleg-Raibstein D. (2019). Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proceedings of the National Academy of Sciences of the United States of America 116, 10547–10556. 10.1073/pnas.1820810116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Ren L, Sun X, Zhang Z, Liu J, Xin Y, Yu J, Jia Y, Sheng J, Hu GF, et al. (2021). Angiogenin mediates paternal inflammation-induced metabolic disorders in offspring through sperm tsRNAs. Nature communications 12, 6673. 10.1038/s41467-021-26909-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alata Jimenez N, Castellano M, Santillan EM, Boulias K, Boan A, Arias Padilla LF, Fernandino JI, Greer EL, Tosar JP, Cochella L, and Strobl-Mazzulla PH (2023). Paternal methotrexate exposure affects sperm small RNA content and causes craniofacial defects in the offspring. Nature communications 14, 1617. 10.1038/s41467-023-37427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Q, Yan W, and Duan E. (2016). Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 17, 733–743. 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo Y, Bai D, Liu W, Liu Y, Zhang Y, Kou X, Chen J, Wang H, Teng X, Zuo J, and Gao S. (2021). Altered sperm tsRNAs in aged male contribute to anxiety-like behavior in offspring. Aging Cell 20, e13466. 10.1111/acel.13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Zhang Y, Shi J, Zhang H, Cao Z, Gao X, Ren W, Ning Y, Ning L, Cao Y, et al. (2014). Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. Journal of molecular cell biology 6, 172–174. 10.1093/jmcb/mjt052. [DOI] [PubMed] [Google Scholar]
- 70.Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Boffelli D, Mote P, and Martin DI (2013). 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC genomics 14, 298. 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hogg MC, Raoof R, El Naggar H, Monsefi N, Delanty N, O’Brien DF, Bauer S, Rosenow F, Henshall DC, and Prehn JH (2019). Elevation in plasma tRNA fragments precede seizures in human epilepsy. The Journal of clinical investigation 129, 2946–2951. 10.1172/JCI126346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu W, Shi J, Liu H, Zhang X, Zhou JJ, Li M, Zhou D, Li R, Lv J, Wen G, et al. (2020). Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol Cancer 19, 159. 10.1186/s12943-020-01280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pawar K, Shigematsu M, Sharbati S, and Kirino Y. (2020). Infection-induced 5’-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol 18, e3000982. 10.1371/journal.pbio.3000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J, Ma G, Ge H, Han X, Mao X, Wang X, Veeramootoo JS, Xia T, Liu X, and Wang S. (2021). Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. NPJ Breast Cancer 7, 4. 10.1038/s41523-020-00211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H, Huang R, Li L, Zhu J, Li Z, Peng C, Zhuang X, Lin H, Shi S, and Huang P. (2021). CPA-seq reveals small ncRNAs with methylated nucleosides and diverse termini. Cell Discov 7, 25. 10.1038/s41421-021-00265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu S, et al. (2021). PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol 23, 424–436. 10.1038/s41556-021-00652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Su Z, Monshaugen I, Wilson B, Wang F, Klungland A, Ougland R, and Dutta A. (2022). TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nature communications 13, 2165. 10.1038/s41467-022-29790-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M, et al. (2018). Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 173, 1204–1216 e1226. 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 79.Guzzi N, Muthukumar S, Ciesla M, Todisco G, Ngoc PCT, Madej M, Munita R, Fazio S, Ekstrom S, Mortera-Blanco T, et al. (2022). Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat Cell Biol 24, 299–306. 10.1038/s41556-022-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuscu C, Kumar P, Kiran M, Su Z, Malik A, and Dutta A. (2018). tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105. 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, and Dalla-Favera R. (2013). tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America 110, 1404–1409. 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren B, Wang X, Duan J, and Ma J. (2019). Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922. 10.1126/science.aav8907. [DOI] [PubMed] [Google Scholar]
- 83.Luo S, He F, Luo J, Dou S, Wang Y, Guo A, and Lu J. (2018). Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic acids research 46, 5250–5268. 10.1093/nar/gky189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, and Meister G. (2008). A human snoRNA with microRNA-like functions. Molecular cell 32, 519–528. 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 85.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, and Lyko F. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19, 900–905. 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 86.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, and Lyko F. (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24, 1590–1595. 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. (2014). Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33, 2020–2039. 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Grone HJ, et al. (2015). The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 34, 2350–2362. 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Durdevic Z, Mobin MB, Hanna K, Lyko F, and Schaefer M. (2013). The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep 4, 931–937. 10.1016/j.celrep.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 90.Cosentino C, Toivonen S, Diaz Villamil E, Atta M, Ravanat JL, Demine S, Schiavo AA, Pachera N, Deglasse JP, Jonas JC, et al. (2018). Pancreatic beta-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic acids research 46, 10302–10318. 10.1093/nar/gky839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rashad S, Han X, Sato K, Mishima E, Abe T, Tominaga T, and Niizuma K. (2020). The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA biology 17, 1092–1103. 10.1080/15476286.2020.1779492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, Lu Z, Zheng Z, Dai Q, and Wang H. (2019). Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic acids research 47, 2533–2545. 10.1093/nar/gky1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vitali P, and Kiss T. (2019). Cooperative 2’-O-methylation of the wobble cytidine of human elongator tRNA(Met)(CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes & development 33, 741–746. 10.1101/gad.326363.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W, Schwartz MH, and Pan T. (2018). Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313. 10.1261/rna.067033.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hernandez-Alias X, Katanski CD, Zhang W, Assari M, Watkins CP, Schaefer MH, Serrano L, and Pan T. (2022). Single-read tRNA-seq analysis reveals coordination of tRNA modification and aminoacylation and fragmentation. Nucleic Acids Res. 10.1093/nar/gkac1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu J, Esberg A, Huang B, and Bystrom AS (2008). Kluyveromyces lactis gamma-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 36, 1072–1080. 10.1093/nar/gkm1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu J, Huang B, Esberg A, Johansson MJ, and Bystrom AS (2005). The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 11, 1648–1654. 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, and Kirino Y. (2015). Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A 112, E3816–3825. 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Z, Kim HK, Xu J, Jing Y, and Kay MA (2021). The 3’tsRNAs are aminoacylated: Implications for their biogenesis. PLoS genetics 17, e1009675. 10.1371/journal.pgen.1009675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. (2017). A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62. 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim HK, Xu J, Chu K, Park H, Jang H, Li P, Valdmanis PN, Zhang QC, and Kay MA (2019). A tRNA-Derived Small RNA Regulates Ribosomal Protein S28 Protein Levels after Translation Initiation in Humans and Mice. Cell Rep 29, 3816–3824 e3814. 10.1016/j.celrep.2019.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Couvillion MT, Sachidanandam R, and Collins K. (2010). A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 24, 2742–2747. 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hasler D, Lehmann G, Murakawa Y, Klironomos F, Jakob L, Grasser FA, Rajewsky N, Landthaler M, and Meister G. (2016). The Lupus Autoantigen La Prevents Mis-channeling of tRNA Fragments into the Human MicroRNA Pathway. Molecular cell 63, 110–124. 10.1016/j.molcel.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 104.Ganser LR, Kelly ML, Herschlag D, and Al-Hashimi HM (2019). The roles of structural dynamics in the cellular functions of RNAs. Nature reviews. Molecular cell biology 20, 474–489. 10.1038/s41580-019-0136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cruz JA, and Westhof E. (2009). The dynamic landscapes of RNA architecture. Cell 136, 604609. 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 106.Al-Hashimi HM, and Walter NG (2008). RNA dynamics: it is about time. Curr Opin Struct Biol 18, 321–329. 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ganser LR, Chu CC, Bogerd HP, Kelly ML, Cullen BR, and Al-Hashimi HM (2020). Probing RNA Conformational Equilibria within the Functional Cellular Context. Cell Rep 30, 2472–2480 e2474. 10.1016/j.celrep.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hentze MW, Castello A, Schwarzl T, and Preiss T. (2018). A brave new world of RNAbinding proteins. Nat Rev Mol Cell Biol 19, 327–341. 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 109.Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, and Collins K. (2009). Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev 23, 2016–2032. 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Couvillion MT, Bounova G, Purdom E, Speed TP, and Collins K. (2012). A Tetrahymena Piwi bound to mature tRNA 3’ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 48, 509–520. 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, et al. (2014). Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34, 2450–2463. 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Keam SP, Sobala A, Ten Have S, and Hutvagner G. (2017). tRNA-Derived RNA Fragments Associate with Human Multisynthetase Complex (MSC) and Modulate Ribosomal Protein Translation. J Proteome Res 16, 413–420. 10.1021/acs.jproteome.6b00267. [DOI] [PubMed] [Google Scholar]
- 113.Sobala A, and Hutvagner G. (2013). Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol 10, 553–563. 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mleczko AM, Celichowski P, and Bakowska-Zywicka K. (2018). Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim Biophys Acta Gene Regul Mech. 10.1016/j.bbagrm.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 115.Pecoraro V, Rosina A, and Polacek N. (2022). Ribosome-Associated ncRNAs (rancRNAs) Adjust Translation and Shape Proteomes. Noncoding RNA 8. 10.3390/ncrna8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ogawa T, Takahashi K, Ishida W, Aono T, Hidaka M, Terada T, and Masaki H. (2021). Substrate recognition mechanism of tRNA-targeting ribonuclease, colicin D, and an insight into tRNA cleavage-mediated translation impairment. RNA Biol 18, 1193–1205. 10.1080/15476286.2020.1838782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zywicki M, Bakowska-Zywicka K, and Polacek N. (2012). Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res 40, 4013–4024. 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bakowska-Zywicka K, Kasprzyk M, and Twardowski T. (2016). tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS yeast research 16. 10.1093/femsyr/fow077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lalande S, Merret R, Salinas-Giege T, and Drouard L. (2020). Arabidopsis tRNA-derived fragments as potential modulators of translation. RNA Biol 17, 1137–1148. 10.1080/15476286.2020.1722514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gonskikh Y, Gerstl M, Kos M, Borth N, Schosserer M, Grillari J, and Polacek N. (2020). Modulation of mammalian translation by a ribosome-associated tRNA half. RNA Biol 17, 1125–1136. 10.1080/15476286.2020.1744296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krishna S, Yim DG, Lakshmanan V, Tirumalai V, Koh JL, Park JE, Cheong JK, Low JL, Lim MJ, Sze SK, et al. (2019). Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep 20, e47789. 10.15252/embr.201947789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gebetsberger J, Wyss L, Mleczko AM, Reuther J, and Polacek N. (2017). A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14, 1364–1373. 10.1080/15476286.2016.1257470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fricker R, Brogli R, Luidalepp H, Wyss L, Fasnacht M, Joss O, Zywicki M, Helm M, Schneider A, Cristodero M, and Polacek N. (2019). A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun 10, 118. 10.1038/s41467-018-07949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, and Tavazoie SF (2015). Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 161, 790–802. 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, et al. (2019). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol 21, 978–990. 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 126.Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, and Hughes TR (2009). Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol 27, 667–670. 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 127.Falconi M, Giangrossi M, Zabaleta ME, Wang J, Gambini V, Tilio M, Bencardino D, Occhipinti S, Belletti B, Laudadio E, et al. (2019). A novel 3’-tRNA(Glu)-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. FASEB J 33, 13228–13240. 10.1096/fj.201900382RR. [DOI] [PubMed] [Google Scholar]
- 128.Liu X, Mei W, Padmanaban V, Alwaseem H, Molina H, Passarelli MC, Tavora B, and Tavazoie SF (2022). A pro-metastatic tRNA fragment drives Nucleolin oligomerization and stabilization of its bound metabolic mRNAs. Molecular cell 82, 2604–2617 e2608. 10.1016/j.molcel.2022.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ivanov P, Emara MM, Villen J, Gygi SP, and Anderson P. (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43, 613–623. 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, and Ivanov P. (2017). Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun 8, 1127. 10.1038/s41467-017-01278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lyons SM, Kharel P, Akiyama Y, Ojha S, Dave D, Tsvetkov V, Merrick W, Ivanov P, and Anderson P. (2020). eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic acids research 48, 6223–6233. 10.1093/nar/gkaa336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lyons SM, Achorn C, Kedersha NL, Anderson PJ, and Ivanov P. (2016). YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic acids research 44, 6949–6960. 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, and Anderson P. (2010). Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. The Journal of biological chemistry 285, 10959–10968. 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ivanov P, O’Day E, Emara MM, Wagner G, Lieberman J, and Anderson P. (2014). G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A 111, 18201–18206. 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, and Cairns BR (2007). Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 21, 261–266. 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Genenncher B, Durdevic Z, Hanna K, Zinkl D, Mobin MB, Senturk N, Da Silva B, Legrand C, Carre C, Lyko F, and Schaefer M. (2018). Mutations in Cytosine-5 tRNA Methyltransferases Impact Mobile Element Expression and Genome Stability at Specific DNA Repeats. Cell Rep 22, 1861–1874. 10.1016/j.celrep.2018.01.061. [DOI] [PubMed] [Google Scholar]
- 137.Chen X, and Wolin SL (2023). Transfer RNA halves are found as nicked tRNAs in cells: evidence that nicked tRNAs regulate expression of an RNA repair operon. RNA 29, 620–629. 10.1261/rna.079575.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Costa B, Li Calzi M, Castellano M, Blanco V, Cuevasanta E, Litvan I, Ivanov P, Witwer K, Cayota A, and Tosar JP (2023). Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proceedings of the National Academy of Sciences of the United States of America 120, e2216330120. 10.1073/pnas.2216330120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Honda S, and Kirino Y. (2016). SHOT-RNAs: A novel class of tRNA-derived functional RNAs expressed in hormone-dependent cancers. Mol Cell Oncol 3, e1079672. 10.1080/23723556.2015.1079672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Honda S, Morichika K, and Kirino Y. (2016). Selective amplification and sequencing of cyclic phosphate-containing RNAs by the cP-RNA-seq method. Nature protocols 11, 476–489. 10.1038/nprot.2016.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tosar JP, Gambaro F, Darre L, Pantano S, Westhof E, and Cayota A. (2018). Dimerization confers increased stability to nucleases in 5’ halves from glycine and glutamic acid tRNAs. Nucleic acids research 46, 9081–9093. 10.1093/nar/gky495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Levitt M. (1969). Detailed molecular model for transfer ribonucleic acid. Nature 224, 759–763. 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 143.Levitt M. (1973). Orientation of double-helical segments in crystals of yeast phenylalanine transfer RNA. J Mol Biol 80, 255–263. 10.1016/0022-2836(73)90171-x. [DOI] [PubMed] [Google Scholar]
- 144.Suddath FL, Quigley GJ, McPherson A, Sneden D, Kim JJ, Kim SH, and Rich A. (1974). Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0angstroms resolution. Nature 248, 20–24. 10.1038/248020a0. [DOI] [PubMed] [Google Scholar]
- 145.Li B, Cao Y, Westhof E, and Miao Z. (2020). Advances in RNA 3D Structure Modeling Using Experimental Data. Front Genet 11, 574485. 10.3389/fgene.2020.574485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Michel F, and Westhof E. (1990). Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216, 585–610. 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 147.Hou YM, and Schimmel P. (1988). A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333, 140–145. 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 148.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T. (2015). Efficient and quantitative high-throughput tRNA sequencing. Nature methods 12, 835–837. 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, and Lowe TM (2015). ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nature methods 12, 879–884. 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Peach SE, York K, and Hesselberth JR (2015). Global analysis of RNA cleavage by 5’hydroxyl RNA sequencing. Nucleic Acids Res 43, e108. 10.1093/nar/gkv536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu H, Yao J, Wu DC, and Lambowitz AM (2019). Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction. Scientific reports 9, 7953. 10.1038/s41598-019-44457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Upton HE, Ferguson L, Temoche-Diaz MM, Liu XM, Pimentel SC, Ingolia NT, Schekman R, and Collins K. (2021). Low-bias ncRNA libraries using ordered two-template relay: Serial template jumping by a modified retroelement reverse transcriptase. Proceedings of the National Academy of Sciences of the United States of America 118. 10.1073/pnas.2107900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Scheepbouwer C, Aparicio-Puerta E, Gomez-Martin C, Verschueren H, van Eijndhoven M, Wedekind LE, Giannoukakos S, Hijmering N, Gasparotto L, van der Galien HT, et al. (2023). ALL-tRNAseq enables robust tRNA profiling in tissue samples. Genes & development 37, 243–257. 10.1101/gad.350233.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Behrens A, Rodschinka G, and Nedialkova DD (2021). High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Molecular cell 81, 1802–1815 e1807. 10.1016/j.molcel.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang W, Foo M, Eren AM, and Pan T. (2022). tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Molecular cell 82, 891–906. 10.1016/j.molcel.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hernandez R, Shi J, Liu J, Li X, Wu J, Zhao L, Zhou T, Chen Q, and Zhou C. (2023). PANDORA-seq unveils the hidden small non-coding RNA landscape in atherosclerosis of LDL receptor-deficient mice. J Lipid Res, 100352. 10.1016/j.jlr.2023.100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang X, Cozen AE, Liu Y, Chen Q, and Lowe TM (2016). Small RNA Modifications: Integral to Function and Disease. Trends in molecular medicine 22, 1025–1034. 10.1016/j.molmed.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bjorkbom A, Lelyveld VS, Zhang S, Zhang W, Tam CP, Blain JC, and Szostak JW (2015). Bidirectional Direct Sequencing of Noncanonical RNA by Two-Dimensional Analysis of Mass Chromatograms. J Am Chem Soc 137, 14430–14438. 10.1021/jacs.5b09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.X, Y. (2021). MLC-Seq: de novo Sequencing of Full-Length tRNAs and Quantitative Mapping of Multiple RNA Modifications. Doi: 10.21203/rs.3.rs-1090754/v1. [DOI] [Google Scholar]
- 160.Wang Y, Zhao Y, Bollas A, Wang Y, and Au KF (2021). Nanopore sequencing technology, bioinformatics and applications. Nature biotechnology 39, 1348–1365. 10.1038/s41587-021-01108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lucas MC, Pryszcz LP, Medina R, Milenkovic I, Camacho N, Marchand V, Motorin Y, Ribas de Pouplana L, and Novoa EM (2023). Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nature biotechnology. 10.1038/s41587-023-01743-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gruber AR, Lorenz R, Bernhart SH, Neubock R, and Hofacker IL (2008). The Vienna RNA websuite. Nucleic acids research 36, W70–74. 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Markham NR, and Zuker M. (2008). UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453, 3–31. 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 164.Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research 31, 3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Parisien M, and Major F. (2008). The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452, 51–55. 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 166.Reuter JS, and Mathews DH (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129. 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sakakibara Y, Brown M, Hughey R, Mian IS, Sjolander K, Underwood RC, and Haussler D. (1994). Stochastic context-free grammars for tRNA modeling. Nucleic acids research 22, 511–25120. 10.1093/nar/22.23.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wj Anderson J, Tataru P, Staines J, Hein J, and Lyngso R. (2012). Evolving stochastic context--free grammars for RNA secondary structure prediction. BMC Bioinformatics 13, 78. 10.1186/1471-2105-13-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang J. (2023). Probing RNA Structures and Interactions Using Fluorescence Lifetime Analyses. Methods Mol Biol 2568, 13–23. 10.1007/978-1-0716-2687-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, and Assmann SM (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700. 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 171.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, and Arkin AP (2011). Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proceedings of the National Academy of Sciences of the United States of America 108, 11063–11068. 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rouskin S, Zubradt M, Washietl S, Kellis M, and Weissman JS (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701705. 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Siegfried NA, Busan S, Rice GM, Nelson JA, and Weeks KM (2014). RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nature methods 11, 959–965. 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Luo QJ, Zhang J, Li P, Wang Q, Zhang Y, Roy-Chaudhuri B, Xu J, Kay MA, and Zhang QC (2021). RNA structure probing reveals the structural basis of Dicer binding and cleavage. Nature communications 12, 3397. 10.1038/s41467-021-23607-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yamagami R, Sieg JP, Assmann SM, and Bevilacqua PC (2022). Genome-wide analysis of the in vivo tRNA structurome reveals RNA structural and modification dynamics under heat stress. Proceedings of the National Academy of Sciences of the United States of America 119, e2201237119. 10.1073/pnas.2201237119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schorn AJ, Gutbrod MJ, LeBlanc C, and Martienssen R. (2017). LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 170, 61–71 e11. 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aw JG, Shen Y, Wilm A, Sun M, Lim XN, Boon KL, Tapsin S, Chan YS, Tan CP, Sim AY, et al. (2016). In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Molecular cell 62, 603–617. 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 178.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279. 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sharma E, Sterne-Weiler T, O’Hanlon D, and Blencowe BJ (2016). Global Mapping of Human RNA-RNA Interactions. Molecular cell 62, 618–626. 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 180.Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu ZY, Kwok CK, Qin CF, MacRae IJ, et al. (2018). COMRADES determines in vivo RNA structures and interactions. Nature methods 15, 785–788. 10.1038/s41592-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang M, Li K, Bai J, Velema WA, Yu C, van Damme R, Lee WH, Corpuz ML, Chen JF, and Lu Z. (2021). Optimized photochemistry enables efficient analysis of dynamic RNA structuromes and interactomes in genetic and infectious diseases. Nature communications 12, 2344. 10.1038/s41467-021-22552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cai Z, Cao C, Ji L, Ye R, Wang D, Xia C, Wang S, Du Z, Hu N, Yu X, et al. (2020). RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 582, 432–437. 10.1038/s41586-020-2249-1. [DOI] [PubMed] [Google Scholar]
- 183.Helwak A, Kudla G, Dudnakova T, and Tollervey D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shen EZ, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding YH, Dai SY, Weng Z, and Mello CC (2018). Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 172, 937–951 e918. 10.1016/j.cell.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kumar P, Anaya J, Mudunuri SB, and Dutta A. (2014). Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12, 78. 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guan L, Karaiskos S, and Grigoriev A. (2020). Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA biology 17, 1070–1080. 10.1080/15476286.2019.1676633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guan L, and Grigoriev A. (2021). Computational meta-analysis of ribosomal RNA fragments: potential targets and interaction mechanisms. Nucleic acids research 49, 4085–4103. 10.1093/nar/gkab190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang J, Fei Y, Sun L, and Zhang QC (2022). Advances and opportunities in RNA structure experimental determination and computational modeling. Nature methods 19, 1193–1207. 10.1038/s41592-022-01623-y. [DOI] [PubMed] [Google Scholar]
- 189.Marklund E, Ke Y, and Greenleaf WJ (2023). High-throughput biochemistry in RNA sequence space: predicting structure and function. Nature reviews. Genetics. 10.1038/s41576-02200567-5. [DOI] [PubMed] [Google Scholar]
- 190.Townshend RJL, Eismann S, Watkins AM, Rangan R, Karelina M, Das R, and Dror RO (2021). Geometric deep learning of RNA structure. Science 373, 1047–1051. 10.1126/science.abe5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, et al. (2023). Visualizing RNA conformational and architectural heterogeneity in solution. Nature communications 14, 714. 10.1038/s41467-023-36184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sajini AA, Choudhury NR, Wagner RE, Bornelov S, Selmi T, Spanos C, Dietmann S, Rappsilber J, Michlewski G, and Frye M. (2019). Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nature communications 10, 2550. 10.1038/s41467-019-10020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fagan SG, Helm M, and Prehn JHM (2021). tRNA-derived fragments: A new class of noncoding RNA with key roles in nervous system function and dysfunction. Prog Neurobiol 205, 102118. 10.1016/j.pneurobio.2021.102118. [DOI] [PubMed] [Google Scholar]
- 196.Fu M, Gu J, Wang M, Zhang J, Chen Y, Jiang P, Zhu T, and Zhang X. (2023). Emerging roles of tRNA-derived fragments in cancer. Mol Cancer 22, 30. 10.1186/s12943-023-01739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mao M, Chen W, Huang X, and Ye D. (2023). Role of tRNA-derived small RNAs(tsRNAs) in the diagnosis and treatment of malignant tumours. Cell Commun Signal 21, 178. 10.1186/s12964-023-01199-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mathew BA, Katta M, Ludhiadch A, Singh P, and Munshi A. (2023). Role of tRNA-Derived Fragments in Neurological Disorders: a Review. Mol Neurobiol 60, 655–671. 10.1007/s12035022-03078-w. [DOI] [PubMed] [Google Scholar]
- 199.Pandey KK, Madhry D, Ravi Kumar YS, Malvankar S, Sapra L, Srivastava RK, Bhattacharyya S, and Verma B. (2021). Regulatory roles of tRNA-derived RNA fragments in human pathophysiology. Mol Ther Nucleic Acids 26, 161–173. 10.1016/j.omtn.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xu D, Qiao D, Lei Y, Zhang C, Bu Y, and Zhang Y. (2022). Transfer RNA-derived small RNAs (tsRNAs): Versatile regulators in cancer. Cancer Lett 546, 215842. 10.1016/j.canlet.2022.215842. [DOI] [PubMed] [Google Scholar]
- 201.Chu X, He C, Sang B, Yang C, Yin C, Ji M, Qian A, and Tian Y. (2022). Transfer RNAsderived small RNAs and their application potential in multiple diseases. Front Cell Dev Biol 10, 954431. 10.3389/fcell.2022.954431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wilson B, and Dutta A. (2022). Function and Therapeutic Implications of tRNA Derived Small RNAs. Front Mol Biosci 9, 888424. 10.3389/fmolb.2022.888424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yu X, Xie Y, Zhang S, Song X, Xiao B, and Yan Z. (2021). tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 11, 461–469. 10.7150/thno.51963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Weng Q, Wang Y, Xie Y, Yu X, Zhang S, Ge J, Li Z, Ye G, and Guo J. (2022). Extracellular vesicles-associated tRNA-derived fragments (tRFs): biogenesis, biological functions, and their role as potential biomarkers in human diseases. J Mol Med (Berl) 100, 679–695. 10.1007/s00109-022-02189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zheng LL, Xu WL, Liu S, Sun WJ, Li JH, Wu J, Yang JH, and Qu LH (2016). tRF2Cancer: A web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic acids research 44, W185–193. 10.1093/nar/gkw414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Childs-Disney JL, Yang X, Gibaut QMR, Tong Y, Batey RT, and Disney MD (2022). Targeting RNA structures with small molecules. Nat Rev Drug Discov 21, 736–762. 10.1038/s41573-022-00521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, and Al-Hashimi HM (2011). Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol 7, 553–559. 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wei J, Chen S, Zong L, Gao X, and Li Y. (2022). Protein-RNA interaction prediction with deep learning: structure matters. Brief Bioinform 23. 10.1093/bib/bbab540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Arora V, and Sanguinetti G. (2022). Challenges for machine learning in RNA-protein interaction prediction. Stat Appl Genet Mol Biol 21. 10.1515/sagmb-2021-0087. [DOI] [PubMed] [Google Scholar]
- 210.Sun L, Xu K, Huang W, Yang YT, Li P, Tang L, Xiong T, and Zhang QC (2021). Predicting dynamic cellular protein-RNA interactions by deep learning using in vivo RNA structures. Cell Res 31, 495–516. 10.1038/s41422-021-00476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dolgin E. (2022). tRNA therapeutics burst onto startup scene. Nature biotechnology 40, 283–286. 10.1038/s41587-022-01252-y. [DOI] [PubMed] [Google Scholar]
- 212.Lueck JD, Yoon JS, Perales-Puchalt A, Mackey AL, Infield DT, Behlke MA, Pope MR, Weiner DB, Skach WR, McCray PB Jr., and Ahern CA (2019). Engineered transfer RNAs for suppression of premature termination codons. Nature communications 10, 822. 10.1038/s41467-019-08329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang J, Zhang Y, Mendonca CA, Yukselen O, Muneeruddin K, Ren L, Liang J, Zhou C, Xie J, Li J, et al. (2022). AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature 604, 343–348. 10.1038/s41586-022-04533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang X, Trebak F, Souza LAC, Shi J, Zhou T, Kehoe PG, Chen Q, and Feng Earley Y. (2020). Small RNA modifications in Alzheimer’s disease. Neurobiol Dis 145, 105058. 10.1016/j.nbd.2020.105058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, and Jeang KT (2009). Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 37, 6575–6586. 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Anderson P, and Ivanov P. (2014). tRNA fragments in human health and disease. FEBS letters 588, 4297–4304. 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Magee R, and Rigoutsos I. (2020). On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic acids research 48, 9433–9448. 10.1093/nar/gkaa657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Diallo I, Ho J, Lambert M, Benmoussa A, Husseini Z, Lalaouna D, Masse E, and Provost P. (2022). A tRNA-derived fragment present in E. coli OMVs regulates host cell gene expression and proliferation. PLoS Pathog 18, e1010827. 10.1371/journal.ppat.1010827. [DOI] [PMC free article] [PubMed] [Google Scholar]