Abstract
Fusion is essential for mitochondrial function in a great variety of eukaryotic cell types. Yeast cells defective in mitohondrial fusion are respiration-deficient, human cells use complementation of fused mitochondria as a defence against the accumulation of oxidative damage during cellular aging and fusion is required to build an intracellular mitochondrial continuum that allows the dissipation of energy in the cell. Moreover, developmental processes such as spermatogenesis in Drosophila require regulated mitochondrial fusion. Some of the molecular mediators of mitochondrial membrane fusion have been identified in recent years. An evolutionarily conserved large GTPase in the outer membrane is essential for mitochondrial fusion, and genetic screens in yeast are revealing an increasing number of additional important genes. Mechanistic studies have provided the first insights into how the problem of faithfully fusing a double membrane-bounded organelle in a coordinated manner is solved.
Introduction
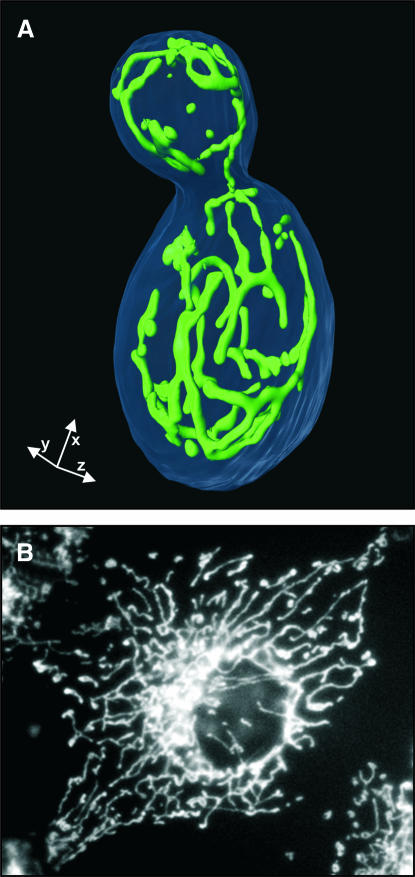
Mitochondria play essential and diverse roles in the metabolism and physiology of eukaryotic cells. They provide energy generated by oxidative phosphorylation, are the compartment for numerous essential metabolic reactions, and play central roles in apoptosis and aging (Scheffler, 2001). The cellular function of mitochondria is reflected in their structure. They are complex, double membrane-bounded organelles with a morphology and intracellular distribution characteristic for each eukaryotic cell type (Bereiter-Hahn, 1990; Warren and Wickner, 1996; Yaffe, 1999; Griparic and van der Bliek, 2001). In the classic electron micrographs that can be found in many text books, mitochondria appear as solitary organelles because these images show single cross-sections through a cell. However, this view is oversimplified. In many organisms, mitochondria build a giant interconnected reticulum. In budding yeast, for example, they form a branched tubular network located immediately below the cell cortex (Figure 1A), and mammalian fibroblast cells harbor interconnected mitochondrial filaments that extend throughout the entire cytosol (Figure 1B). The mitochondrial reticulum is a highly dynamic structure. It is maintained by permanent opposing but balanced membrane fusion and fission events (Nunnari et al., 1997; Bleazard et al., 1999; Sesaki and Jensen, 1999; Santel and Fuller, 2001). There is accumulating evidence that mitochondrial dynamics are important for many aspects of cellular function. Our current view of the mitochondrial division apparatus has been reviewed recently by Osteryoung (2001) and Yoon and McNiven (2001). This review describes what is known about the role of mitochondrial membrane fusion in different cell types and highlights recent studies that have identified and functionally characterized some of the molecular components involved in this process.
Fig. 1. Intracellular mitochondrial networks. (A) Branched mitochondrial network in a budding yeast cell. A yeast strain expressing mitochondria-targeted green fluorescent protein was grown on the non-fermentable carbon source glycerol to logarithmic growth phase. The cell wall was stained with calcofluor white. A surface-rendered three-dimensional stack recorded with an MMM-4Pi confocal fluorescence microscope is shown. The length of each arrow corresponds to 1 µm. Reproduced from Egner et al. (2002) with permission. © 2002 National Academy of Sciences, USA. (B) Extended and interconnected mitochondrial filaments in a COS-7 cell (african green monkey fibroblast). Mitochondria were stained by indirect immunofluorescence against cytochrome c. The image was kindly provided by Ansgar Santel, Stanford University, CA.
Cellular role of mitochondrial fusion
Fusion of mitochondria serves to mix and unify mitochondrial compartments. This is of particular importance for the inheritance and maintenance of the mitochondrial genome, which encodes several polypeptides required for respiratory function. Mitochondrial fusion is a prerequisite for the faithful segregation of mitochondrial DNA (mtDNA) in budding yeast, in both the sexual and the vegetative life cycles. Upon mating of two yeast cells, the mitochondrial compartments of both parents merge into a continuous reticulum, and organellar proteins rapidly redistribute throughout the zygote. Subsequently, mtDNA is sorted into the newly formed bud by an active non-random segregation mechanism. Buds arising from the middle of the zygote receive mtDNA from both parents, which allows recombination at high frequency (Nunnari et al., 1997; Okamoto et al., 1998; Berger and Yaffe, 2000). During mitotic growth, yeast cells defective in mitochondrial fusion lose mtDNA rapidly, indicating that the integrity of the mitochondrial compartment is important for transmission of mitochondrial chromosomes from the mother to the bud cell (Hermann et al., 1998; Rapaport et al., 1998; Wong et al., 2000; Sesaki and Jensen, 2001).
Complementation of mitochondrial gene products in fused mitochondria has also been described for human cells (Ono et al., 2001), although it does not occur with equal efficiency in all experimental systems (Enriquez et al., 2000). Ono et al. (2001) established two respiration-deficient HeLa cell lines, each carrying a pathogenic mutation in a different mitochondrial tRNA gene. Hybrids obtained by fusion of these cells showed restoration of normal respiratory activity within a few days. This can only be explained by fusion and exchange of mitochondrial content. Strong support for an inter-mitochondrial complementation mechanism in the living organism was obtained from the analysis of a transgenic mouse model that contained a mixture of intact and mutant mtDNA (Nakada et al., 2001). Irrespective of the ratio of mitochondria with different genotypes, mitochondria in various tissues showed a homogeneous pattern of respiratory activity at the subcellular level, as measured by electron microscopy of stained active cytochrome c oxidase complexes. The observed equilibration of mitochondrial gene products throughout the entire mitochondrial network of a cell indicates organellar fusion. Intriguingly, the extensive intermixing of mitochondria even prevented the animals from expressing disease phenotypes associated with mtDNA mutations (Nakada et al., 2001; Turnbull and Lightowlers, 2001). These findings suggest that mitochondrial fusion counteracts the manifestation of mtDNA-linked diseases. Moreover, it has physiological significance in individuals born with intact mitochondrial genomes. The highly oxidative metabolism of mitochondria increases the risk for mtDNA damage. Thus, lesions and point mutations of mtDNA accumulate during aging and result in the loss of bioenergetic function (Wallace, 1992; Nagley and Wei, 1998; Raha and Robinson, 2000). Fusion of mitochondria might contribute to the maintenance of respiratory activity by allowing trans-complementation of somatic mutations that accumulate in mitochondrial chromosomes over time (Ono et al., 2001), and thus mitochondrial fusion may be a crucial defence mechanism against cellular aging.
The establishment and maintenance of an intracellular mitochondrial continuum by continuous membrane fusion and fission events bears relevance for the dissipation of energy in the cell. Experiments with human fibroblasts using fluorescent labeling techniques proved that the mitochondrial membrane potential (Δψ) is equal over the whole length of a mitochondrion. This was demonstrated by damaging the mitochondrial membranes locally by irradiation with a laser beam, which led to an immediate collapse of Δψ in an extended network of interconnected filaments (Amchenkova et al., 1988). This suggests that mitochondrial filaments represent electrically united systems that facilitate the delivery of energy to remote parts of the cell (Skulachev, 2001). In muscle cells, for example, a multilayer of mitochondria is found below the outer cell membrane (sarcolemma) where the oxygen supply is high. These organelles are connected by mitochondrial filaments to mitochondria located in the oxygen-poor core part of the muscle fiber. It has been proposed that the mitochondrial network in muscle cells serves to transmit the membrane potential generated by subsarcolemmal mitochondria to intermyofibrillar organelles, which can then use oxygen from the cell periphery to produce ATP in the core of the muscle fiber (Skulachev, 2001). Morphological observations (Bakeeva et al., 1983) are in agreement with the view that mitochondrial fusion is required for the establishment of the intermyofibrillar network and maintenance of muscle function.
An essential role of mitochondrial fusion in cell development has been demonstrated for Drosophila spermatids (Hales and Fuller, 1997). Spermatogenesis requires a dramatic rearrangement of cell organelles. During this process, mitochondria are positioned next to the flagellum where they provide energy for the vigorous movement of the sperm cell. During normal spermatogenesis in the fly, mitochondria aggregate next to spermatid nuclei and fuse into two giant organelles that wrap around each other. This spherical structure, termed Nebenkern, resembles an onion slice when viewed in cross-section by electron microscopy. During further development, Nebenkern mitochondria unfurl from each other and elongate beside the flagellar base. Hales and Fuller (1997) showed that fuzzy onions mutant males are sterile because spermatogenesis is blocked at the stage of mitochondrial fusion. This is an intriguing example of the essential role that mitochondrial morphogenesis can play in a cell-developmental process.
Role of Fzo family members/mitofusins in mitochondrial fusion
Molecular genetic analysis of the fuzzy onions mutant identified the first known molecular mediator of mitochondrial fusion, Fzo (Hales and Fuller, 1997). This protein is the founding member of a novel evolutionarily conserved family of large GTPases in the mitochondrial outer membrane. Functional characterization of the yeast Fzo homolog, Fzo1p, established that this protein is a central player in mitochondrial fusion. Yeast mutants lacking Fzo1p contain fragmented mitochondria because fusion is blocked while fission continues. As a secondary consequence of aberrant mitochondrial morphology, fzo1 mutants lose mtDNA and become respiration-deficient (Hermann et al., 1998; Rapaport et al., 1998). Direct evidence for a role of Fzo1p in mitochondrial fusion was obtained by employing an elegant in vivo assay that was developed by Nunnari et al. (1997). Mitochondria from yeast cells of opposite mating type were labeled with different fluorescent markers. Upon mating of wild-type cells, the fluorescent labels immediately intermixed in zygotes, indicating fusion (Nunnari et al., 1997). In zygotes obtained from fzo1 mutant cells, mitochondria still entered the newly formed bud. However, the fluorescent labels from the parental cells did not intermix, indicating a block in fusion (Hermann et al., 1998).
Interestingly, higher metazoa have more than one fzo-related gene. The originally described fuzzy onions protein of Drosophila is expressed only in spermatids, just at the time when mitochondria fuse to build the Nebenkern, and disappears soon after fusion is complete (Hales and Fuller, 1997). A second, as yet uncharacterized, Fzo-related protein (genome annotation number CG3869) might play a more general role in regulating mitochondrial morphology in the fly. Similarly, two different human Fzo family members, termed mitofusins (Mfn1 and Mfn2), have been identified. Both mitofusins have been shown to associate with mitochondria when expressed by transient transfection in tissue culture cells. Overexpression of Mfn2 induced extensive perinuclear clustering of mitochondria in the transfected cells, suggesting a role in mitochondrial fusion. Intriguingly, coexpression of Mfn2 with a dominant-negative mutant of the predicted mitochondrial fission protein, Drp1, led to the formation of an interconnected network of thin mitochondrial tubules that was probably generated by extensive mitochondrial fusion unopposed by mitochondrial fission (Santel and Fuller, 2001). It is not known whether the human mfn1 and mfn2 genes are differentially expressed. Thus, it remains a challenge for the future to examine the roles of different Fzo-related proteins in determining mitochondrial shape in highly differentiated tissues.
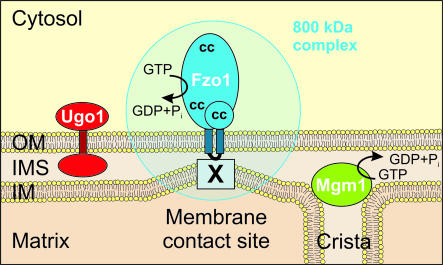
So far, we can only speculate about the molecular mechanisms underlying mitochondrial membrane fusion. There are some indications that Fzo1p might act directly as a fusogen for the outer membrane. First, Fzo1p is found in the correct location. It is anchored in the outer membrane, with both its large N-terminal domain and its smaller C-terminal tail exposed to the cytosol (Hermann et al., 1998; Rapaport et al., 1998; Fritz et al., 2001). This topology would allow interactions with partner proteins on the surface of an approaching mitochondrion. Electron micrographs of fusing mitochondria suggest that fusion is initiated at contact sites, i.e. at sites of close apposition of the mitochondrial outer and inner membranes (Bereiter-Hahn and Vöth, 1994). Intriguingly, biochemical subfractionation of mitochondria has localized Fzo1p to contact sites (Hermann et al., 1998; Fritz et al., 2001), a finding that has implications for a possible fusion mechanism. As double membrane-bounded organelles, mitochondria face the challenge of faithfully fusing four membranes. Thus, it is conceivable that a connection between the mitochondrial membranes by the fusion machinery is mechanistically important. Indeed, mutation of the intermembrane space loop of Fzo1p has been observed to abolish its contact with the inner membrane, and this results in a loss of function of the mutant protein. This suggests that the confinement of Fzo1p to contact sites is important for coordination of the mitochondrial membranes during the fusion reaction (Fritz et al., 2001).
A second indication that Fzo1p might be involved directly in fusion is that it possesses all of the features predicted to be required for a fusogen. The presence of membrane anchors and extensive coiled-coil domains is a characteristic of other membrane fusogens, such as SNARE proteins or viral fusion proteins (Skehel and Wiley, 1998; Weber et al., 1998). As already stated above, Fzo protein family members are transmembrane proteins. They also contain several regions that are predicted to form coiled-coil structures that could potentially mediate inter- or intramolecular protein–protein interactions (Hales and Fuller, 1997; Hermann et al., 1998). Furthermore, the GTPase domain may provide biomechanical energy that can be used for membrane fusion. As in vitro assays for mitochondrial fusion are not readily available, however, a direct function for the Fzo proteins as fusogens for the mitochondrial outer membrane remains to be proven.
The mechanism behind mitochondrial fusion must be rather complex, since this process appears to be coordinated with the antagonistic process of fission. This is suggested by two lines of evidence. First, counting fusion and fission events over time revealed that the rates of both processes are approximately equivalent in single yeast cells (Nunnari et al., 1997). Secondly, fragmentation of mitochondria in mutants defective in fusion can be suppressed by mutations of components of the mitochondrial division machinery (Bleazard et al., 1999; Sesaki and Jensen, 1999, 2001; Santel and Fuller, 2001). Thus, mechanisms must exist that coordinate fusion events with fission events to maintain the continuity of the mitochondrial network. Furthermore, the fusion machinery of the outer membrane has to be connected to that of the inner membrane (Fritz et al., 2001), and fusion proteins probably need to be recycled after completion of one round of fusion. Therefore, one might predict that more than one protein is involved (see Figure 2 for an arrangement of the components known currently). Biochemical evidence shows that Fzo1p assembles—possibly together with other as yet unknown components—into a high molecular weight complex (Rapaport et al., 1998), and genetic screens in yeast are revealing an increasing number of additional important genes, as discussed below.
Fig. 2. Molecular components of mitochondrial fusion. The topology and arrangement of proteins important for mitochondrial fusion in yeast are presented schematically. See text for details. OM, mitochondrial outer membrane; IMS, intermembrane space; IM, mitochondrial inner membrane; cc, coiled coils; X, hypothetical component(s) of the inner membrane fusion machinery.
Other components involved in mitochondrial fusion
The finding that growth defects due to mitochondrial fragmentation and consecutive loss of mtDNA can be suppressed by a block of mitochondrial fission was employed for an elegant genetic screen for novel components of the mitochondrial fusion machinery. Sesaki and Jensen (2001) isolated the ugo1 mutant (ugo is Japanese for fusion) by selection for strains that maintained mtDNA only in the absence of functional Dnm1p, a component essential for mitochondrial fission. Deletion of the UGO1 gene leads to a phenotype very similar to that exhibited by fzo1 mutants. Cells lacking Ugo1p harbor highly fragmented mitochondria, they lose mtDNA, and their mitochondria fail to fuse and mix their matrix contents in zygotes. Ugo1p is a protein of the outer membrane, and has one membrane-spanning segment. However, it does not localize to contact sites, and evidence for an interaction with Fzo1p could not be obtained, suggesting that Ugo1p and Fzo1p mediate distinct steps in mitochondrial fusion (Sesaki and Jensen, 2001).
Mgm1p is a dynamin-like protein that has long been known to be required for mitochondrial genome maintenance (Jones and Fangman, 1992) and normal mitochondrial morphology (Guan et al., 1993). Wong et al. (2000) isolated temperature-sensitive mgm1 mutants and observed that mitochondria fragment rapidly upon shifting to the non-permissive temperature. Furthermore, mitochondrial fusion was found to be blocked in mgm1 cells during mating, suggesting that Mgm1p is required for maintenance of fusion-competent mitochondria. In contrast to the situation in fzo1 and ugo1 mutants (Bleazard et al., 1999; Sesaki and Jensen, 1999; Sesaki and Jensen, 2001), fusion activity can be restored in mgm1 mutants by blocking mitochondrial fission in a dnm1 mutant background (Wong et al., 2000). This demonstrates that Mgm1p does not play a direct role in fusion. Based on its homology to dynamin-like proteins and its localization in the intermembrane space, it was proposed that Mgm1p instead plays a role in remodeling events of the inner membrane, such as cristae formation and/or inner membrane fission (Wong et al., 2000). As the fusions of the outer and inner membranes are intimately linked (Fritz et al., 2001), it seems possible that mutation of MGM1 renders the organelle fusion incompetent by affecting the structure of the inner membrane.
A systematic genome-wide screen of a deletion mutant library covering the non-essential genes of Saccharomyces cerevisiae revealed recently a number of additional, novel mutants defective in mitochondrial distribution and morphology (Dimmer et al., 2002). Some of these mutants display fragmented mitochondria, suggesting that the corresponding gene products are important for mitochondrial fusion. Of particular interest is Mdm30p, a protein of unknown function that contains an F-box. As F-box proteins are known to be involved in targeting proteins for ubiquitin-dependent degradation (Patton et al., 1998), Mdm30p might regulate the turnover of components of the mitochondrial fusion and fission machinery, and thereby regulate these processes (Dimmer et al., 2002). Functional analysis of novel yeast mutants isolated in this and other screens should soon improve our understanding of the molecular machinery of mitochondrial membrane fusion.
Perspectives
Genetic studies in yeast have proven to be extremely valuable for the identification and characterization of some key components of mitochondrial fusion, and we are likely to obtain further exciting insights from these studies in the coming years. Moreover, there are many challenging problems that go beyond the elucidation of the basic molecular mechanisms of mitochondrial fusion. For example, it has been reported that a splice variant of a SNARE protein, which is normally required for membrane fusion in the secretory pathway, is localized on mitochondria (Isenmann et al., 1998). This suggests an involvement of mitochondria-localized SNARE proteins in fusion events with heterologous membranes, such as the endoplasmic reticulum, and this could be important, for example, in facilitating transfer of lipids between the organelles. Furthermore, there is good evidence that mitochondrial fusion in the true slime mold Physarum polycephalum depends on a mitochondrial plasmid that encodes proteins unrelated to those known from yeast (Takano et al., 1997). Thus, it appears that evolution developed more than one way to solve the challenging problem of fusing mitochondria. Finally, the recent demonstration that inhibition of mitochondrial fission blocks apoptosis (Frank et al., 2001) suggests that mitochondrial fusion also influences this process. Clearly, we have only started to answer many open questions in a fascinating area of cell biology.

Benedikt Westermann
Acknowledgments
Acknowledgements
I thank the members of my laboratory for their invaluable work, Walter Neupert for continuous support, Hannes Herrmann for many stimulating discussions and for critical comments on the manuscript, Alexander Egner, Stefan Jakobs, Stefan W. Hell and Ansgar Santel for contributing images, and the Deutsche Forschungsgemeinschaft for support through grant WE 2174/2-3.
References
- Amchenkova A.A., Bakeeva, L.E., Chentsov, Y.S., Skulachev, V.P. and Zorov, D.B. (1988) Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J. Cell Biol., 107, 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakeeva L.E., Chentsov, Y.S. and Skulachev, V.P. (1983) Intermitochondrial contacts in myocardiocytes. J. Mol. Cell. Cardiol., 15, 413–420. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J. (1990) Behavior of mitochondria in the living cell. Int. Rev. Cytol., 122, 1–63. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J. and Vöth, M. (1994) Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech., 27, 198–219. [DOI] [PubMed] [Google Scholar]
- Berger K.H. and Yaffe, M.P. (2000) Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol., 8, 508–513. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J. and Shaw, J. (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol., 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer K.S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W. and Westermann, B. (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell, 13, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner A., Jakobs, S. and Hell, S.W. (2002) Fast 100 nm resolution 3D-microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl Acad. Sci. USA, 99, 3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J.A., Cabezas-Herrera, J., Bayona-Bafaluy, M.P. and Attardi, G. (2000) Very rare complementation between mitochondria carrying different mitochondrial DNA mutations points to intrinsic genetic autonomy of the organelles in cultured human cells. J. Biol. Chem., 275, 11207–11215. [DOI] [PubMed] [Google Scholar]
- Frank S., Gaume, B., Bergmann-Leitner, E.S., Leitner, W.W., Robert, E.G., Catez, F., Smith, C.L. and Youle, R.J. (2001) The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell, 1, 515–525. [DOI] [PubMed] [Google Scholar]
- Fritz S., Rapaport, D., Klanner, E., Neupert, W. and Westermann, B. (2001) Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J. Cell Biol., 152, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L. and van der Bliek, A.M. (2001) The many shapes of mitochondrial membranes. Traffic, 2, 235–244. [DOI] [PubMed] [Google Scholar]
- Guan K., Farh, L., Marshall, T.K. and Deshenes, R.J. (1993) Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet., 24, 141–148. [DOI] [PubMed] [Google Scholar]
- Hales K.G. and Fuller, M.T. (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell, 90, 121–129. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher, J.W., Mills, J.P., Hales, K.G., Fuller, M.T., Nunnari, J. and Shaw, J.M. (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol., 143, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann S., Khew-Goodall, Y., Gamble, J., Vadas, M. and Wattenberg, B.W. (1998) A splice isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol. Biol. Cell, 9, 1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.A. and Fangman, W.L. (1992) Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev., 6, 380–389. [DOI] [PubMed] [Google Scholar]
- Nagley P. and Wei, Y.H. (1998) Ageing and mammalian mitochondrial genetics. Trends Genet., 14, 513–517. [DOI] [PubMed] [Google Scholar]
- Nakada K., Inoue, K., Ono, T., Isobe, K., Ogura, A., Goto, Y.I., Nonaka, I. and Hayashi, J.I. (2001) Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med., 7, 934–940. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W. and Walter, P. (1997) Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell, 8, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Perlman, P.S. and Butow, R.A. (1998) The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol., 142, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Isobe, K., Nakada, K. and Hayashi, J.I. (2001) Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet., 28, 272–275. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W. (2001) Organelle fission in eukaryotes. Curr. Opin. Microbiol., 4, 639–646. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Willems, A.R. and Tyers, M. (1998) Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet., 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Raha S. and Robinson, B.H. (2000) Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem. Sci., 25, 502–508. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner, M., Neupert, W. and Westermann, B. (1998) Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem., 273, 20150–20155. [DOI] [PubMed] [Google Scholar]
- Santel A. and Fuller, M.T. (2001) Control of mitochondrial morphology by a human mitofusin. J. Cell Sci., 114, 867–874. [DOI] [PubMed] [Google Scholar]
- Scheffler I.E. (2001) A century of mitochondrial research: achievements and perspectives. Mitochondrion, 1, 3–31. [DOI] [PubMed] [Google Scholar]
- Sesaki H. and Jensen, R.E. (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol., 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H. and Jensen, R.E. (2001) UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol., 152, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J. and Wiley, D.C. (1998) Coiled coils in both intracellular vesicle and viral membrane fusion. Cell, 95, 871–874. [DOI] [PubMed] [Google Scholar]
- Skulachev V.P. (2001) Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci., 26, 23–29. [DOI] [PubMed] [Google Scholar]
- Takano H., Kuroiwa, T. and Kawano, S. (1997) Mitochondrial fusion promoting plasmid. Cell Struct. Funct., 22, 299–308. [DOI] [PubMed] [Google Scholar]
- Turnbull D.M. and Lightowlers, R.N. (2001) Might mammalian mitochondria merge? Nat. Med., 7, 895–896. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. (1992) Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science, 256, 628–632. [DOI] [PubMed] [Google Scholar]
- Warren G. and Wickner, W. (1996) Organelle inheritance. Cell, 84, 395–400. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman, B.V., McNew, J.A., Westermann, B., Gmachl, M., Parlati, F., Söllner, T.H. and Rothman, J.E. (1998) SNAREpins: minimal machinery for membrane fusion. Cell, 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner, J.A., Gorsich, S.W., McCaffery, J.M., Shaw, J.M. and Nunnari, J. (2000) The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol., 151, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M.P. (1999) The machinery of mitochondrial inheritance and behavior. Science, 283, 1493–1497. [DOI] [PubMed] [Google Scholar]
- Yoon Y. and McNiven, M.A. (2001) Mitochondrial division: new partners in membrane pinching. Curr. Biol., 11, R67–R70. [DOI] [PubMed] [Google Scholar]