Abstract
Background and Objectives:
Reproducibility is a major challenge in developing machine learning (ML)-based solutions in computational pathology (CompPath). The NCI Imaging Data Commons (IDC) provides >120 cancer image collections according to the FAIR principles and is designed to be used with cloud ML services. Here, we explore its potential to facilitate reproducibility in CompPath research.
Methods:
Using the IDC, we implemented two experiments in which a representative ML-based method for classifying lung tumor tissue was trained and/or evaluated on different datasets. To assess reproducibility, the experiments were run multiple times with separate but identically configured instances of common ML services.
Results:
The results of different runs of the same experiment were reproducible to a large extent. However, we observed occasional, small variations in AUC values, indicating a practical limit to reproducibility.
Conclusions:
We conclude that the IDC facilitates approaching the reproducibility limit of CompPath research (i) by enabling researchers to reuse exactly the same datasets and (ii) by integrating with cloud ML services so that experiments can be run in identically configured computing environments.
Keywords: reproducibility, computational pathology, FAIR, cloud computing, machine learning, artificial intelligence
1. Introduction
Computational pathology (CompPath) is a new discipline that investigates the use of computational methods for the interpretation of heterogeneous data in clinical and anatomical pathology to improve health care in pathology practice. A major focus area of CompPath is the computerized analysis of digital tissue images [1]. These images show thin sections of surgical specimens or biopsies that are stained to highlight relevant tissue structures. To cope with the high level of complexity and variability of tissue images, virtually all state-of-the-art methods use sophisticated machine learning (ML) algorithms such as Convolutional Neural Networks (CNN) [2].
Because CompPath is applicable in a wide variety of use cases, there has been an explosion of research on ML-based tissue analysis methods [3, 4]. Many methods are intended to assist pathologists in routine diagnostic tasks such as the recognition of tissue patterns for disease classification [5–9]. Beyond that, CompPath methods have also shown promise for deriving novel biomarkers from tissue patterns that can predict outcome, genetic mutations, or therapy response [3].
1.1. Reproducibility challenges
In recent years, it has become increasingly clear that reproducing the results of published ML studies is challenging [10–13]. Reproducibility is commonly defined as the ability to obtain “consistent results using the same input data, computational steps, methods, and conditions of analysis” [14]. Difficulties related to reproducibility prevent other researchers from verifying and reusing published results and are a critical barrier to translating solutions into clinical practice [15]. In most cases, reproducibility problems seem to stem not from a lack of scientific rigor, but from challenges to convey all details and set-up of complex ML methods [12, 15, 16]. In the following, we provide an overview of the main challenges related to ML reproducibility and the existing approaches to address them.
The first challenge is the specification of the analysis method itself. ML algorithms have many variables, such as the network architecture, hyperparameters, and performance metrics [16–18]. ML workflows usually consist of multiple processing steps, e.g., data selection, preprocessing, training, evaluation [18]. Small variations in these implementation details can have significant effects on performance. To make all these details transparent, it is crucial to publish the underlying source code [15]. Workflows should be automated as much as possible to avoid errors when performing steps manually. Particular care must be taken to ensure that all operations are deterministic, e.g., by seeding pseudo-random operations, fixing initial network weights and controlling parallelism [13]. Jupyter notebooks have emerged as the de facto standard to implement and communicate ML workflows [19]. By combining software code, intermediate results and explanatory texts into “computational narratives” [20] that can be interactively run and validated, notebooks make it easier for researchers to reproduce and understand the work of others [19].
The second challenge to reproducibility is the specification and setup of the computing environment. ML workflows require significant computational resources including, e.g., graphics or tensor processing units (GPUs or TPUs). In addition, they often have many dependencies on specific software or library versions. Minor variations in the computing environment can significantly affect the results [13]. Setting up a consistent computational environment can be very expensive and time consuming. This challenge can be partially solved by embedding ML workflows in virtual machines or software containers like Docker [21]. Both include all required software dependencies so that ML workflows can be shared and run without additional installation effort. Cloud ML services, like Google Vertex AI, Amazon SageMaker, or Microsoft Azure Machine Learning, provide an even more comprehensive solution. By offering preconfigured computing environments for ML research in combination with the required highperformance hardware, such services can further reduce the setup effort and enable the reproduction of computationally intensive ML workflows even if one does not own the required hardware. They also typically provide web-based graphical user interfaces through which Jupyter notebooks can be run and shared directly in the cloud, making it easy for others to reproduce, verify, and reuse ML workflows [21].
The third challenge related to ML reproducibility is the specification of data and its accessibility. The performance of ML methods depends heavily on the composition of their training, validation and test sets [13, 22]. For current ML studies, it is rarely possible to reproduce this composition exactly as studies are commonly based on specific, hand-curated datasets which are only roughly described rather than explicitly defined [17, 23]. Also, the datasets are often not made publicly available [15], or the criteria/identifiers used to select subsets from publicly available datasets are missing. Stakeholders from academia and industry have defined the Findability, Accessibility, Interoperability, and Reusability (FAIR) principles [24], a set of requirements to facilitate discovery and reuse of data. FAIR data provision is now considered a “must” to make ML studies reproducible and the FAIR principles are adopted by more and more public data infrastructure initiatives and scientific journals [25].
Reproducing CompPath studies is particularly challenging. To reveal fine cellular details, tissue sections are imaged at microscopic resolution, resulting in gigapixel whole-slide images (WSI) [26]. Due to the complexity and variability of tissue images [27], it takes many—often thousands—of example WSI to develop and test reliable ML models. Processing and managing such large amounts of data requires extensive computing power, storage resources, and network bandwidth. Reproduction of CompPath studies is further complicated by the large number of proprietary and incompatible WSI file formats that often impede data access and make it difficult to combine heterogeneous data from different studies or sites. The Digital Imaging and Communications in Medicine (DICOM) standard [28] is an internationally accepted standard for storage and communication of medical images. It is universally used in radiology and other medical disciplines, and has great potential to become the uniform standard for pathology images as well [29]. However, until now, there have been few pathology data collections provided in DICOM format.
1.2. NCI Imaging Data Commons
The National Cancer Institute (NCI) Imaging Data Commons (IDC) is a new cloud-based repository within the US national Cancer Research Data Commons (CRDC) [30]. A central goal of the IDC is to improve the reproducibility of data-driven cancer imaging research. For this purpose, the IDC provides large public cancer image collections according to the FAIR principles.
Besides pathology images (brightfield and fluorescence) and their metadata, the IDC includes radiology images (e.g., CT, MR, and PET) together with associated image analysis results, image annotations, and clinical data providing context about the images. At the time of writing this article, the IDC contained 128 data collections with more than 63,000 cases and more than 38,000 WSI from different projects and sites. The collections cover common tumor types, including carcinomas of the breast, colon, kidney, lung, and prostate, as well as rarer cancers such as sarcomas or lymphomas. Most of the WSI collections originate from The Cancer Genome Atlas (TCGA) [31] and Clinical Proteomic Tumor Analysis Consortium (CPTAC) [32] projects and were curated by The Cancer Imaging Archive (TCIA) [33]. These collections are commonly used in the development of CompPath methods [7, 34–36].
The IDC implements the FAIR principles as follows:
Interoperability:
While the original WSIs were provided in proprietary, vendor-specific formats, the IDC harmonized the data and converted them into the open, standard DICOM format [29]. DICOM defines data models and services for storage and communication of medical image data and metadata, as well as attributes for different real-world entities (e.g., patient, study) and controlled terminologies for their values. In DICOM, a WSI corresponds to a “series” of DICOM image objects that represent the digital slide at different resolutions. Image metadata are stored as attributes directly within the DICOM objects.
Accessibility:
The IDC is implemented on the Google Cloud Platform (GCP), enabling cohort selection and analysis directly in the cloud. Since IDC data are provided as part of the Google Public Datasets Program, it can be freely accessed from cloud or local computing environments. In the IDC, DICOM objects are stored as individual DICOM files in Google Cloud Storage (GCS) buckets and can be retrieved using open, free, and universally implementable tools.
Findability:
Each DICOM file in the IDC has a persistent universally unique identifier (UUID) [37]. DICOM files in storage buckets are referenced through GCS URLs, consisting of the bucket URL and the UUID of the file. Images in the IDC are described with rich metadata, including patient (e.g., age, sex), disease (e.g., subtype, stage), study (e.g., therapy, outcome), and imaging-related data (e.g., specimen handling, scanning). All DICOM and non-DICOM metadata are indexed in a BigQuery database [38] that can be queried programmatically using standard Structured Query Language (SQL) statements (see section “IDC data access”), allowing for an exact and persistent definition of cohorts for subsequent analysis.
Reusability:
All image collections are associated with detailed provenance information but stripped of patient-identifiable information. Most collections are released under data usage licenses that allow unrestricted use in research studies.
1.3. Objective
This paper explores how the IDC and cloud ML services can be used in combination for CompPath studies and how this can facilitate reproducibility. This paper is also intended as an introduction to how the IDC can be used for reproducible CompPath research. Therefore, important aspects such as data access are described in more detail in the Methods section.
2. Methods
2.1. Overview
We implemented two CompPath experiments using data collections from the IDC and common ML services (Figure 1). Since the computing environments provided by cloud ML services are all virtualized, two identically configured instances may run different host hardware and software (e.g., system software versions, compiler settings) [13]. To investigate if and how this affects reproducibility, both experiments were executed multiple times, each in a new instance of the respective ML service.
Figure 1:
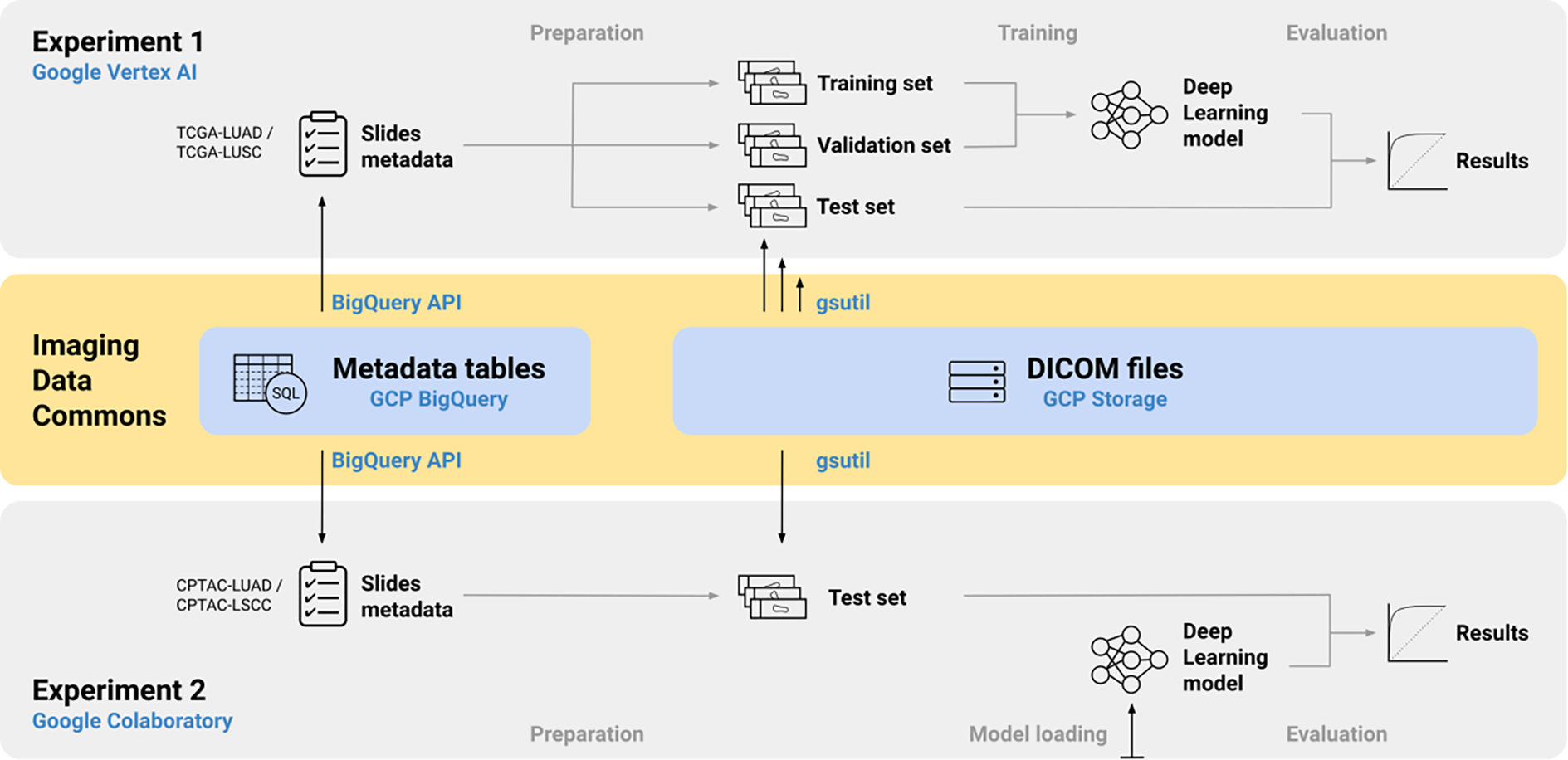
Overview of the workflows of both experiments and their interactions with the IDC.
The experiments are based on a basic CompPath analysis method that addresses a use case representative of common CompPath tasks [5–9]: the automatic classification of entire WSI of hematoxylin and eosin (H&E)-stained lung tissue sections into either non-neoplastic (normal), lung adenocarcinoma (LUAD), or lung squamous cell carcinoma (LSCC/LUSC).
Experiment 1 replays the entire development process of the method, including model training and validation. Experiment 2 performs inference with a trained model on independent data. The model trained in Experiment 1 was used as the basis for Experiment 2. The two experiments were conducted with different collections in the IDC: TCGA-LUAD/LUSC [39, 40] and CPTAC-LUAD/LSCC [41, 42], respectively. While both the TCGA and the CPTAC collections cover H&E-stained lung tissue sections of the three classes considered (Figure 2), they were created by different clinical institutions using different slide preparation techniques.
Figure 2:
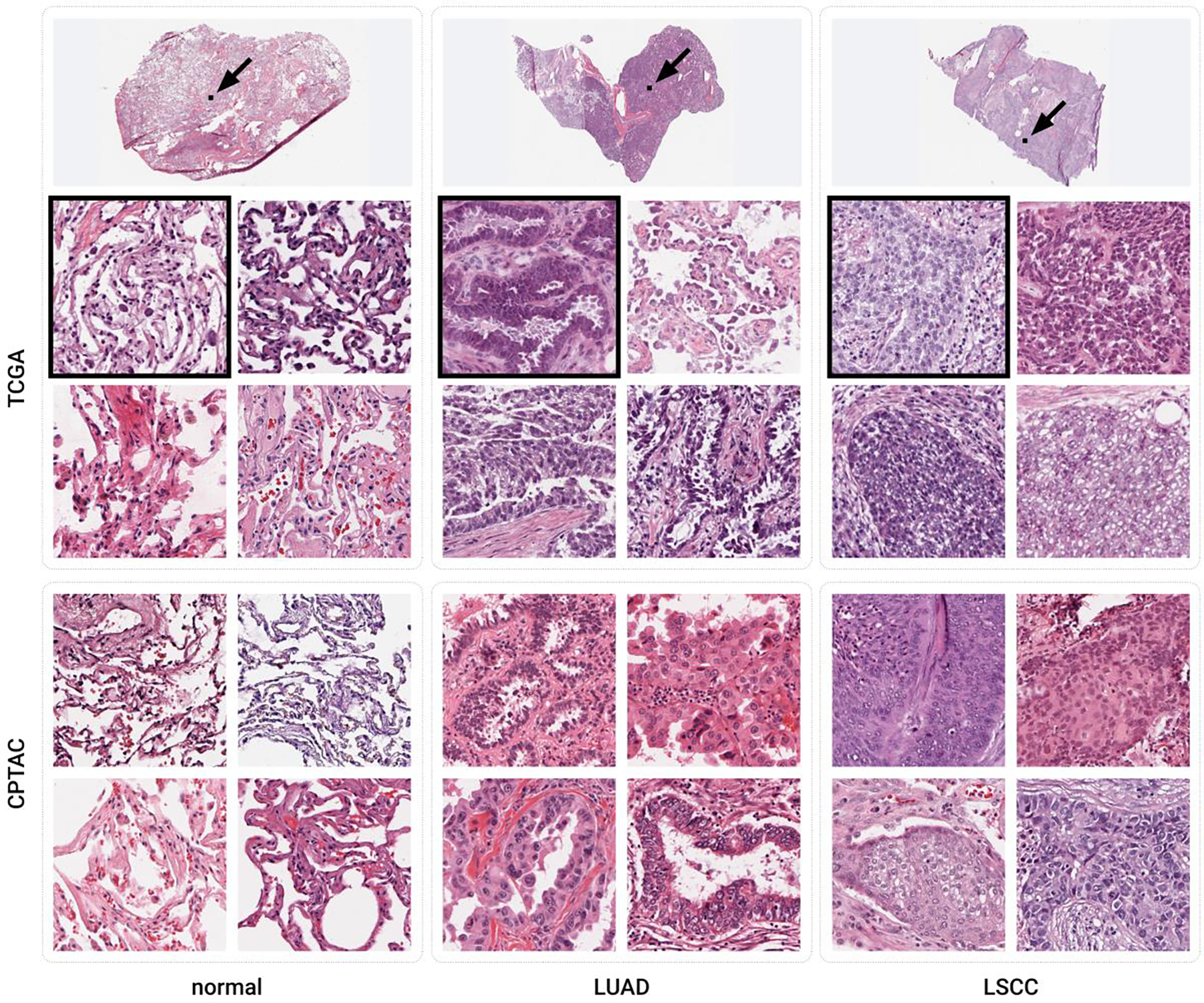
Example tiles of the three classes considered from the TCGA and CPTAC datasets. The width of each tile is 256 μm. The black boxes marked with arrows in the whole slide images on top show the boundaries of the upper left tiles of the TCGA data set.
2.2. Implementation
Both experiments were implemented as standalone Jupyter notebooks that are available open source [43]. We followed best practices to make operations deterministic [44], e.g., by seeding pseudo-random operations, fixing initial network weights between runs, requesting the use of deterministic algorithms within the TensorFlow framework, and by iterating over unordered container types in a defined order. Library versions were specified as part of the Cloud ML service configuration or explicitly specified in the notebooks. Utility functionality was designed as generic classes and functions that can be reused for similar use cases.
As the analysis method itself is not the focus of this paper, we adopted the algorithmic steps and evaluation design of a lung tumor classification method described in a widely cited study by Coudray et al. [7]. Although more modern approaches exist [45], this method was chosen because it is representative of proven CompPath applications and easy to understand. Our implementation processed images at a lower resolution, which is significantly less computationally expensive.
In our analysis workflow, a WSI was subdivided into non-overlapping rectangular tiles, each measuring 256 pixels at a resolution of 1 μm/px. Tiles containing less than 50% tissue, as determined by pixel value statistics, were discarded. Each tile was assigned class probabilities by performing multi-class classification using an InceptionV3 CNN [46] pretrained on ImageNet [47]. The per-tile results were finally aggregated to a single classification of the entire slide. The workflow is visualized in Figure 3 and a detailed description of the approach and hyperparameters is provided in the respective notebooks.
Figure 3:
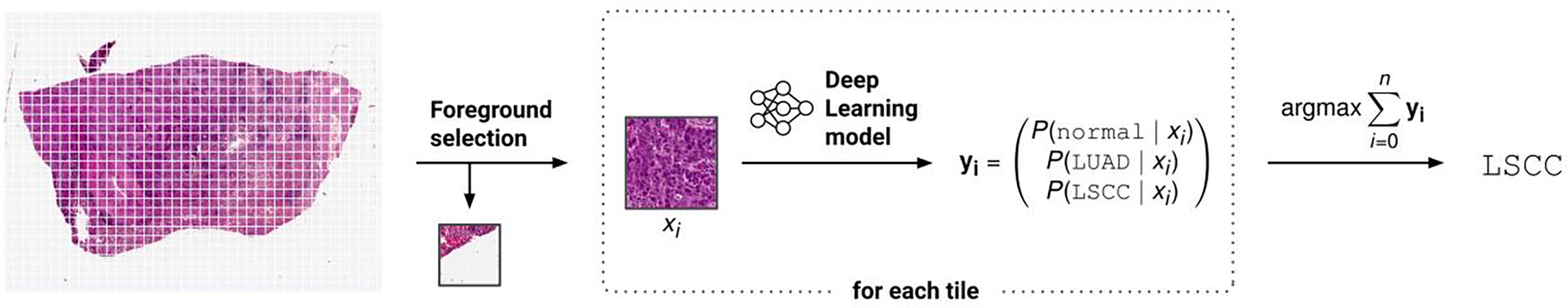
Illustration of the CompPath analysis method. Slides were subdivided into non-overlapping rectangular tiles discarding those with more background than tissue. Each tile was assigned class probabilities using a neural network performing multi-class classification. Slide-based class values were determined by aggregating the tile-based results.
In Experiment 1, the considered slides were divided into training, validation, and test sets with proportions of 70%, 15%, and 15%, respectively. The respective subsets were exactly the same in each run of the notebook. To keep the sets independent and avoid overoptimistic performance estimates [48], slides from a given patient were assigned to only one set, which resulted in 705, 151 and 153 patients per subset. The data collections used did not contain annotations of tumor regions, but only one reference class value per WSI. Following the procedure used by Coudray et al., all tiles were considered to belong to the reference class of their respective slide. Training was performed using a categorical cross-entropy loss between the true class labels and the predicted class probabilities, and the RMSProp optimizer with minimal adjustments to the default hyperparameter values [49]. The epoch with the highest area under the receiver operating characteristic (ROC) curve (AUC) on the validation set was chosen for the final model.
2.3. IDC data access
For most CompPath studies, one of the first steps is to select relevant slides using appropriate metadata. In the original data collections, parts of the metadata were stored in the image files and other parts in separate files of different formats (e.g., CSV, JSON files). In order to select relevant slides, the image and metadata first had to be downloaded in their entirety and then the metadata had to be processed using custom tools. With the IDC, data selection can be done by filtering a rich set of DICOM attributes with standard BigQuery SQL statements (Figure 4). The results are tables in which rows represent DICOM files and columns represent selected metadata attributes. As this facilitates the accurate and reproducible definition of the data subsets used in the analysis, these statements are described in more detail below.
Figure 4:
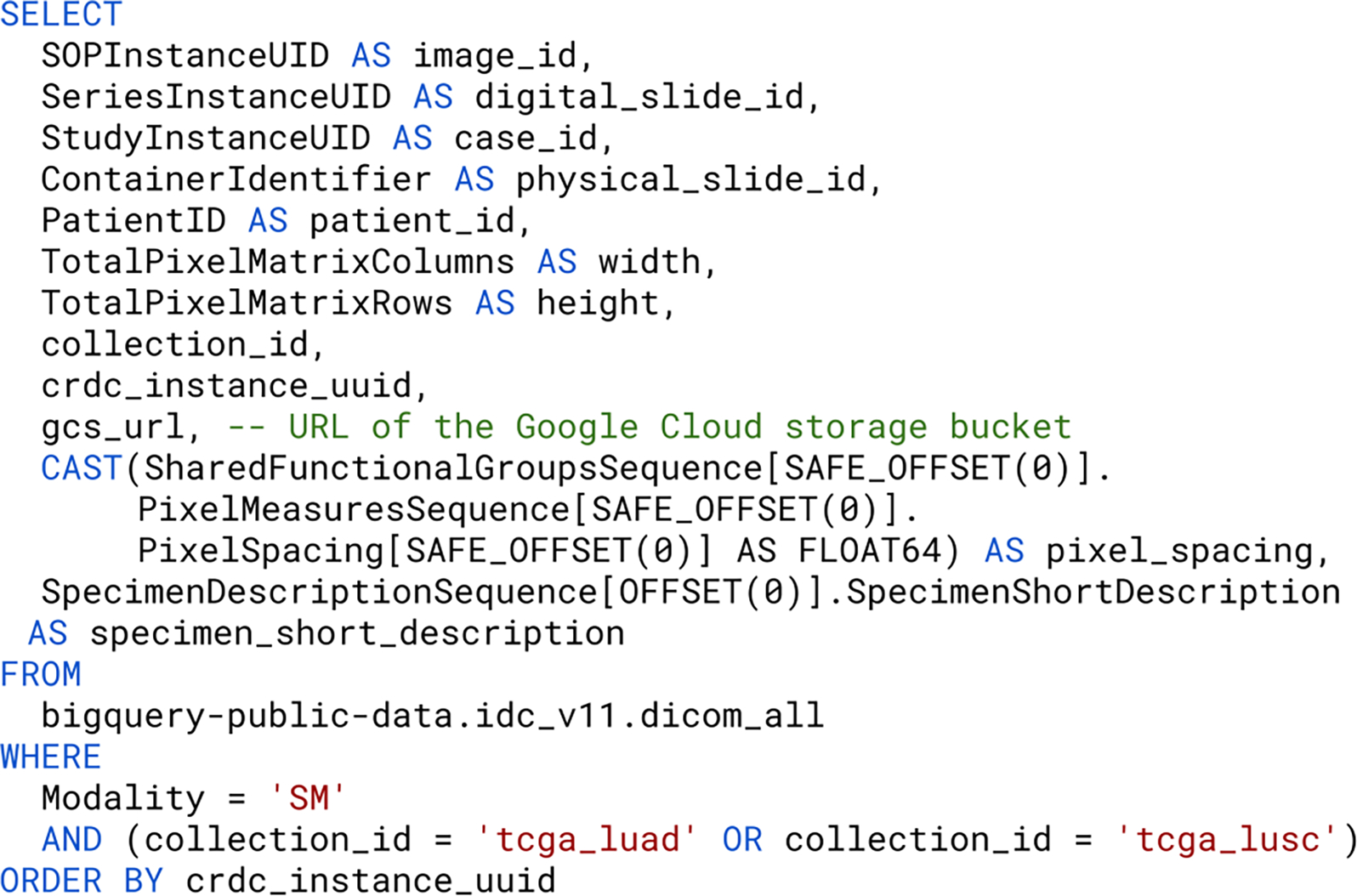
Generic example of a BigQuery SQL statement for compiling slide metadata. The result set is limited to slide microscopy images, as indicated by the value “SM” of the DICOM attribute “Modality”, from the collections “TCGA-LUAD” and “TCGA-LUSC”.
An SQL query for selecting WSI in the IDC generally consists of at least a SELECT, a FROM and a WHERE clause. The SELECT clause specifies the metadata attributes to be returned. The IDC provides a wealth of metadata attributes, including image-, patient-, disease-, and study-level properties. The attribute “gcs_url” is usually selected because it stores the GCS URL needed to access the DICOM file. The FROM clause refers to a central table “dicom_all” which summarizes all DICOM attributes of all DICOM files. This table can be joined with other tables containing additional project-specific metadata. Crucial to reproducibility is that all IDC data are versioned: Each new release of the IDC is represented as a new BigQuery dataset, keeping the metadata for the previous release and the corresponding DICOM files accessible even if they are modified in the new release. The version to use is specified via the dataset specifier in fully qualified table names. All experiments in this manuscript were conducted against IDC data version 11, i.e., the BigQuery table “bigquery-public-data.idc_v11.dicom_all”. The WHERE clause defines which DICOM files are returned by imposing constraints for certain metadata attributes. To guarantee reproducibility, it is essential to not use SQL statements that are non-deterministic (e.g., those that utilize ANY_VALUE) and conclude the statement with an ORDER BY clause, which ensures that results are returned in a sorted order.
The two experiments considered in this paper also begin with the execution of a BigQuery SQL statement to select appropriate slides and required metadata from the IDC. A detailed description of the statements is given in the respective notebooks. Experiment 1 queries specific H&E-stained tissue slides from the TCGA-LUAD/LUSC collections, resulting in 2163 slides (591 normal, 819 LUAD, 753 LSCC). Experiment 2 uses a very similar statement to query the slides from the CPTAC-LUAD/LSCC collections, resulting in 2086 slides (743 normal, 681 LUAD, 662 LSCC).
Once their GCS URLs are known, the selected DICOM files in the IDC can be accessed efficiently using the open source tool “gsutil” [50] or any other tool that supports the Simple Storage Service (S3) API. During training in Experiment 1, image tiles of different WSI had to be accessed repeatedly in random order. To speed up this process, all considered slides were preprocessed and the resulting tiles were extracted from the DICOM files and cached as individual PNG files on disk before training. In contrast, simply applying the ML method in Experiment 2 required only a single pass over the tiles of each WSI in sequential order. Therefore, it was feasible to access the respective DICOM files and iterate over individual tiles at the time they were needed for the application of the ML method.
2.4. Cloud ML services
The two experiments were conducted with two different cloud ML services of the GCP—Vertex AI and Google Colaboratory. Both services offer virtual machines (VMs) preconfigured with common ML libraries and a JupyterLab-like interface that allows editing and running notebooks from the browser. They are both backed with extensive computing resources including state-of-the-art GPUs or TPUs. The costs of both services scale with the type and duration of use for the utilized compute and storage resources. To use any of them with the IDC, a custom Google Cloud project must be in place for secure authentication and billing, if applicable.
Since training an ML model is much more computationally intensive than performing inference, we conducted Experiment 1 with Vertex AI and Experiment 2 with Google Colaboratory. Vertex AI can be attached to efficient disks for storage of large amounts of input and output data, making it more suitable for memory-intensive and long-running experiments. Colaboratory, on the other hand, offers several less expensive payment plans, with limitations in the provided computing resources and guaranteed continuous usage times. Colaboratory can even be used completely free of charge, with a significantly limited guaranteed GPU usage time (12 hours at the time of writing). This makes Colaboratory better suited for smaller experiments or exploratory research.
2.5. Evaluation
Experiment 1 was performed using a common Vertex AI VM configuration (8 vCPU, 30 GB memory, NVIDIA T4 GPU, Tensorflow Enterprise 2.8 distribution). Experiment 2 was performed with Colaboratory runtimes (2–8 vCPU, 12–30 GB memory). When using Google Colaboratory for Experiment 2, we were able to choose between different GPU types, including NVIDIA T4 and NVIDIA P100 GPUs. Since it has been suggested that the particular type of GPU can affect results [51], all runs of Experiment 2 were repeated on both GPUs, respectively. Runs with NVIDIA T4 were performed with the free version of Colaboratory, while runs with NVIDIA P100 were performed in combination with a paid GCE Marketplace VM, which was necessary for guaranteed use of this GPU.
For each run of an experiment, classification accuracy was assessed in terms of class-specific, one vs. rest AUC values based on the slide-level results. In addition, 95% confidence intervals (CI) of the AUC values were computed by 1000-fold bootstrapping over the slide-level results.
To speed up Experiment 2, only a random subset of 300 of the selected slides (100 normal, 100 LUAD, 100 LSCC) was considered in the analysis, which was approximately the size of the test set in Experiment 1.
3. Results
The evaluation results of both experiments are summarized in Table 1. In Experiment 1, 9 out of 10 runs produced identical results. However, in one run, class-specific AUC values deviated between 0.004 and 0.061. The results of Experiment 2 were identical in 14 out of 20 runs, regardless of the GPU type used. The remaining runs had deviations in AUC values of up to 0.001. These deviations occurred more frequently with the free version of Colaboratory (T4) than with the GCE Marketplace VM (P100). In both experiments, the CIs varied in a similar order of magnitude as the respective class-specific AUC values.
Table 1:
Class-specific, slide-based AUC values and 95% confidence intervals (CI) obtained through multiple runs of both experiments. Deviations are indicated in bold.
| Experiment | ML Service (GPU) | Run | normal AUC [CI] | LUAD AUC [CI] | LSCC AUC [CI] |
|---|---|---|---|---|---|
|
| |||||
| Experiment 1 | Vertex AI (T4) | 1 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] |
| 2 | 0.99014 [ 0.98125 , 0.99651 ] | 0 87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 3 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 4 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 5 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 6 | 0.99442 [0.98841, 0.99876] | 0.93344 [0.90058, 0.96134] | 0.92875 [0.89293, 0.95844] | ||
| 7 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 8 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 9 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| 10 | 0.99014 [ 0.98125 , 0.99651 ] | 0.87259 [ 0.82324 , 0.91476 ] | 0.90489 [ 0.85869 , 0.94591 ] | ||
| Experiment 2 | Colaboratory Free (T4) | 1 | 0.81100 [0.75148, 0.87246] | 0.69780 [0.63447, 0.75574] | 0.85050 [0.80244, 0.89452] |
| 2 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 3 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 4 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 5 | 0.81095 [0.74802, 0.86944] | 0.69775 [0.63239, 0.75791] | 0.85055 [0.80178, 0.89628] | ||
| 6 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 7 | 0.81130 [0.74908, 0.87176] | 0.69680 [0.63242, 0.75899] | 0.85035 [0.80094, 0.89642] | ||
| 8 | 0.81100 [0.75148, 0.87246] | 0.69780 [0.63447, 0.75574] | 0.85050 [0.80244, 0.89452] | ||
| 9 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 10 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| Experiment 2 | Colaboratory GCE (P100) | 1 | 0.81000 [0.74686, 0.86909] | 0.69660 [0.63062, 0.76014] | 0.85055 [0.80115, 0.89595] |
| 2 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 3 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 4 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 5 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 6 | 0.81005 [0.74948, 0.87058] | 0.69665 [0.63308, 0.75578] | 0.85055 [0.80213, 0.89457] | ||
| 7 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 8 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 9 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
| 10 | 0.81095 [ 0.74732 , 0.87279 ] | 0.69795 [ 0.62736 , 0.75974 ] | 0.85050 [ 0.80170 , 0.89663 ] | ||
The classification accuracy of the method trained in Experiment 1 appears satisfactory when evaluated on the TCGA test set but is somewhat inferior compared to the results of a similar study based on the same TCGA collections [7]. When applied to the CPTAC test set in Experiment 2, the same model performed substantially worse (Figure 5).
Figure 5:
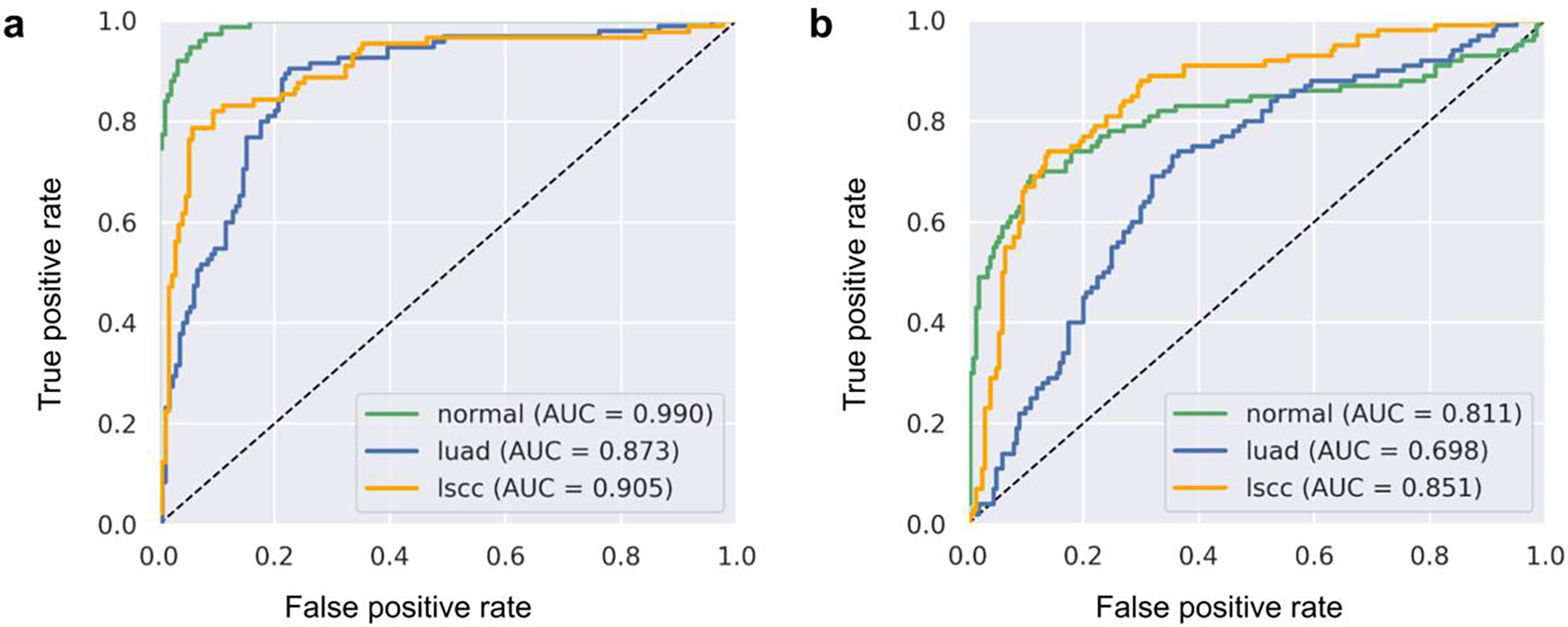
One-vs-rest ROC curves for the multi-class classification as obtained in (a) the first run of Experiment 1 using Vertex AI and (b) the second run of Experiment 2 using Colaboratory (T4).
Experiment 1 took an order of magnitude longer to complete (mean runtime of 2 d 9 h ±13 h) than Experiment 2 (mean runtime of 1 h 43min ±29 min with NVIDIA T4 and mean runtime of 1 h 19 min ±10 min with NVIDIA P100). The ML service usage charges for Experiment 1 were approximately US$ 34 per run. With the free version of Colaboratory, Experiment 2 was performed at no cost, while runs with the GCE Marketplace VM cost approximately US$ 2 per run.
4. Discussion
As described in the Introduction section, three challenges need to be addressed in combination to maximize the reproducibility of ML studies. To address the first two challenges, namely specifying the analysis method and the computing environment, we implemented the experiments as open source Jupyter notebooks, ensuring that operations were deterministic, and we ran the experiments using identically configured cloud ML services. We used the IDC to address the third challenge: specifying and accessing the data.
We found that both experiments were reproducible to a large extent. In 9 out of 10 runs of Experiment 1, the results were exactly the same. In all runs of Experiment 2, the AUC values were identical when rounded to the second decimal place, which should be sufficient for benchmarking ML results. As the individual runs of the experiments were conducted in different, newly created instances of ML services, other researchers should be able to reproduce them in the same way.
Nevertheless, both experiments were not perfectly reproducible and there were small deviations in the results. By calculating checksums, we could verify that image and metadata were always successfully retrieved from the IDC and that the respective tiles used for training, validation or testing were identical in all runs of the same experiment. Therefore, we assume that challenge 3 has been successfully met and that the deviations are due to challenge 1 or 2.
We followed best practices to make our code deterministic (see section “Implementation”). However, it can be extremely difficult to achieve true determinism [13], and we cannot rule out the possibility that there are still non-deterministic operations in our code or the libraries used.
Strikingly, the deviations were only occasional. Therefore, we consider it likely that they are due to differences in the computing environment. We ensured that the respective ML services used in Experiments 1 and 2 were always configured identically. However, their environments are all virtualized and the underlying host hardware and software is typically not within the user’s control. As this is known to affect results [13], there appears to be a practical limit to reproducibility when using cloud-based ML services. The free version of Colaboratory offers less control over the computing environment than Vertex AI or GCE Marketplace VMs, which may explain why the deviations occurred more frequently there. It should be noted that in practice it is rarely possible to use exactly the same host hardware and software as other researchers. Therefore, we expect the reproducibility limit to also apply to other computing environments, both in the cloud and locally.
4.1. Advantages of the IDC
The IDC helped us overcome the third reproducibility challenge with regard to the special requirements of CompPath. By providing imaging data collections according to the FAIR principles, the IDC supported the precise definition of the datasets used in the analysis and ensured that the exact same data can be reused in follow-up studies. The IDC also facilitated the use of cloud ML services by making terabytes of WSI data efficiently accessible in the cloud. We consider our experiments to be representative of common CompPath applications and believe that the IDC can similarly support the reproducibility of other CompPath studies.
While other repositories provide whole-slide images in vendor proprietary file formats, all data collections in the IDC are uniformly represented in the open DICOM format. This greatly simplifies data access using open software tools. When using images from other repositories, important metadata on image acquisition and processing is often represented in some textual or structured form and managed separately from the images, if it is available at all. For the data collections in the IDC, such metadata can be included as standardized DICOM attributes alongside the pixel data, so that the risk of data confusion is greatly reduced.
It is common practice to define the datasets used in ML studies as a set of image references stored, for example, as CSV files. To enable the reuse of the datasets, it must be ensured that the references remain valid in the long term. As described in section “IDC data access”, the IDC makes it possible to define datasets as BigQuery statements. The returned image references are always guaranteed to be valid, and by specifying an IDC version identifier, it can be ensured that the result set remains the same. BigQuery statements also make it clear what criteria or constraints were used to compile a dataset and can be easily adapted to newer versions of the IDC, for instance to assess reproducibility on extended datasets.
The results of Experiment 2 also reveal the transferability of the model trained in Experiment 1 to independent data. Although the majority of slides were correctly classified, AUC values were significantly lower, indicating that the model is only transferable to a limited extent and additional training is needed. Since all IDC data collections (both the image pixel data and the associated metadata) are harmonized into a standardized DICOM representation, testing transferability to a different dataset required only minor adjustments to our BigQuery SQL statement. In the same way, the IDC makes it straightforward to use multiple datasets in one experiment or to transfer an experimental design to other applications.
4.2. Limitations
Using cloud ML services comes with certain trade-offs. Conducting computationally intensive experiments requires setting up a payment account and paying a fee based on the type and duration of the computing resources used. Furthermore, although the ML services are widely used and likely to be supported for at least the next few years, there is no guarantee that they will be supported in the long term and support the specific configuration of the computing environment used (e.g., software version, libraries). Those who do not want to make these compromises can also access IDC data collections without using ML services, both in the cloud and locally. Even if this means losing the previously mentioned advantages with regard to the first two reproducibility challenges, the IDC can still help to specify the data used in a clear and reproducible manner.
Independent of the implementation, a major obstacle to the reproducibility of CompPath methods remains their high computational cost. A full training run can take up to several days, making reproduction by other scientists tedious. Performing model inference is generally faster and less resource intensive when compared to model training. Therefore, Experiment 2 runs well even with the free version of Google Colaboratory, enabling others to reproduce it without spending money. The notebook also provides a demo mode, which completes in a few minutes, so anyone can easily experiment with applying the inference workflow to arbitrary images from IDC.
At the moment, the IDC exclusively hosts public data collections. New data must undergo rigorous curation to de-identify (done by TCIA or data submitter) and harmonize images into standard representation (done by IDC), which can require a significant effort. Therefore, only data collections that are of general relevance and high quality are included in the IDC. As a result, the data in the IDC were usually acquired for other purposes than a particular CompPath application and cannot be guaranteed to be representative and free of bias [52]. Compiling truly representative CompPath datasets is very challenging [48]. Nevertheless, the data collections in the IDC can provide a reasonable basis for exploring and prototyping CompPath methods.
4.3. Outlook
The IDC is under continuous development and its technical basis is constantly being refined, e.g., to support new data types or to facilitate data selection and access. Currently, DICOM files in the IDC can only be accessed as a whole from their respective storage buckets. This introduces unnecessary overhead when only certain regions of a slide need to be processed, and it may make it necessary to temporarily cache slides to efficiently access multiple image regions (see section “IDC data access”). Future work should therefore aim to provide efficient random access to individual regions within a WSI. For maximum portability, such access should ideally be possible via standard DICOM network protocols such as DICOMweb [29, 53].
The IDC is continuously being expanded to support even more diverse CompPath applications. For instance, images collected by the Human Tumor Atlas Network (HTAN) that provide rich, multispectral information on subcellular processes [54] have recently been added. The IDC is integrated with other components of the CRDC, such as the Genomic Data Commons [55] or the Proteomic Data Commons [56]. This opens up many more potential CompPath applications involving tissue images and different types of molecular cancer data [57].
4.4. Conclusion
We demonstrated how the IDC can facilitate the reproducibility of CompPath studies. Implementing future studies in a similar way can help other researchers and peer reviewers to understand, validate and advance the analysis approach.
Highlights.
The Imaging Data Commons (IDC) is a new repository of FAIR cancer image collections.
Introduction to using the IDC for reproducible research in computational pathology.
The IDC and cloud-based machine learning services facilitate reproducibility in complementary ways.
Evaluation results indicate a practical reproducibility limit.
Categorization of key reproducibility challenges of computational pathology studies.
Acknowledgements
The authors thank Lars Ole Schwen for advice on deterministic implementations of machine learning algorithms, Tim-Rasmus Kiehl for advice on tissue morphology, and Vamsi Krishna Thiriveedhi for advice on reproducible use of cloud services.
The results published here are in whole or part based upon data generated by the TCGA Research Network and the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC).
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Task Order No. HHSN26110071 under Contract No. HHSN261201500003l.
Footnotes
Declaration of Competing Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Louis DN, Feldman M, Carter AB, Dighe AS, Pfeifer JD, Bry L, et al. Computational pathology: A path ahead. Archives of Pathology & Laboratory Medicine. 2015;140:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. The Lancet Oncology. 2019;20:e253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: A new generation of clinical biomarkers. British Journal of Cancer. 2020;124:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui M, Zhang DY. Artificial intelligence and computational pathology. Laboratory Investigation. 2021;101:412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih NNC, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A deep learning approach for quantifying tumor extent. Scientific Reports. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Silva VWK, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nature Medicine. 2019;25:1301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from nonsmall cell lung cancer histopathology images using deep learning. Nature Medicine. 2018;24:1559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Chen H, Gan C, Lin H, Dou Q, Tsougenis E, et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Transactions on Cybernetics. 2020;50:3950–62. [DOI] [PubMed] [Google Scholar]
- 9.Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Scientific Reports. 2020;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fell C, Mohammadi M, Morrison D, Arandjelovic O, Caie P, Harris-Birtill D. Reproducibility of deep learning in digital pathology whole slide image analysis. PLOS Digital Health. 2022;1:e0000145. doi: 10.1371/journal.pdig.0000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutson M Artificial intelligence faces reproducibility crisis. Science. 2018;359:725–6. [DOI] [PubMed] [Google Scholar]
- 12.Raff E A step toward quantifying independently reproducible machine learning research. In: Wallach HM, Larochelle H, Beygelzimer A, d’Alché-Buc F, Fox EB, Garnett R, editors. Advances in neural information processing systems 32: Annual conference on neural information processing systems 2019, NeurIPS 2019, december 8–14, 2019, vancouver, BC, canada. 2019. p. 5486–96. https://proceedings.neurips.cc/paper/2019/hash/c429429bf1f2af051f2021dc92a8ebea-Abstract.html. [Google Scholar]
- 13.Gundersen OE, Shamsaliei S, Isdahl RJ. Do machine learning platforms provide out-of-the-box reproducibility? Future Generation Computer Systems. 2022;126:34–47. [Google Scholar]
- 14.National Academies of Sciences, Engineering, and Medicine. Reproducibility and replicability in science. Washington, DC: The National Academies Press; 2019. doi: 10.17226/25303. [DOI] [PubMed] [Google Scholar]
- 15.Haibe-Kains B, Adam GA, Hosny A, Khodakarami F, Shraddha T, Kusko R, et al. Transparency and reproducibility in artificial intelligence. Nature. 2020;586:E14–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pineau J, Vincent-Lamarre P, Sinha K, Lariviere V, Beygelzimer A, d’Alche-Buc F, et al. Improving reproducibility in machine learning research(a report from the NeurIPS 2019 reproducibility program). Journal of Machine Learning Research. 2021;22:1–20. http://jmlr.org/papers/v22/20-303.html. [Google Scholar]
- 17.Hartley M, Olsson TSG. dtoolAI: Reproducibility for deep learning. Patterns. 2020;1:100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renard F, Guedria S, Palma ND, Vuillerme N. Variability and reproducibility in deep learning for medical image segmentation. Scientific Reports. 2020;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkel JM. Why jupyter is data scientists’ computational notebook of choice. Nature. 2018;563:145–6. [DOI] [PubMed] [Google Scholar]
- 20.Rule A, Birmingham A, Zuniga C, Altintas I, Huang S-C, Knight R, et al. Ten simple rules for writing and sharing computational analyses in jupyter notebooks. PLOS Computational Biology. 2019;15:e1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkel JM. Make code accessible with these cloud services. Nature. 2019;575:247–8. [DOI] [PubMed] [Google Scholar]
- 22.Maier-Hein L, Eisenmann M, Reinke A, Onogur S, Stankovic M, Scholz P, et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nature Communications. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundersen OE, Kjensmo S. State of the art: Reproducibility in artificial intelligence. Proceedings of the AAAI Conference on Artificial Intelligence. 2018;32. [Google Scholar]
- 24.Wilkinson MD, Dumontier M, Aalbersberg IjJ, Appleton G, Axton M, Baak A, et al. The FAIR guiding principles for scientific data management and stewardship. Scientific Data. 2016;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheffler M, Aeschlimann M, Albrecht M, Bereau T, Bungartz H-J, Felser C, et al. FAIR data enabling new horizons for materials research. Nature. 2022;604:635–42. [DOI] [PubMed] [Google Scholar]
- 26.Patel A, Balis UGJ, Cheng J, Li Z, Lujan G, McClintock DS, et al. Contemporary whole slide imaging devices and their applications within the modern pathology department: A selected hardware review. Journal of Pathology Informatics. 2021;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCann MT, Ozolek JA, Castro CA, Parvin B, Kovacevic J. Automated histology analysis: Opportunities for signal processing. IEEE Signal Processing Magazine. 2015;32:78–87. [Google Scholar]
- 28.Bidgood WD, Horii SC, Prior FW, Syckle DEV. Understanding and using DICOM, the data interchange standard for biomedical imaging. Journal of the American Medical Informatics Association. 1997;4:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrmann MD, Clunie DA, Fedorov A, Doyle SW, Pieper S, Klepeis V, et al. Implementing the DICOM standard for digital pathology. Journal of Pathology Informatics. 2018;9:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedorov A, Longabaugh WJR, Pot D, Clunie DA, Pieper S, Aerts HJWL, et al. NCI imaging data commons. Cancer Research. 2021;81:4188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Cancer Genome Atlas Program. https://www.cancer.gov/tcga. Accessed 30 Jan 2023.
- 32.The National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium. https://proteomics.cancer.gov/programs/cptac. Accessed 30 Jan 2023. [Google Scholar]
- 33.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. Journal of Digital Imaging. 2013;26:1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Reports. 2018;23:181–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine. 2018;27:317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noorbakhsh J, Farahmand S, pour AF, Namburi S, Caruana D, Rimm D, et al. Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images. Nature Communications. 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leach P, Mealling M, Salz R. A universally unique IDentifier (UUID) URN namespace. RFC Editor; 2005. [Google Scholar]
- 38.Google Cloud - Cloud Healthcare API - Understanding the BigQuery DICOM schema. https://cloud.google.com/healthcare/docs/how-tos/dicom-bigquery-schema. Accessed 30 Jan 2023.
- 39.Albertina B, Watson M, Holback C, Jarosz R, Kirk S, Lee Y, et al. The Cancer Genome Atlas Lung Adenocarcinoma Collection (TCGA-LUAD). 2016. doi: 10.7937/K9/TCIA.2016.JGNIHEP5. [DOI] [Google Scholar]
- 40.Kirk S, Lee Y, Kumar P, Filippini J, Albertina B, Watson M, et al. The Cancer Genome Atlas Lung Squamous Cell Carcinoma Collection (TCGA-LUSC). 2016. doi: 10.7937/K9/TCIA.2016.TYGKKFMQ. [DOI] [Google Scholar]
- 41.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Lung Adenocarcinoma Collection (CPTAC-LUAD). 2018. doi: 10.7937/K9/TCIA.2018.PAT12TBS. [DOI] [Google Scholar]
- 42.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Lung Squamous Cell Carcinoma Collection (CPTAC-LSCC). 2018. doi: 10.7937/K9/TCIA.2018.6EMUB5L2. [DOI] [Google Scholar]
- 43.Classification of lung tumor slide images with the NCI Imaging Data Commons. https://github.com/ImagingDataCommons/idc-comppath-reproducibility.git. Accessed 15 Sep 2023. [Google Scholar]
- 44.TensorFlow Determinism. https://github.com/NVIDIA/framework-reproducibility/blob/master/doc/d9m/tensorflow.md. [Google Scholar]
- 45.Hosseini MS, Bejnordi BE, Trinh VQ-H, Hasan D, Li X, Kim T, et al. Computational pathology: A survey review and the way forward. 2023. https://arxiv.org/abs/2304.05482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. 2015. [Google Scholar]
- 47.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. Ieee; 2009. p. 248–55. [Google Scholar]
- 48.Homeyer A, Geißler C, Schwen LO, Zakrzewski F, Evans T, Strohmenger K, et al. Recommendations on compiling test datasets for evaluating artificial intelligence solutions in pathology. Modern Pathology. 2022;35:1759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.RMSprop class. https://keras.io/api/optimizers/rmsprop. Accessed 30 Jan 2023. [Google Scholar]
- 50.Gsutil tool. https://cloud.google.com/storage/docs/gsutil. Accessed 30 Jan 2023.
- 51.Nagarajan P, Warnell G, Stone P. Deterministic implementations for reproducibility in deep reinforcement learning. 2018. [Google Scholar]
- 52.Varoquaux G, Cheplygina V. Machine learning for medical imaging: Methodological failures and recommendations for the future. npj Digital Medicine. 2022;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DICOMweb. https://www.dicomstandard.org/using/dicomweb. Accessed 30 Jan 2023.
- 54.Rozenblatt-Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, et al. The human tumor atlas network: Charting tumor transitions across space and time at single-cell resolution. Cell. 2020;181:236–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. New England Journal of Medicine. 2016;375:1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Proteomic data commons. https://pdc.cancer.gov. Accessed 30 Jan 2023. [Google Scholar]
- 57.Schneider L, Laiouar-Pedari S, Kuntz S, Krieghoff-Henning E, Hekler A, Kather JN, et al. Integration of deep learning-based image analysis and genomic data in cancer pathology: A systematic review. European Journal of Cancer. 2022;160:80–91. [DOI] [PubMed] [Google Scholar]


