Abstract
Background:
Levocarnitine (L-carnitine) has shown promise as a metabolic-therapeutic for septic shock, where mortality approaches 40%. However, high-dose (≥ 6 grams) intravenous supplementation results in a broad range of serum concentrations.
Objectives:
We sought to describe the population pharmacokinetics (PK) of high-dose L-carnitine, test various estimates of kidney function, and assess the correlation of PK parameters with pre-treatment metabolites in describing drug response for patients with septic shock.
Methods:
We leveraged serum samples and metabolomics data from a phase II trial of L-carnitine in vasopressor-dependent septic shock. Patients were adaptively randomized to receive intravenous L-carnitine (6 grams, 12 grams, or 18 grams) or placebo. Serum was collected at baseline (T0); end-of-infusion (T12); and 24, 48, and 72 hours after treatment initiation. Population PK analysis was done with baseline normalized concentrations using nonlinear mixed effect models in the modeling platform Monolix. Various estimates of kidney function, patient demographics, dose received, and organ dysfunction were tested as population covariates.
Results:
The final dataset included 542 serum samples from 130 patients randomized to L-carnitine. A two-compartment model with linear elimination and a fixed volume of distribution (17.1 liters) best described the data and served as a base structural model. Kidney function estimates as a covariate on the elimination rate constant (k) reliably improved model fit. Estimated glomerular filtration rate (eGFR), based on the 2021 Chronic Kidney Disease Epidemiology collaboration (CKD-EPI) equation with creatinine and cystatin C, outperformed creatinine clearance (Cockcroft-Gault) and older CKD-EPI equations that use an adjustment for self-identified race.
Conclusions:
High-dose L-carnitine supplementation is well-described by a two-compartment population PK model in patients with septic shock. Kidney function estimates that leverage cystatin C provided superior model fit. Future investigations into high-dose L-carnitine supplementation should consider baseline metabolic status and dose adjustments based on renal function over a fixed or weight-based dosing paradigm.
Keywords: critical illness, precision medicine, drug dosing, metabolomics
1.2. Introduction
Sepsis is a clinical syndrome defined by life-threatening organ dysfunction and a dysregulated host response to infection.1 In 2017, nearly 50 million cases of sepsis were identified worldwide, and mortality in the most severe form, septic shock, approaches 40%.2 Beyond antimicrobials, treatment for sepsis remains largely non-specific and supportive with a litany of failed clinical trials for more targeted interventions.3
Sepsis pathophysiology is complex but is partly characterized by a hypermetabolic state and mitochondrial dysfunction, both of which are associated with greater mortality.4, 5 Given the lack of targeted metabolic pharmacotherapy, L-carnitine, an endogenous metabolite that serves a key bioenergetic role in the mobilization of fatty acids for mitochondrial beta-oxidation, was recently tested in patients with sepsis. In a phase I, randomized, double-blind trial, high-dose L-carnitine was found to be safe in 31 patients with septic shock and demonstrated a modest, but significant improvement in patient mortality versus placebo.6 A follow-up phase IIb trial did not find evidence that L-carnitine significantly improved patient mortality or organ dysfunction7, as measured by the Sequential Organ Failure Assessment (SOFA) score.8 However, pharmacometabolomic analyses of the phase I trial demonstrated significant interpatient variability in post-treatment L-carnitine concentrations that correlated with mortality.9, 10 Subsequent work showed that variations in the genetics of the organic cation transporter novel family member 2 (OCTN2), body size, and kidney function may also be important drivers of the observed variability and possibly, therapeutic response.11 Furthermore, a significant mortality benefit from supplemental L-carnitine was observed in the phase IIb trial in patients with elevated acylcarnitines including acetylcarnitine.12 Taken together, these findings suggest heterogeneity in the pharmacokinetics (PK) and effectiveness (pharmacodynamics, PD) of high-dose L-carnitine in septic shock.
The overall goal of our study was to construct a population PK model of high-dose, intravenous, L-carnitine in an acutely ill cohort of patients with septic shock to better understand the factors that drive L-carnitine blood concentration variability. Given that L-carnitine is extensively cleared by kidney elimination13, we recognized an additional opportunity to leverage trial data and contribute to the ongoing conversation regarding the ideal approach to estimate kidney function in critically ill patients. These patients are prone to acute kidney injury for which serum creatinine is not routinely a reliable measure of renal function.14 As such, we tested different equations to estimate kidney function based on serum creatinine (Scr), serum cystatin C (Scys), and self-identified race in critically ill patients using high-dose L-carnitine. In addition, we sought to determine if other widely available patient covariates improved the model’s predictions. As an exploratory aim, we assessed the relationship between individual patient PK parameters with baseline metabolic status and genomic variability in OCTN2.
1.3. Methods
1.3.1. Study design and participants
Our work was a secondary analysis of the Rapid Administration of Carnitine in Sepsis (RACE) clinical trial (NCT01665092).7 The RACE study was a multicenter, placebo-controlled, phase IIb clinical trial that adaptively randomized patients with septic shock to saline placebo or one of three dosing arms for intravenous L-carnitine: 6 grams, 12 grams, or 18 grams. The Bayesian adaptive randomization scheme selected the highest dose as the most efficacious.15 Study drug or an equivalent volume of saline placebo was given as an intravenous bolus (33% of dose) immediately followed by a 12-hour infusion. The trial was conducted in accordance with the Declaration of Helsinki, where all patients or their legal representatives provided informed consent and all sites were approved by their local Institutional Review Board.
Adult patients were eligible for the trial if they were: i) enrolled within 24-hours of the identification of septic shock; ii) required high-dose vasopressors; iii) presented with moderate organ dysfunction (SOFA ≥ 6); and iv) had a blood lactate of at least 18 mg/dL (2 mmol/L). Patients who were pregnant, breastfeeding, immunocompromised, or had a history of seizures were excluded. Serum samples for drug and other metabolomics analysis were collected at baseline (T0), end-of-infusion (T12), and 24 hours (T24), 48 hours (T48), and 72 hours (T72) after treatment initiation. Full inclusion and exclusion criteria, as well as detailed sample collection and processing have been previously described;7, 12, 16 some additional trial details can be found in the supporting information.
1.3.2. Drug and Metabolite Quantification
Carnitine and acylcarnitines
We used an existing metabolomics data set including time-series measurements of L-carnitine and pre-treatment (baseline) measurements of acylcarnitines. Analytes were measured in serum samples collected in the RACE trial by reverse phase, liquid-chromatography mass-spectrometry (LC-MS) at the Michigan Regional Comprehensive Metabolomics Resource Core at the University of Michigan as previously described.9, 12 Acylcarnitines are esters formed from the conjugation of L-carnitine and fatty acids of various carbon chain lengths.17 Absolute quantification for L-carnitine and several acylcarnitines (C2, C3, C4, C5, C8, C14, and C16) was achieved through stable isotope internal standards at a known concentration (NSK-B Cambridge Isotope Laboratories). An additional eight acylcarnitines were relatively quantified by peak area.
Small polar molecules
We also used an existing metabolomics data set of polar compounds that was acquired from pre-treatment serum samples using proton nuclear magnetic resonance spectroscopy (1H-NMR). This assay, which was conducted at the University of Michigan’s Biochemical Core, is detailed elsewhere12, 16, 18 and identified and quantified 27 low-molecular weight metabolites. Metabolites included several amino acids, intermediates of the tricarboxylic acid (TCA) cycle, and other bioenergetic compounds.
1.3.3. Kidney Function Estimates
Quantification of serum creatinine and cystatin C
Serum creatinine was measured clinically as part of the RACE study, and baseline measures were abstracted from the trial’s research electronic data capture (REDCap) database.19 Cystatin C was measured using biobanked residual serum samples using a standard, commercially available enzyme-linked immunoassay (ELISA) assay according to the manufacturer’s instructions (R & D Systems, Minneapolis, MN, catalog number DSCTC0).
Equations to estimate kidney function
The Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) has established equations to estimate glomerular filtration rate (eGFR) based on Scr and/or Scys, patient age, sex, and race. Given the increasing controversy about inclusion of patient race as a variable20 and the drawbacks of Scr as a kidney biomarker21, we estimated eGFR using four iterations of the CKD-EPI equation: the 2009 CKD-EPI equation22 (includes patient race and Scr); the 2021 CKD-EPI equation23 (uses Scr but drops patient race); the 2012 CKD-EPI equation 24 (uses Scys); and the 2021 CKD-EPI equation23 (includes both Scr and Scys without an adjustment for race). All eGFR calculations were calculated according to standard body surface area and are in the units of mL/min per 1.73 m2. We also tested the estimated creatinine clearance (CrCl) using the Cockcroft-Gault equation.25
1.3.4. Transporter Genotyping
L-carnitine is transported into the cell through the OCTN2 transporter, which is also responsible for its renal tubular reabsorption.13 Given L-carnitine’s critical role in metabolic homeostasis, loss of function variants in the gene encoding OCTN2 (SLC22A5) are rare and result in inborn errors of metabolism. Nonetheless, a single nuclear polymorphism (rs2631367, −207C>G) has been associated with lower mRNA levels in previous studies and in the Genotype-Tissue Expression (GTEx) Project.26–28 We isolated DNA from buffy coat collected in the RACE trial and genotyped patients at the rs2631367 loci using a commercially available TaqMan genotyping assay (ThermoFisher®, assay ID C__26479161_30).
1.3.5. Pharmacokinetic modeling
We restricted our secondary analysis to patients who were randomized to receive study drug and who had a baseline and at least one post-treatment serum sample available. For population PK analysis, post-treatment L-carnitine concentrations were baseline normalized in accordance with United States Food and Drug Administration guidance for modeling endogenous molecules.29 Baseline normalization was done on the individual level such that each post-treatment L-carnitine concentration was subtracted from the individual’s pre-treatment (baseline) measurement. Post-treatment concentrations below baseline were assigned a value of zero.
All data were cleaned in RStudio, and population PK analysis was performed in Monolix modeling platform (Version 2021R1, Lixoft SAS, Antony, France). Given the sparse sampling scheme of the RACE trial in relationship to the drug infusion time, we opted for a fixed population parameter for the volume of distribution (Vd) based on the median weight of the cohort and previous PK reports that Vd for intravenous L-carnitine is 0.2 to 0.3 L/kg.13
To determine the optimal structural PK model, we built a series of models with one, two, or three compartments and a linear elimination rate constant (k). We selected the model based on the Akaike information criterion (AIC) and model diagnostic plots. For the best performing structural model, we assessed the impact of different kidney function parameters as a covariate on the elimination rate constant. We tested the performance of eGFR as estimated by the various CKD-EPI equations described above; the CrCl according to Cockcroft-Gault; and Scr and Scys as standalone biomarkers. In addition, we considered the sarcopenia index, calculated as 100*( Scr / Scys ), which is biomarker of muscle mass rather than true kidney function.30
Next, we considered additional clinical and demographic patient variables as covariates using the available automated stepwise covariate model (SCM) building algorithm in Monolix. Patient demographics included age, sex, weight, and self-identified black race. We also considered the dose of L-carnitine received, organ dysfunction as measured by the SOFA score (with the kidney function score removed), and the sarcopenia index.
1.3.6. Statistical analysis of individual PK parameters
Once the final covariate model was selected, we explored the relationship between the predicted individual patient PK parameters and baseline metabolites, OCTN2 genotype, and patient mortality. Specifically, we computed the Spearman’s coefficient between individual’s predicted values for k, the rate constant out of compartment one (k12), and the rate constant out of compartment two (k21) and measured concentration of baseline acylcarnitines and small, polar metabolites measured by NMR. The correlation for comparisons were plotted for relationships with a p-value less than 0.05. We also compared the model predicted individual parameters stratified by OCTN2 genotype and 28-day patient mortality using the Kruskal–Wallis test and the Wilcoxon signed-rank test, respectively. All statistical analyses were performed using R Studio (RStudio: Integrated Development for R. RStudio, PBC, Boston, MA; http://www.rstudio.com/).
1.4. Results
1.4.1. Patients and pharmacokinetic data
Of the 175 patients randomized to receive L-carnitine in the RACE trial, 130 patients had a baseline and follow-up serum sample available for population PK analysis. In these patients, we measured drug concentrations in 542 serum samples. Observations at T12 and T72 were underrepresented (Table 1), as these samples were only collected during the initial ‘burn-in’ phase of the trial, where the first 40 patients were randomized equally to all trial arms.15 As such, 60% of the cohort in this secondary analysis was randomized to the 18-g treatment arm, as it was selected as the most efficacious by the Bayesian adaptive design.
Table 1.
Patient demographics and clinical characteristics of individuals included in population pharmacokinetic modeling.
| Patient characteristics | Total Patients, N = 1301 |
|---|---|
|
| |
| L-carnitine dose received | |
| 6 grams | 27 (21%) |
| 12 grams | 25 (19%) |
| 18 grams | 78 (60%) |
|
| |
| Sex (N) | |
| Female | 50 (38%) |
| Male | 80 (62%) |
|
| |
| Age (years) | 62 (53, 70) |
|
| |
| Weight (kg) | 85 (70, 102) |
|
| |
| Self-Identified Race | |
| Black | 41 (32%) |
| White, Asian, or Other | 89 (68%) |
|
| |
| Serum Creatinine (mg/dL) | 1.93 (1.29, 2.79) |
|
| |
| Serum Cystatin C (mg/L) | 2.44 (1.57, 3.66) |
|
| |
| Baseline SOFA Score (points) | 10.0 (8.0, 13.0) |
|
| |
| OCTN2 genotype at rs2631367 (N) | |
| CC | 23 (18%) |
| CG | 50 (38%) |
| GG | 37 (28%) |
| Unknown | 20 (15%) |
|
| |
| Serum samples analyzed for pharmacokinetics | |
| Baseline, T0 | 130 (100%) |
| End-of-infusion, T12 | 23 (18%) |
| 24-hours after treatment initiation, T24 | 127 (98%) |
| 48-hours after treatment initiation, T48 | 114 (88%) |
| 72-hours after treatment initiation, T72 | 18 (14%) |
Data shown as n (%); Median (IQR)
SOFA: Sequential Organ Failure Assessment score; OCTN2: Organic cation transporter novel family 2 (gene name: solute carrier family 22 member 5, SLC22A5)
Baseline Scr was available for all patients. Four patients did not have a sufficient volume of residual baseline serum to measure Scys, and values were imputed from a simple linear model using Scr and patient age, sex, and weight as predictors (Supplementary Figure 1). Buffy coat for DNA isolation and thus genetic information at the rs2631367 loci of the OCTN2 transporter was only available in a subset of the cohort (N=110, Table 1).
1.4.2. Population pharmacokinetic modeling
The two-compartment and three-compartment structural models provided significant improvements in model fit over the one-compartment model (Table 2, ΔAIC = −204.31 and −206.74 points, respectively). Although the three-compartment model could be considered a superior model based on AIC reduction alone, this model was plagued by high residual standard errors (R.S.E.) for both population parameters and random effects (Table 2). Thus, we opted to proceed with the simpler, more stable two-compartment structural model. Model diagnostic plots, including the observed versus predicted concentrations, the distribution of residuals, and the visual predictive check (VPC), are provided for the 2-compartment structural model in the supplement (Supplementary Figures 2A, 3A, and 4A).
Table 2. Comparison of structural pharmacokinetic models.
Both two- and three-compartment models provided substantial improvement in model performance based on the reduction in AIC. The simpler, two-compartment model was selected based on model diagnostic plots and the high residual standard errors in the three-compartment model.
| Model | 1-compartment linear elimination (base) | 2-compartment linear elimination | 3-compartment linear elimination |
|---|---|---|---|
| Model comparison | |||
| AIC | 2685.77 | 2481.46 | 2479.03 |
| ΔAIC | - | −204.31 | −206.74 |
| Fixed-effect parameters | |||
| V_pop (L) | 17.1 | 17.1 | 17.1 |
| k_pop (h−1) | 0.053 (8.18%) |
0.22 (6.27%) |
0.18 (23.7%) |
| k12_pop (h−1) | - | 0.55 (14.4%) |
0.84 (16.3%) |
| k21_pop (h−1) | - | 0.19 (11.8%) |
0.60 (45.6%) |
| k13_pop (h−1) | - | - | 0.13 (24.9%) |
| k31_pop (h−1) | - | - | 0.087 (77.0%) |
| Random-effect parameters | |||
| Standard deviation of inter-individual variability (IIV) | |||
| ωV | 2.1 (7.28%) |
0.48 (24.7%) |
0.28 (44.5%) |
| ωk | 0.75 (7.76%) |
0.52 (10.9%) |
0.57 (19.6%) |
| ωk12 | - | 0.24 (69.2%) |
0.31 (51.6%) |
| ωk21 | - | 0.48 (20.3%) |
0.54 (28.5%) |
| ωk13 | - | - | 0.49 (315%) |
| ωk31 | - | - | 2.25 (34%) |
| Residual variability (RV) | |||
| b | 0.36 (6.16%) |
0.36 (7.24%) |
0.34 (6.78%) |
CKD-EPI: Chronic Kidney Disease Epidemiology Collaboration; eGFR: estimated glomerular filtration rate using equations that leverage self-identified black race (race), serum creatinine (cr), and/or serum cystatin C (cys); AIC: Akaike information criteria; V_pop: fixed population parameter for volume of distribution; k_pop: fit population parameter for the elimination rate; k12_pop, k21_pop, k13_pop, k31_pop : fit population parameter for the rate into/out of compartments; ωV, ωk, ωk12, ωk21, ωk13, ωk31: standard deviation of random effects for population parameters; b: estimated value from proportional error model.
Kidney function as a covariate of the elimination rate constant reliably improved model fit regardless of the equation or biomarker used. Table 3 shows the impact on the AIC after including various kidney function estimates as a covariate on the elimination rate constant. The eGFRcr-cys, estimated according to the 2021 CKD-EPI equation using both Scr and Scys, provided the largest reduction in AIC (−48.74 points). The eGFRcr (2021 CKD-EPI using only Scr), eGFRcys (2012 CKD-EPI using only Scys), and CrCl provided a similar improvement over the base structural model (ΔAIC = −39.56, −40.9, and −39.68 points, respectively). A similar AIC reduction (−40.04 points) was seen for eGFRrace-cr (2009 CKD-EPI using both self-identified race and Scr) compared to eGFR estimates without the race factor. Inclusion of Scys concentration as a covariate of the elimination rate (Δ AIC = −37.42 points) outperformed Scr (Δ AIC = −31.71 points) as a single kidney function biomarker. The sarcopenia index, a measure of muscle loss rather than true kidney function, provided a worse model fit compared to the structural base model (Δ AIC = +3.29 points).
Table 3. Population pharmacokinetic models with different renal function estimates as a covariate of the elimination rate, k.
Results shown represent the population parameter estimate and residual standard error (%).
| Model | Structural Model | 2009 CKD-EPI eGFRrace-cr | 2021 CKD-EPI eGFRcr | 2012 CKD-EPI eGFRcys | 2021 CKD-EPI eGFRcr-cys | Creatinine Clearance | Serum Creatinine | Serum Cystatin C | Sarcopenia Index |
|---|---|---|---|---|---|---|---|---|---|
| Model comparison | |||||||||
| AIC | 2481.46 | 2441.42 | 2441.9 | 2440.56 | 2432.72 | 2441.78 | 2449.67 | 2444.04 | 2484.75 |
| ΔAIC | - | -40.04 | -39.56 | -40.9 | -48.74 | -39.68 | -31.79 | -37.42 | 3.29 |
| Fixed-effect parameters | |||||||||
| V_pop (L) | 17.1 | 17.1 | 17.1 | 17.1 | 17.1 | 17.1 | 17.1 | 17.1 | 17.1 |
| k_pop (h−1) | 0.22 (6.27%) |
0.2 (5.34%) |
0.19 (6.71%) |
0.23 (5.78%) |
0.21 (5.3%) |
0.17 (5.92%) |
0.22 (5.52%) |
0.20 (5.11%) |
0.19 (7.93%) |
| k12_pop (h−1) | 0.55 (14.4%) |
0.56 (12.1% |
0.41 (28.7%) |
0.39 (17.0%) |
0.34 (17.4%) |
0.43 (10.8%) |
0.55 (15.9%) |
0.32 (10.8%) |
0.30 (33.8%) |
| k21_pop (h−1) | 0.19 (11.8%) |
0.2 (12.2%) |
0.17 (17.0%) |
0.17 (12.2%) |
0.16 (13.4%) |
0.18 (10.8%) |
0.20 (15.5%) |
0.15 (9.54%) |
0.15 (19.7%) |
| βrenal on k|| | - | 0.44 (15.9%) |
0.45 (16.3%) |
0.45 (18.2%) |
0.5 (13.1%) |
0.48 (14.6%) |
-0.50 (15.8%) |
-0.54 (15.5%) |
-0.08 (156%) |
| Random-effect parameters | |||||||||
| Standard deviation of inter-individual variability (IIV) | |||||||||
| ωV | 0.48 (24.7%) |
0.31 (17.6%) |
0.23 (38.9%) |
0.18 (197%) |
0.25 (23.6%) |
0.22 (27.8%) |
0.43 (18.1%) |
0.20 (42.5%) |
0.27 (37.4%) |
| ωk | 0.52 (10.9%) |
0.45 (10.0%) |
0.44 (9.87%) |
0.47 (14%) |
0.43 (9.62%) |
0.46 (8.97%) |
0.41 (11.4%) |
0.48 (9.26%) |
0.55 (9.29%) |
| ωk12 | 0.24 (69.2%) |
0.27 (29.2%) |
0.39 (39.2%) |
0.4 (30.5%) |
0.24 (63.1%) |
0.36 (25.9%) |
0.29 (49.1%) |
0.41 (28.9%) |
0.57 (23.0%) |
| ωk21 | 0.48 (20.3%) |
0.51 (17.5%) |
0.47 (27.8%) |
0.54 (26.6%) |
0.63 (15.7%) |
0.64 (14.6%) |
0.56 (18.0%) |
0.53 (14.0%) |
0.60 (21.7%) |
| Residual variability (RV) | |||||||||
| b | 0.36 (7.24%) |
0.36 (6.54%) |
0.38 (6.9%) |
0.36 (8.43%) |
0.36 (6.4%) |
0.36 (6.74%) |
0.35 (6.32%) |
0.37 (6.75%) |
0.36 (6.75%) |
k = k_pop × (renal estimate/constant) β. For eGFR and creatinine clearance the constant was 30. For serum creatinine, cystatin C, and the sarcopenia index the constant was set equal to the median observed value (1.9, 2.3, and 81.3, respectively).
CKD-EPI: Chronic Kidney Disease Epidemiology Collaboration; eGFR: estimated glomerular filtration rate using equations that leverage self-identified black race (race), serum creatinine (cr), and/or serum cystatin C (cys); AIC: Akaike information criteria; V_pop: fixed population parameter for volume of distribution; k_pop: fit population parameter for the elimination rate; k12_pop and k21_pop: fit population parameter for the rate into/out of second compartment; ωV, ωk, ωk12, ωk21: standard deviation of random effects for population parameters; b: estimated value from proportional error model.
With eGFRcr-cys as a covariate of k, we ran the automated SCM algorithm in Monolix, which considered additional patient factors as covariates of k, k12, and k21 in a stepwise fashion. Age as an additional patient covariate of the elimination rate constant was selected in the final model. Population parameters for this ‘best’ performing model are shown in Table 4 and model diagnostic plots are shown in Supplementary Figures 2B, 3B, and 4B.
Table 4. Population pharmacokinetic model selected by the automated stepwise covariate model (SCM) building algorithm in Monolix.
Renal function, estimated as eGFR by the 2021 CKD-EPI equation using both serum cystatin C and serum creatinine, and age were selected as covariates of the elimination rate, k.
| Final Model | 2-compartment, linear elimination eGFRcys, Scr & Age as covariates on k |
|---|---|
| Model comparison | |
| AIC | 2427.31 |
| ΔAIC vs. structural model+ | −54.15 |
| Fixed-effect parameters | |
| V_pop (L) | 17.1 |
| k_pop (h−1) | 0.22 (6.47%) |
| k12_pop (h−1) | 0.46 (20.4%) |
| k21_pop (h−1) | 0.18 (11.8%) |
| βeGFR on k|| | 0.45 (16.5%) |
| βAge on k|| | −0.52 (39.8%) |
| Random-effect parameters | |
| Standard deviation of inter-individual variability (IIV) | |
| ωV | 0.3 (22.8%) |
| ωk | 0.43 (13.7%) |
| ωk12 | 0.15 (123%) |
| ωk21 | 0.62 (31.6%) |
| Residual variability (RV) | |
| b | 0.36 (8.06%) |
Comparison is to the 2-compartment structural model described in Table 2.
k = k_pop × (eGFR/30) βeGFR × (Age/60) βAge.
CKD-EPI: Chronic Kidney Disease Epidemiology Collaboration; eGFR: estimated glomerular filtration rate using equations that leverage self-identified black race (race), serum creatinine (cr), and/or serum cystatin C (cys); AIC: Akaike information criteria; V_pop: fixed population parameter for volume of distribution; k_pop: fit population parameter for the elimination rate; k12_pop and k21_pop: fit population parameter for the rate into/out of second compartment; ωV, ωk, ωk12, ωk21: standard deviation of random effects for population parameters; b: estimated value from proportional error model.
1.4.3. Other patient factors and individual variation in pharmacokinetics
From the final model, we determined the individual population PK parameters for the elimination rate constant (k), the rate constant out of compartment one (k12), and the rate constant out of compartment two (k21). Figure 1A compares individual estimates for the elimination rate to the eGFRcr-cys stratified by self-identified race. This demonstrated a strong, positive relationship between the elimination rate of L-carnitine and kidney function that is consistent across the two groups. In contrast to kidney function, the relationship between patient weight and the elimination rate was negligible (Supplementary Figure 5, R2 = 0.01).
Figure 1. Association of individual patient parameters with patient characteristics.
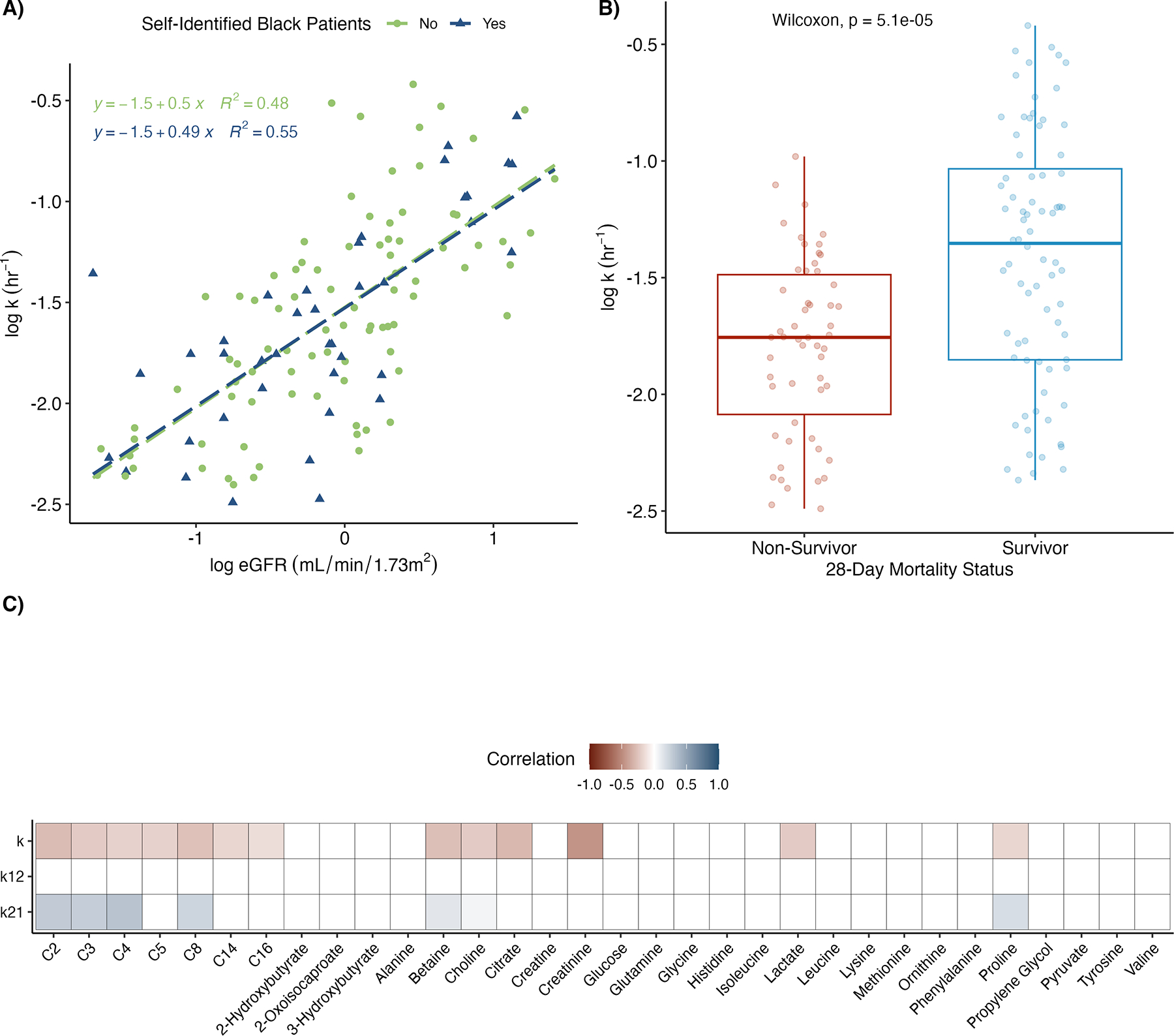
(A) Scatter plot and line of best fit for the predicted elimination rate constant (k) from the final model versus eGFR. Self-identified black (in blue) and non-black (in green) patients are plotted separately. eGFR was estimated using the 2021CKD-EPI equation with serum creatinine and cystatin c. Patient parameters were log-transformed prior to plotting. (B) Boxplots of estimated elimination rate constant (k) stratified by 28-day mortality status. (C) Heatmap of Spearman correlation coefficients between conditional mode estimated individual parameters and baseline (pre-treatment) metabolite levels as measured by LC-MS (acylcarnitines, C2-C16) and 1H-NMR spectroscopy. Individual parameters considered were k and rate in-to (k12) and out-of (k21) tissue. eGFR: estimated glomerular filtration rate; CKD-EPI: Chronic Kidney Disease Epidemiology Collaboration; LC-MS: liquid chromatography-mass spectroscopy; 1H-NMR: proton nuclear magnetic resonance.
Individual PK parameters were also compared to OCTN2 genotypes (rs2631367), baseline metabolite concentrations, and patient mortality at 28-days. Twenty-three patients were wildtype (CC) at rs2631367, while 87 patients contained either one (CG, 50 patients) or two (GG, 37 patients) copies of the G allele, which has been associated with greater transporter expression. There was no evidence of a relationship between OCTN2 genotype and any individual PK parameters (by the Kruskal–Wallis rank test, p>0.05). Patients who died before 28-days had a lower predicted value for k (Wilcoxon signed rank test, p=5.1e-05, Figure 1B), but similar values for k12 and k21. Figure 1C shows the correlation between individual PK parameters and baseline metabolites measured by LC-MS or NMR. Baseline acylcarnitines tended to be negatively correlated to k and positively correlated to k21. Lactate and creatinine were also negatively correlated to k.
1.5. Discussion
The host-response to infection and pharmacotherapy in sepsis is highly heterogeneous.31 A phase I trial of intravenous L-carnitine in patients with septic shock demonstrated a high degree of interindividual variability in the response to the candidate metabolic-therapeutic.6, 9 This high degree of variability was also evident in post-treatment blood concentrations of L-carnitine. In this secondary population PK analysis of the subsequent phase IIb trial7, a two-compartment model with a fixed population parameter for the Vd and eGFR as covariates of the elimination rate constant best fit the observed data. Importantly, we also found that patient mortality and baseline metabolic status, but not transporter genomics, were related to individual drug response. Pre-treatment Scr was the metabolite most strongly associated with L-carnitine elimination (Figure 1C); other metabolites, those attributable to energy metabolism, were also inversely associated with the estimated L-carnitine elimination rate constant. In aggregate, these findings suggest that while renal function is a primary driver of the variability in L-carnitine blood concentrations following high-dose administration in sepsis patients, pre-treatment energy metabolism also contributes.
In addition to the found broad dynamic range of measured L-carnitine blood concentrations, the lack of PK data for L-carnitine given at high-doses in patients with septic shock served as the primary justification for our analysis. We also leveraged prior knowledge to inform our analysis. In the phase I trial of high-dose L-carnitine in septic shock there was considerable interpatient variability in carnitine and acylcarnitine concentrations post-treatment, with elevated levels associated with mortality.6, 9 Here, we see a similar broad-dynamic range in concentrations following treatment, with non-survivors characterized by lower individual values for the elimination rate and higher concentrations (Figure 1B). Similarly, all acylcarnitines measured were negatively correlated with individual parameters for the elimination rate as were other energy-related metabolites (Figure 1C). Adverse drug reactions due to L-carnitine were assessed in the phase I6 and II7 trials of L-carnitine but known toxicity to the compound, including an increased potential for seizures and gastrointestinal side effects, were not widely reported. This suggests the higher mortality in patients with elevated concentrations is not directly attributable to L-carnitine toxicity, however this cannot be completely ruled out. Rather, we speculate that the patients with elevated concentrations had worse kidney function and greater metabolic dysfunction over the course of the study. Importantly, we and others have shown that elevations in acylcarnitines in patients with sepsis who have not received supplemental L-carnitine are associated with increased disease severity and mortality.32–34
Previous reports of L-carnitine PK utilized lower doses that are routinely used in the clinic and in patients who are not acutely ill.13, 35 Administration of radiolabeled L-carnitine demonstrated a renally eliminated drug that can be represented as a 3- compartment model with a central pool (approximating extracellular fluid), a faster equilibrating compartment (likely generalizing to kidney and liver), and a slowly equilibrating compartment (i.e., skeletal muscle).36 Moreover, endogenous L-carnitine is extensively (>98%) reabsorbed in the renal tubules, and single intravenous doses demonstrate saturation of this process and increased clearance of the compound.37, 38 Although we tested more complex population PK models with nonlinear elimination and multiple compartments, these models were characterized by a higher AIC and poor predictions compared to the 2-compartment model with linear elimination. However, our work does strongly support the importance of kidney function in the elimination of high-dose exogenous L-carnitine, as each kidney function estimate we considered as a covariate of k dramatically lowered the AIC compared to the base model (Table 3). This strengthens our justification in using L-carnitine to test alternative equations that estimate kidney function in critically ill patients.
Current clinical and drug development standards rely on the measurement of Scr as a biomarker of kidney function. Although wide use and the international standardization of the analytical method to quantify Scr are strengths of this paradigm, there are increasing calls to adopt alternative kidney function biomarkers, particularly in critically ill patients.14 Another endogenous biomarker, Scys, has demonstrated modest improvement in estimating kidney function for renally eliminated drugs.39 In our analysis, we found that Scys outperformed Scr as an individual kidney function biomarker and covariate of the elimination rate of a renally cleared compound. We also found eGFR equations that leverage Scys provided superior model performance and that inclusion of race to estimate eGFR weakened model fit. Our work adds to growing calls to reconsider the approach to estimating kidney function in clinical practice and drug development.
Finally, we assessed genetic variability at rs2631367 in the OCTN2 transporter to determine its contribution to L-carnitine blood concentration variability and because we had previously found that it was associated with peak concentrations of L-carnitine.11 The G allele has been associated with increased mRNA expression of the transporter in eQTL analysis, potentially granting systemic tissue a greater ability to sequester exogenous L-carnitine.11 Our results here found that the elimination rate and the rates into and out of tissue were not meaningfully related to OCTN2 genotype. Given that OCTN2 is a highly conserved transporter, owing to its critical role in host bioenergetics, it is possible the impact of altered gene transcription was insufficient to impact drug response in a heterogeneous, acutely ill clinical cohort. Moreover, we lacked detailed concomitant medications in the RACE trial and are unable to account for drug-transporter interactions that could impact tissue sequestration of L-carnitine.
Our study has several strengths and limitations that warrant further consideration. We employed rigorous metabolomics and PK methods to build a well-performing population model of high-dose, intravenous L-carnitine in the setting of septic shock. In building the population model, we chose to test the impact of only patient covariates commonly available in the clinical setting. However, as mentioned above, this did not include an accounting of concomitant medication use, in particular those known to adversely impact mitochondrial function such as propofol and valproic acid.40 Nevertheless, we had a unique opportunity to assess the relationship between less commonly available patient information including OCTN2 transporter genotype and baseline metabolic status. We were also able to assess different approaches for estimating kidney function in critical illness using a therapeutic candidate drug that is extensively cleared by the kidneys. However, we acknowledge since the study was not designed to model L-carnitine PK, the blood sampling scheme for the trial was rather sparse, particularly early during the drug’s infusion, which superseded our ability to fit a population parameter for the Vd. In addition, we opted to use baseline normalization when considering drug concentrations post-treatment, as L-carnitine is an endogenous molecule and the investigative product administered was not radio-labeled. Our estimates of kidney function are also indexed to a standard body surface area (i.e., eGFR estimates in mL/min/1.73 m2). Missingness in patient height data precluded us from individualizing these estimates in absolute units (mL/min). Finally, our measurement of Scys was done using residual, biobanked serum and a commercially available ELISA kit rather than a clinical measurement from a fresh patient sample. As such, our results regarding the optimal method for estimating eGFR must be interpreted as exploratory and requires rigorous further validation using additional cohorts of critically ill patients and probe drug molecules.
In conclusion, we found that high-dose intravenous L-carnitine in patients with septic shock can be reliably modeled at the population level using a two-compartment model with linear elimination. Kidney function as a covariate of the elimination rate dramatically improved model performance, with methods that incorporate Scys, but not patient race, providing the greatest improvement. We also found that patient mortality and baseline metabolites were strongly related to individual patient PK parameters. Future assessment of high-dose L-carnitine as a therapeutic for septic shock could include a more tailored dosing approach that considers renal function and pre-treatment metabolic status. We have previously shown that pre-treatment acetylcarnitine serum concentration is predictive of therapeutic benefit from L-carnitine treatment.34 Consideration of these patient features could aid in moving sepsis, a field that presently has few therapeutic options, towards a precision medicine approach.
Supplementary Material
Acknowledgements:
This study is supported by the National Institute of General Medical Sciences (NIGMS) via R01GM103799 (A.E.J.), K23GM113041 (M.A.P.), R01GM111400 (K.A.S.), and R35GM136312 (K.A.S.). T.S.J. has received support from the American Foundation of Pharmaceutical Education. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or the National Institutes of Health. The concentration data for L-carnitine and the metabolomics data described in this manuscript will be publicly available on the NIH’s Metabolomics Workbench site (https://www.metabolomicsworkbench.org/).
Footnotes
Conflicts of Interest: The authors declare no conflicts of interests
1.5 References
- 1.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). Jama 2016;8:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. The Lancet 2020;10219:200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaillon J-M, Singer M, Skirecki T. Sepsis therapies: learning from 30 years of failure of translational research to propose new leads. EMBO Molecular Medicine 2020;4:e10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singer M The role of mitochondrial dysfunction in sepsis-induced multi-organ failure. Virulence 2014;1:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pravda J Metabolic theory of septic shock. World journal of critical care medicine 2014;2:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puskarich MA, Kline JA, Krabill V, Claremont H, Jones AE. Preliminary safety and efficacy of L-carnitine infusion for the treatment of vasopressor-dependent septic shock: a randomized control trial. JPEN Journal of parenteral and enteral nutrition 2014;6:736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AE, Puskarich MA, Shapiro NI, et al. Effect of Levocarnitine vs Placebo as an Adjunctive Treatment for Septic Shock: The Rapid Administration of Carnitine in Sepsis (RACE) Randomized Clinical Trial. JAMA Network Open 2018;8:e186076–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive care medicine 1996;7:707–10. [DOI] [PubMed] [Google Scholar]
- 9.Puskarich MA, Evans CR, Karnovsky A, Das AK, Jones AE, Stringer KA. Septic Shock Nonsurvivors Have Persistently Elevated Acylcarnitines Following Carnitine Supplementation. Shock (Augusta, Ga) 2018;4:412–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puskarich MA, Finkel MA, Karnovsky A, et al. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Annals of the American Thoracic Society 2015;1:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennaro TS, Puskarich MA, McCann MR, et al. Using L-Carnitine as a Pharmacologic Probe of the Interpatient and Metabolic Variability of Sepsis. Pharmacotherapy 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puskarich MA, Jennaro TS, Gillies CE, et al. Pharmacometabolomics identifies candidate predictor metabolites of an L-carnitine treatment mortality benefit in septic shock. Clinical and translational science 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans AM, Fornasini G. Pharmacokinetics of L-carnitine. Clinical pharmacokinetics 2003;11:941–67. [DOI] [PubMed] [Google Scholar]
- 14.McMahon BA, Galligan M, Redahan L, et al. Biomarker Predictors of Adverse Acute Kidney Injury Outcomes in Critically Ill Patients: The Dublin Acute Biomarker Group Evaluation Study. Am J Nephrol 2019;1:19–28. [DOI] [PubMed] [Google Scholar]
- 15.Lewis RJ, Viele K, Broglio K, Berry SM, Jones AE. An adaptive, phase II, dose-finding clinical trial design to evaluate L-carnitine in the treatment of septic shock based on efficacy and predictive probability of subsequent phase III success. Critical care medicine 2013;7:1674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillies CE, Jennaro TS, Puskarich MA, et al. A Multilevel Bayesian Approach to Improve Effect Size Estimation in Regression Modeling of Metabolomics Data Utilizing Imputation with Uncertainty. Metabolites 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dambrova M, Makrecka-Kuka M, Kuka J, et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol Rev 2022;3:506–51. [DOI] [PubMed] [Google Scholar]
- 18.McHugh CE, Flott TL, Schooff CR, et al. Rapid, Reproducible, Quantifiable NMR Metabolomics: Methanol and Methanol: Chloroform Precipitation for Removal of Macromolecules in Serum and Whole Blood. Metabolites 2018;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics 2009;2:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado C, Baweja M, Crews DC, et al. A Unifying Approach for GFR Estimation: Recommendations of the NKF-ASN Task Force on Reassessing the Inclusion of Race in Diagnosing Kidney Disease. Am J Kidney Dis 2022;2:268–88.e1. [DOI] [PubMed] [Google Scholar]
- 21.Pai MP. Antimicrobial Dosing in Specific Populations and Novel Clinical Methodologies: Kidney Function. Clinical pharmacology and therapeutics 2021;4:952–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levey AS, Stevens LA, Schmid CH, et al. A New Equation to Estimate Glomerular Filtration Rate. Annals of Internal Medicine 2009;9:604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inker LA, Eneanya ND, Coresh J, et al. New Creatinine- and Cystatin C–Based Equations to Estimate GFR without Race. New England Journal of Medicine 2021;19:1737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inker LA, Schmid CH, Tighiouart H, et al. Estimating Glomerular Filtration Rate from Serum Creatinine and Cystatin C. New England Journal of Medicine 2012;1:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron 1976;1:31–41. [DOI] [PubMed] [Google Scholar]
- 26.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;6:580–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahara H, Yee SW, Urban TJ, et al. Functional genetic variation in the basal promoter of the organic cation/carnitine transporters OCTN1 (SLC22A4) and OCTN2 (SLC22A5). Journal of pharmacology and experimental therapeutics 2009;1:262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grube M, Meyer zu Schwabedissen HE, Prager D, et al. Uptake of cardiovascular drugs into the human heart: expression, regulation, and function of the carnitine transporter OCTN2 (SLC22A5). Circulation 2006;8:1114–22. [DOI] [PubMed] [Google Scholar]
- 29.U.S. Food and Drug Administration. Bioequivalence Studies With Pharmacokinetic Endpoints for Drugs Submitted Under an Abbreviated New Drug Application, 08/21/2021. Available from https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioequivalence-studies-pharmacokinetic-endpoints-drugs-submitted-under-abbreviated-new-drug. Accessed 01/05/2023, [Google Scholar]
- 30.Barreto EF, Poyant JO, Coville HH, et al. Validation of the sarcopenia index to assess muscle mass in the critically ill: A novel application of kidney function markers. Clinical Nutrition 2019;3:1362–67. [DOI] [PubMed] [Google Scholar]
- 31.Leligdowicz A, Matthay MA. Heterogeneity in sepsis: new biological evidence with clinical applications. Critical Care 2019;1:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung KP, Chen GY, Chuang TY, et al. Increased Plasma Acetylcarnitine in Sepsis Is Associated With Multiple Organ Dysfunction and Mortality: A Multicenter Cohort Study. Critical care medicine 2019;2:210–18. [DOI] [PubMed] [Google Scholar]
- 33.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013;195:195ra95–95ra95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jennaro TS, Puskarich MA, Evans CR, et al. Sustained Perturbation of Metabolism and Metabolic Subphenotypes Are Associated With Mortality and Protein Markers of the Host Response. Crit Care Explor 2023;4:e0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rebouche CJ. Kinetics, pharmacokinetics, and regulation of L-carnitine and acetyl-L-carnitine metabolism. Annals of the New York Academy of Sciences 2004;30–41. [DOI] [PubMed] [Google Scholar]
- 36.Rebouche CJ, Engel AG. Kinetic compartmental analysis of carnitine metabolism in the human carnitine deficiency syndromes. Evidence for alterations in tissue carnitine transport. J Clin Invest 1984;3:857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahajwalla CG, Helton ED, Purich ED, Hoppel CL, Cabana BE. Comparison of L-carnitine pharmacokinetics with and without baseline correction following administration of single 20-mg/kg intravenous dose. J Pharm Sci 1995;5:634–9. [DOI] [PubMed] [Google Scholar]
- 38.Harper P, Elwin CE, Cederblad G. Pharmacokinetics of bolus intravenous and oral doses of L-carnitine in healthy subjects. Eur J Clin Pharmacol 1988;1:69–75. [DOI] [PubMed] [Google Scholar]
- 39.Barreto EF, Rule AD, Murad MH, et al. Prediction of the Renal Elimination of Drugs With Cystatin C vs Creatinine: A Systematic Review. Mayo Clin Proc 2019;3:500–14. [DOI] [PubMed] [Google Scholar]
- 40.Jennaro TS, Puskarich MA, McCann MR, et al. Using l-Carnitine as a Pharmacologic Probe of the Interpatient and Metabolic Variability of Sepsis. Pharmacotherapy 2020;9:913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


