Abstract
Based on genetic and bioinformatic analysis, 80 proteins from the newly sequenced Schizosaccharomyces pombe genome appear to be splicing factors. The fission yeast splicing factors were compared to those of Homo sapiens and Saccharomyces cerevisiae in order to determine the extent of conservation or divergence that has occurred over the billion years of evolution that separate these organisms. Our results indicate that many of the factors present in all three organisms have been well conserved throughout evolution. It is calculated that 38% of the fission yeast splicing factors are more similar to the human proteins than to the budding yeast proteins (>10% more similar or similar over a greater region). Many of the factors in this category are required for recognition of the 3′ splice site. Ten fission yeast splicing factors, including putative regulatory factors, have human homologs, but no apparent budding yeast homologs based on sequence data alone. Many of the budding yeast factors that are absent in fission yeast are associated with the U1 and U4/U6.U5 snRNP. Collectively the data presented in this survey indicate that of the two yeasts, S.pombe contains a splicing machinery more closely reflecting the archetype of a spliceosome.
COMPLETION OF THE FISSION YEAST SEQUENCING PROJECT
The removal of introns from pre-mRNAs takes place in a large multi-protein–snRNA complex, called the spliceosome. The basic mechanism of pre-mRNA splicing has been elucidated in vitro using cell-free extracts prepared from mammalian cells and budding yeast. In addition, genetic studies in both fission and budding yeasts have provided functional information on many splicing factors. Gaps remain, however, in our understanding of the assembly process that produces a catalytic spliceosome. The major structural rearrangements of the snRNAs and the pre-mRNA substrate, many of which are mediated by proteins, are beginning to be understood. There are numerous RNA–RNA, RNA–protein and protein–protein interactions that are dynamic and change as splicing proceeds. The modification of some of the proteins by phosphorylation appears to play an important role in regulating splicing. In addition, the function of this complex molecular machine needs to be coordinated spatially and temporally with other physiological events within the nucleus to ensure proper gene expression. This will be the challenge of the coming years and will receive its fuel from the post-genomic era.
The complete genomic sequence of Saccharomyces cerevisiae has been available since 1996. Over 70 spliceosomal protein components have been identified by genetic and biochemical means. So far only 15 protein splicing factors have been genetically identified in Schizosaccharomyces pombe and none have been identified biochemically. The biochemical analysis of splicing factors in fission yeast has been hampered by the fact that an in vitro splicing system currently does not exist in S.pombe, though efforts to establish one continue to move forward. This will not impair our ability to identify new splicing factors in fission yeast, however, since the sequencing of the S.pombe genome is now complete. This will allow us to undertake for the first time an in-depth analysis comparing the splicing machinery of the two yeasts.
In addition to the genome projects, another approach has recently proven very fruitful for the identification of factors involved with splicing. Neubauer et al. have identified 40 proteins putatively associated with the spliceosome in nuclear extracts from HeLa cells using mass spectrometry (1). Nineteen of these proteins have not been identified previously as spliceosome-associated proteins. The more recent methods of mass spectrometry and genome analysis, along with the classical genetic and biochemical approaches, have the potential of unveiling a wealth of new information with regard to factors that are involved with pre-mRNA splicing. In this study we have taken advantage of the newly available fission yeast genome to compare the proteins involved in pre-mRNA splicing in the two yeast species and humans. Our findings show that most of the splicing apparatus has been evolutionarily conserved. There are, however, some factors present in fission yeast and humans that appear to be absent in budding yeast. In addition there appear to be several factors present in humans that do not exist in yeasts, and several factors that may be unique to budding yeast.
IDENTIFICATION OF FISSION YEAST SPLICING FACTORS
Several approaches were used to identify fission yeast splicing factors for this study. Initially we focused on the published S.pombe prp (pre-mRNA processing) genes that were identified genetically and proteins that were identified using the yeast two-hybrid assay (Table 1). We then searched for fission yeast homologs of the factors identified in HeLa splicing extracts biochemically and by mass spectrometry (Table 2). Finally, we searched for homologs of splicing factors identified genetically or biochemically in budding yeast (Table 3). Our searches were facilitated by sorting through the list of putative splicing factors identified by the fission yeast sequencing project. The S.cerevisiae and Homo sapiens sequences were used as query sequences to conduct basic BLAST searches, using the blastp program, to look for proteins that are similar in S.pombe in the non-redundant database. We aligned the fission yeast proteins with those of budding yeast or mammals using the BLAST 2 Sequences blastp program and the 3PAM250 matrix (http://www.ncbi.nlm.nih.gov/gorf/bl2.html ). The percent identity and similarity over the region of greatest match was recorded.
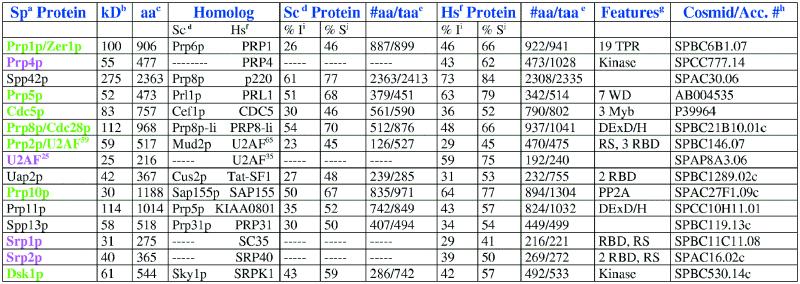
Table 1. Fission yeast splicing factors identified genetically or by protein interactions.
Pink indicates proteins present in fission yeast and humans that have no apparent homolog in budding yeast based on sequence data. Green indicates proteins that are at least 10% more similar or have a region of 100 amino acids more aligned between the fission yeast and human proteins.
aSchizosaccharomyces pombe.
bKilodalton.
cNumber of amino acids present in the fission yeast protein.
dSaccharomyces cerevisiae.
eThe number of amino acids used in the alignment/total number of amino acids in the S.cerevisiae or H.sapiens protein.
fHomo sapiens.
gFeatures: TPR, tetratricopeptide repeats; WD, protein interaction motif characteristic of β subunit of G protein; Myb, related to DNA-binding domain of human c-Myb; DExD/H, ATP-dependent RNA helicase motif; RBD, RNA binding domain; RS, arginine- and serine-rich region; PP2A, similar to the regulatory subunit A of the phosphatase PP2A.
hCosmid number or accession number, which may be accessed at http://www.sanger.ac.uk/Projects/S_pombe/FUNCAT/rna_proc.shtml
iPercent identity.
jPercent similarity.
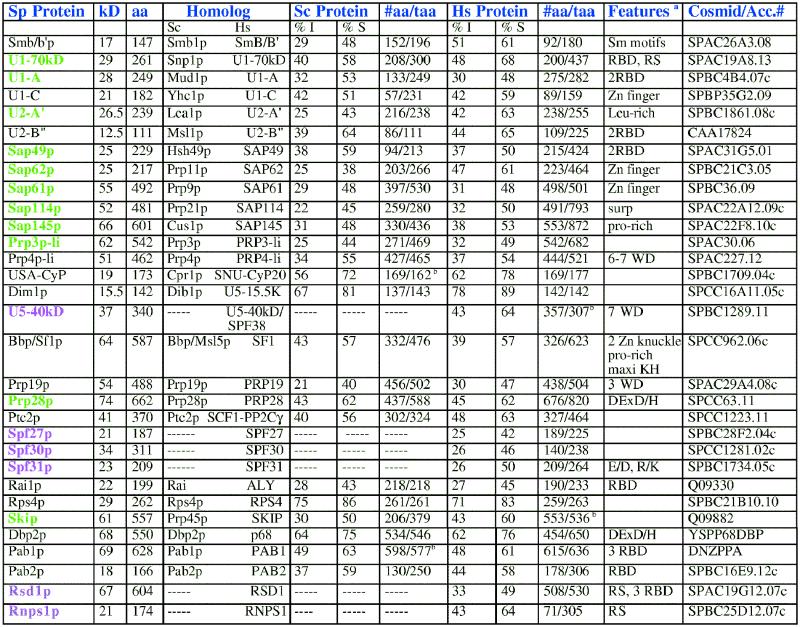
Table 2. Fission yeast proteins that are similar to human spliceosome-associated proteins.
aFeatures the same as for Table 1. In addition: E/D,R/K, glutamic acid/aspartic acid, arginine/lysine-rich regions; pro-rich, proline-rich; surp, potential RNA binding motif; maxi KH, hnRNP H homology implicated in RNA binding.
bThe #aa is greater than the taa because the BLAST 2 Sequence blastp program counts a gap as an amino acid.
Table 3. Fission yeast proteins that are similar to budding yeast splicing factors.
aFeatures the same as in Tables 1 and 2. In addition: Zn knuckle, protein structural motif that binds zinc; WW, protein motif believed to interact with proline-rich regions; Sm motifs, motif present in proteins that bind to snRNAs.
bThe #aa is greater than the taa because the BLAST 2 Sequence blastp program counts a gap as an amino acid.
cThe complete H.sapiens sequence is not known; therefore, only the known sequence was compared with S.pombe.
SPLICING FACTORS IDENTIFIED GENETICALLY OR USING TWO-HYBRID ASSAYS
Several fission yeast splicing factors have been identified genetically including Prp1p/Zer1p, Prp2p, Prp4p, Prp5p, Prp8p/Cdc28p, Prp10p, Prp11p, Cdc5p, Dsk1p, Spp13p and Spp42p (2–10; T.Tani, personal communication). A few factors were identified by their interactions with the splicing factor Prp2p/U2AF59, including U2AF25, Uap2p and Sap155p (Prp10p), using a yeast two-hybrid assay (11–13). Srp1p and Srp2p were identified in a search for factors that have arginine/serine-rich regions, a motif found in several mammalian splicing factors (14,15). In order to determine the extent of similarity between the fission yeast splicing factors and those of budding yeast and humans, we compared the amino acid sequences using the BLAST 2 Sequences program. The results of this analysis are shown in Table 1.
Prp1p/Zer1p is more similar to the human protein than the budding yeast protein based on the percent similarity and identity over an extensive stretch of amino acids (Table 1). In S.cerevisiae the homolog of Prp1p/Zer1p is called Prp6p. Prp6p is associated with the tri-snRNP U4/U6.U5. The protein contains 19 tetratricopeptide repeats. Interestingly, the S.pombe prp1-4 and zer1 strains express a polymorphic phenotype indicating that the gene product is involved in pre-mRNA splicing, poly(A)+ RNA transport and cell cycle progression. The prp13 mutant of fission yeast is synthetic lethal with prp1 (2). The gene that encodes prp13+ has not yet been cloned, but a suppressor, spp13 has recently been cloned (T.Tani, personal communication). The budding yeast homolog of Spp13p is Prp31p, which has been shown to be associated with the U4/U6.U5 tri-snRNP.
Prp1p also interacts genetically with the Prp4p protein kinase and is phosphorylated in vitro by Prp4p protein kinase (2,10; N.F.Käufer, unpublished data). The Prp4p protein kinase shows 62% similarity over 46% of its amino acids with a human kinase, whereas no S.cerevisiae kinase with similarity was identified (14) (Table 1). The region of greatest similarity between the fission yeast and human protein comprises a kinase domain plus approximately 150 amino acids at the N-terminal domain. The additional amino acids in the human protein extend the N-terminal domain. Prp4p is essential for growth, and kinase activity is required for pre-mRNA splicing (4; N.F. Käufer, unpublished data). This study was the first demonstration that a kinase is involved in pre-mRNA splicing.
The Prp4p protein kinase interacts genetically with Spp42p (10; unpublished data). Spp42p is the homolog of Prp8p (Dbf3p, Slt21p) in budding yeast and p220 in mammals. This protein has been shown to be a bona fide splicing factor in budding yeast and mammals (16,17). Spp42p has been highly conserved throughout evolution and Spp42p is slightly more similar to p220 than Prp8p (Table 1). Prp8p is thought to be a major player in the switch from inactive to active spliceosome (18). Surprisingly, Neubauer et al. did not identify p220 as a spliceosomal protein by mass spectrometry (1).
The temperature sensitive (ts) prp5-1 allele is synthetically lethal with the ts prp4-73 allele, suggesting a functional interaction (N.F.Käufer, unpublished data). Prp5p has been evolutionarily conserved between all three organisms we examined, with the fission yeast and human proteins showing the greatest amount of similarity (Table 1). The protein contains seven WD domains. Little is known about the function of Prp5p in splicing, but in S.pombe the prp5-1 allele accumulates pre-mRNA and arrests with a 2C DNA content at the restrictive temperature, suggesting that entry into or completion of mitosis is blocked.
Cdc5+ encodes an myb-related protein and is essential for G2/M progression in fission yeast (8). Because there is good evidence to show that Cdc5p homologs are splicing factors, the phenotype of cdc5-120 was re-examined and the mutant accumulated pre-mRNA at the non-permissive temperature (8). Both Cdc5p and the S.cerevisiae homolog, Cef1p, are part of a large complex that has been shown in budding yeast to join the spliceosome when U4 dissociates. It is noteworthy that cdc5-120 is synthetically lethal with prp5-1 and with prp4-73 (8; N.F.Käufer, unpublished data). Table 1 shows that between the three organisms the protein is evolutionarily conserved with the fission yeast and human proteins showing a greater similarity over a larger region of the protein.
Prp8/cdc28+ of S.pombe encodes a member of the DExD/H family of putative helicases. The human homolog is more similar over a larger portion of the protein than the budding yeast protein (Table 1). In S.pombe this gene was identified independently as a splicing and cell cycle mutant (6). The cdc28 mutant produced elongated cells blocked in G2. The role(s) of Prp8p/Cdc28p in splicing and the cell cycle has not yet been determined.
As discussed above, several of the prp mutants in S.pombe arrest in specific stages of the cell cycle, indicating a link between pre-mRNA splicing and cell cycle progression. This phenotype is not easily explained. In the case of a general splicing factor, one would expect that a stringent mutant phenotype would cause the cells to arrest in all stages of the cell cycle. This is based on the fact that ∼45% of the genes in S.pombe, including major cell cycle regulators such as cdc2+, contain introns. A specific cell cycle arrest might indicate that: (i) the protein has two independent functions in cell cycle and pre-mRNA splicing or (ii) the cell cycle block is a consequence of the splicing defect of a transcript needed for cell cycle transitions. In the latter case the protein may be regulating the splicing efficiency of specific pre-mRNAs. So far no study has been able to distinguish between these possibilities to explain the splicing and cell cycle phenotypes.
Prp2p was identified in one of the original screens for splicing mutants (19). Prp2p encodes the counterpart of the large subunit of U2 auxiliary factor (U2AF65) in mammals (3). Using Prp2p/U2AF59 as bait in a two-hybrid assay, a protein was cloned and named U2AF25 because of its similarity to the small subunit of U2AF, referred to as U2AF35 (11). The large subunits of U2AF are 45% similar and the small subunits are 75% similar over the entire proteins between humans and fission yeast (Table 1). Interestingly, the S.cerevisiae genome apparently does not contain a sequence significantly similar to U2AF59 (based on a Pileup sequence analysis) or U2AF25, though Mud2p appears to be a functional ortholog of the large subunit of U2AF (20) (Table 1). The U2AF complex is involved in 3′ splice site recognition. U2AF65 binds through its RNA binding domains (RBDs) to the polypyrimidine tract facilitating the interaction of the U2 snRNP with the branchpoint sequence (21). In three recent studies, it was shown that the 3′ splice site recognition by the U2AF complex is dependent on the architecture of the 3′ end of the intron. For example: introns with very weak polypyrimidine tracts upstream of the AG at the 3′ splice site, require U2AF35 to be spliced properly and the protein interacts directly with the AG (22–24).
The intron architecture, particularly at the 3′ splice site is very different between the two yeasts. Schizosaccharomyces pombe introns have a degenerate branchpoint consensus sequence CURAY (where R represents purine and Y represents pyrimidine) similar to that present in mammals. In S.cerevisiae the branchpoint sequence UACUAAC is highly conserved setting it apart from humans and fission yeast. Most fission yeast introns between the branch sequence and the 3′ splice site are pyrimidine-rich and have an average length of 7 nt (25). However some fission yeast introns are purine-rich while others exhibit a balanced R/Y content in this region, similar to the variability seen in mammalian introns (25). It is conceivable, therefore, that 3′ splice site recognition in S.pombe reflects the ancestral splicing machinery, whereas in S.cerevisiae this part of the machinery has diverged. This notion is consistent with the fact that the small T-antigen intron of the SV40 early region is spliced properly in fission yeast, but not in budding yeast (26). This intron has the typical architecture of an S.pombe intron. Therefore, we suggest that the introns of S.pombe may reflect the architecture of ancestral introns.
Another splicing factor identified in the two-hybrid screen that used Prp2p/U2AF59 as bait was named Uap2p because it was the second U2AF associated protein identified (12). Uap2p shows 48% similarity with budding yeast Cus2p (Table 1). Cus2p is associated with the U2 snRNA in splicing extracts and is postulated to play a role in the proper folding of U2 into a favorable structure prior to spliceosome assembly (27). Interestingly, although Uap2p and Cus2p have been evolutionarily conserved, the protein–protein interactions between fission and budding yeast were not. Uap2p was identified initially through its interaction with U2AF59, whereas Cus2p does not interact with the ortholog Mud2p (12,27). Uap2p is most similar to the human protein Tat-SF1, especially in the N-terminal domain that contains two RBDs (12,27,28). Tat-SF1 has an acidic C-terminal domain that is reduced considerably in the yeast proteins (27). Tat-SF1 was originally identified as a transcription factor, but its interactions with other components of the U2 snRNP indicate it may also function in splicing (27,28). It has been suggested that the role of Uap2p and its homologs is to mediate the U2AF-dependent association of the U2 snRNP with the intron (27).
Prp10p was identified by functional complementation of the prp10-1 strain that accumulates pre-mRNAs at the restrictive temperature (7). Prp10p is 77% similar to the human protein SAP155 and 67% similar to the budding yeast homolog (Table 1). Both yeast proteins are shorter than the human protein. SAP155 is part of the human U2 snRNP (29). Prp10p has been shown to interact with Prp2p/U2AF59 genetically and in a two-hybrid assay (7,13). Whether or not Prp10p plays a role in the recruitment of the U2 snRNP to the 3′ splice site remains to be determined.
Prp11p appears to be the homolog of the S.cerevisiae helicase Prp5p (T.Tani, personal communication). In S.cerevisiae, Prp5p is needed for the base pairing between the U2 snRNP and the branchpoint sequence. The predicted role of Prp5p is the disruption of a Prp9p/U2 snRNP interaction which allows the RNA to refold into a configuration capable of pairing with the intron during pre-spliceosome assembly.
Srp1p and Srp2p are two proteins belonging to the family of SR splicing factors that have been identified and characterized in S.pombe (14,15). These are the first family members of SR splicing factors found in a unicellular organism, therefore a brief discussion is appropriate. Mammals have nine SR proteins consisting of one or two RBDs at the N-terminus and RS/SR dipeptides of different lengths, referred to as the RS domain, at the C-terminus. SR-splicing factors are involved in constitutive and alternative splicing in mammals (30). All nine SR proteins have at least one RBD containing the submotif RNP-1 that has the highly conserved signature sequence RDAE/DDA. The four SR proteins consisting of two RBDs contain in RBD2 the invariant sequence SWQDLKD. Srp1p and Srp2p contain one and two RBDs, respectively, with the signature sequences. Srp1p has a typical RS/SR domain, whereas Srp2p contains an arginine-rich region including two short SR elements. Overexpression of mutations in the RS/SR domain and the signature sequence RDAE/DDA leads to the accumulation of pre-mRNA indicating an involvement in pre-mRNA processing (14,15). Recently the Dsk1p kinase of fission yeast was shown to phosphorylate the RS domains of Srp1p and Srp2p in vitro (31). Dsk1p is a homolog of SRPK1. SRPK1 phosphorylates SR proteins like its fission yeast homolog. Comparing Srp1p with the databank revealed that it is closely related to SC35. The two proteins share 29% identity and 41% similarity (Table 1). Srp2p is closely related to mammalian SRP40, SRP55 and SRP75 (Table 1).
In addition to these two SR family members we also found a gene rsd1 in the S.pombe databank, encoding a protein which contains an extensive RS domain at the N-terminus followed by three RBDs. This protein has a mammalian counterpart, but no similar protein was found in S.cerevisiae (Table 2). Interestingly, Rsd1p shows the same domain arrangement as Prp2p/U2AF59 and both proteins are phosphorylated in vitro by Dsk1p (31,32). It is presently not known whether Rsd1p plays a role in splicing.
SPLICING FACTORS IDENTIFIED BY THEIR SIMILARITY TO PROTEINS ASSOCIATED WITH THE HUMAN SPLICEOSOME
Table 2 shows fission yeast homologs of factors identified by searching the databases for HeLa spliceosomal components identified biochemically and by mass spectrometry (1). When an S.pombe counterpart was found, the fission yeast sequence was used as the query sequence and the database was searched for homologous counterparts in S.cerevisiae.
From the search for spliceosomal-associated factors, several proteins were identified including the snRNP associated proteins Smb/b′, the U1-associated U1-A, U1-C and 70 kDa proteins, the U2-associated proteins A′, B′′, Sap49p, Sap61p, Sap62p, Sap114p and Sap145p, the U4/U6-associated proteins Prp3p-like, Prp4p-like and USA-Cyp and the U5-associated Dim1p and a 40 kDa protein. Six of the snRNP-associated proteins in this group were equally well conserved among humans and both yeasts, whereas Smb/b′, the 70 kDa protein, U1-A, U2-A′, Sap62p, Sap114p, Sap145p and Prp3p-like proteins are more similar between humans and fission yeast (Table 2). In addition, U1-A, Sap49p, Sap61p, Sap114p, Sap145p and Prp3p-like are similar to the human protein over an extended region. A homolog of the U5 40 kDa (Spf38p) protein does not appear to be present in S.cerevisiae.
A few non-snRNP proteins were identified including Bbp/Sf1p, which binds to the branch point sequence of the intron in mammals and budding yeast (33,34), Prp19p, which may play a role in recombination in addition to its role in splicing (35), the helicase Prp28p, which may be required to disrupt the U1-5′ splice site base pairing (36) and Ptc2p, a serine/threonine phosphatase which is required for early spliceosome formation (37). Bbp/Sf1p, Prp19p and Ptc2p are equally well conserved among the three organisms, whereas Prp28p is conserved over a larger region of the protein in fission yeast and humans (Table 2).
We also identified three fission yeast proteins, Spf27p, Spf30p and Spf31p, with significant sequence similarity to human spliceosome-associated proteins, but no known S.cerevisiae homologs (Table 2). The function of these proteins is currently unknown. Several evolutionarily conserved spliceosome-associated proteins were identified including the RNA annealing protein Rai1p, the ribosomal protein Rps4p, Skip, which has been shown to co-localize with snRNPs, the helicase p68 and the poly(A) binding proteins Pab1p and Pab2p, whose function in splicing remains unknown. The presence of poly(A) binding proteins in the spliceosome suggests that there are additional links between splicing and polyadenylation. Among this group of proteins, Skip is more similar between mammals and fission yeast, whereas the other factors are equally well conserved among all three organisms (Table 2). There are additional proteins identified by Neubauer et al. (1) as spliceosome associated, including SPF45 and Gry-Rbp, which did not have significant sequence similarities to proteins in fission yeast.
Mammalian spliceosome-associated proteins include the hnRNP proteins A1, A2/B2, C, G and M, which play a role in the regulation of splicing. No fission yeast proteins show significant sequence similarity to these proteins (Table 4). PTB1 also plays a role in the regulation of alternative splicing in mammals and there does not appear to be a fission yeast homolog of it either. The absence of fission yeast homologs of these regulatory splicing factors suggests that some aspects of the regulation of splicing are different between humans and fission yeast. A recent study suggests, however, that, as well as the SR proteins, there may be additional regulatory factors conserved between fission yeast and humans. Rnps1p has recently been identified as a general splicing activator (A.Mayeda and T.Tani, personal communication). The BLAST analysis indicates that a short central region of the protein is highly conserved and a closer inspection of the sequence indicates that there is also an arginine/serine-rich region at both the N- and C-termini in both organisms. To determine the extent of conservation of regulatory splicing factors between fission yeast and humans we must await the results of the human sequencing projects.
Table 4. Proteins without an obvious S.pombe homolog.
The data provided indicates the fission yeast protein that has the greatest similarity to the splicing factor based on sequence. In most cases the sequence conservation is within a limited region of the protein as indicated in the column #aa/taa.
aThe helicase in S.pombe that shows the greatest amount of similarity to Prp2p is the protein listed as Prp8p/Cdc28p in Table 1.
FACTORS SIMILAR TO BUDDING YEAST SPLICING FACTORS
Table 3 contains the comparison of protein sequences of S.pombe with those in S.cerevisiae and mammals, which we identified from the S.pombe Genome Project or by searching for homologs of budding yeast factors. Several snRNP proteins were identified by this method including all seven Sm proteins that associate with all the snRNAs and seven Lsm proteins which associate with the U6 snRNA. In addition, several snRNP-specific proteins were identified including Prp39p, Prp40p and Luc7p, which are U1-specific, the U2-associated Prp12p, the U4/U6.U5-associated Snu13p, the U5-associated Snu114p, Prp18p and the helicase Brr2p (Prp44p, Snu246p, Slt22p, Rss1p). The identification of so many new fission yeast splicing factors demonstrates clearly the advantage of the genome project. All of the genes that encode Sm proteins are more similar between fission yeast and humans than between the two yeasts except for Smb/b′ which shows a greater percent similarity over a shorter region of the protein (Tables 2 and 3). A comparison of the Lsm proteins also shows that the human and fission yeast proteins are more similar than the two yeast proteins except for Lsm2p and Lsm7p (Table 3). So far no human homolog of Prp39p has been identified and FBP11, the human protein most similar to Prp40p, aligns only within a limited region. Luc7p, which is also required for commitment complex formation in budding yeast, and Snu13p are equally well conserved among all three organisms, whereas fission yeast Prp12p and Brr2p, which unwinds U4/U6 duplexes (38), are more similar to the human factors. Prp18p, which is required for the second step of splicing (38), is less similar to human than budding yeast, but the region of similarity is more extensive between fission yeast and humans. Because Snu114p has extensive sequence similarity to the ribosomal elongation factor EF-2 which functions as a translocase, it is thought to fulfill a similar function in the spliceosome. The fission yeast homolog of Snu114p is more similar to the human protein. The results presented here, together with those in Table 2, suggest that the snRNP-associated proteins are most similar between fission yeast and humans.
Several non-snRNP proteins were also identified using this method. Four helicases which function in splicing or are associated with splicing factors were identified, including Prp16p, Prp22p, Prp43p and Uap56p. Prp16p is needed prior to cleavage at the 3′ splice site and is thought to play a role in maintaining splicing fidelity (38). Prp22p and Prp43p are required to release mRNA and the intron, respectively, from the spliceosome (38). The function of Uap56p in not known. Each of these putative ATP-dependent RNA helicases is equally well conserved among all three organisms, setting them apart from the U5-associated Brr2p helicase mentioned before.
Fission yeast Slu7p is 51% similar to human and only 36% similar to budding yeast. Slu7p is required for a spliceosomal structural change that occurs between the first and second step of splicing (39). Slu7p is also required for the selection of the proper AG residues at the 3′ splice site and thus plays a role in maintaining the fidelity of the splicing apparatus (40). The greater similarity between fission yeast and human Slu7p may be another reflection of the similarity of factors required for recognition of the 3′ splice site in these organisms.
Prp17p (Slt15p), which is required for the second step of splicing (38), is more similar between the two yeasts. The debranching enzyme Dbr1p appears to be equally conserved among the three organisms when percent similarity and region of similarity are compared. No apparent human homolog has been identified for Syf3p (Clf1p), which is thought to play a role in the association of the U4/U6.U5 snRNP with the pre-spliceosome. Sad1p is needed for U4/U6 biogenesis and it is equally well conserved among all three organisms. Ecm2p (Slt11p) and Isy1p show greater similarity to the human proteins, whereas Syf1p is more similar to the human protein but over a shorter stretch of sequence than the budding yeast protein. The function of these proteins in splicing is not known.
No fission yeast homologs were identified that correspond to the budding yeast Nam8p, Prp42p, Prp38p, Prp2p, Prp24p, Snu56p, Snu71p, Snu23p, Spp381p, Brr1p, Snt309p, Spp2p and Syf2p factors (Table 4). Based on which snRNAs these factors associated with, the absence of these factors in fission yeast suggests that the U1 and U4/U6.U5 snRNP may be very different in the two yeasts. It also suggests that recognition of the 5′ splice site may differ significantly.
CONCLUDING REMARKS
In her article ‘The best yeast?’, Susan Forsburg settles the dust swept up by the battles between the defenders of the two model yeasts, S.cerevisiae and S.pombe, by giving an account of what we have learned from each of these genetically tractable, but evolutionarily divergent organisms, for the understanding of cell cycle control (41). One of the most significant points she makes is that in order to understand problems of cell biology one should study both yeasts (41). The comparative analysis of the splicing machinery presented in this survey is consistent with the notion that between the two yeasts broad similarities exist, but also distinct differences are present which help to understand larger eukaryotes. One of the obvious differences in the case of the splicing machinery is that the factors required for 3′ splice site recognition are more similar between fission yeast and humans. This correlates well with the more loosely conserved recognition sequences at the 3′ end of the intron in these two organisms compared to the invariant sequences present in budding yeast. The second significant difference is the involvement of protein kinases and SR protein family members in both fission yeast and humans that are absent in budding yeast. This suggests that phosphorylation may be important in regulating splicing in humans and fission yeast. Since the SR proteins in humans have been shown to interact with proteins that recognize the 3′ splice site, this difference among the three organisms may be related to the differences in the intron structure mentioned above. In addition, many components of the S.pombe apparatus show a higher degree of similarity with the mammalian counterparts than those of budding yeast. The conclusion one may draw from these results is that studying both genetically pliable yeast species will be important to obtaining a complete in-depth understanding of pre-mRNA splicing.
Comparing the number of interrupted genes in the two species reveals that 5% of the S.cerevisiae and 45% of S.pombe genes contain introns. If one accepts the theory that introns are inserted randomly during evolution, the similar genome size of the two yeasts suggests that there is selective pressure for more introns to be inserted into the fission yeast genome. If, however, introns occurred early during evolution, then it would appear that budding yeast has had more selective pressure to lose introns with time. The greater differences in the splicing machinery in budding yeast might reflect selective pressure to modify the machinery required to recognize less variable introns. In contrast, there was less selective pressure on the fission yeast and human splicing machinery which resulted in the retention of more introns. If one accepts the intron early theory, the fission yeast splicing machinery would more closely reflect the ancestral spliceosome.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the members of the Sanger Centre who worked on the S.pombe genome project. In particular we want to acknowledge the efforts of Rachel McDougal and Valerie Wood who provided answers to our many inquiries about the functional catalog. We also would like to thank Robert Kemp and Mitchell Beales for many fruitful discussions regarding the implications of this study, Deming Xu for assistance with nomenclature and Bertrand Séraphin for help in identifying homologs. In addition we would like to thank our numerous colleagues in the splicing field whose constant questions about the relationship between the human splicing factors compared to those of yeasts provided the inspiration for this study. This work was partially supported by National Institutes of Health grant R01GM47487 awarded to J.P and a Deutsche Forschungsgemeinschaft grant awarded to N.F.K.
REFERENCES
- 1.Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- 2.Urushiyama S., Tani,T. and Ohshima,Y. (1997) Genetics, 147, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potashkin J., Naik,K. and Wentz-Hunter,K. (1993) Science, 262, 573–575. [DOI] [PubMed] [Google Scholar]
- 4.Alahari S.K., Schmidt,H. and Käufer,N.F. (1993) Nucleic Acids Res., 21, 4079–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundgren K., Allan,S., Urushiyama,S., Tani,T., Ohshima,Y., Frendewey,D. and Beach,D. (1996) Mol. Biol. Cell, 7, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Habara Y., Urushiyama,S., Tani,T. and Ohshima,Y. (1998) Nucleic Acids Res., 26, 5662–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns C.G., Ohi,R., Krainer,A.R. and Gould,K.L. (1999) Proc. Natl Acad. Sci. USA, 96, 13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi M. and Yanagida,M. (1993) Mol. Biol. Cell, 4, 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt H., Richert,K., Drakas,R.A. and Käufer,N.F. (1999) Genetics, 153, 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wentz-Hunter K. and Potashkin,J. (1996) Nucleic Acids Res., 24, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney R., Wentz-Hunter,K., Schmidt,H. and Potashkin,J. (1997) Curr. Genet., 32, 323–330. [DOI] [PubMed] [Google Scholar]
- 13.Gozani O., Potashkin,J. and Reed,R. (1998) Mol. Cell Biol., 18, 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groß T., Lützelberger,M., Wiegmann,H., Klingenhoff,A., Shenoy,S. and Käufer,N.F. (1997) Nucleic Acids Res., 25, 1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lützelberger M., Groß,T. and Käufer,N.F. (1999) Nucleic Acids Res., 27, 2618–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson G.J., Bach,M., Lührmann,R. and Beggs,J.D. (1989) Nature, 342, 819–821. [DOI] [PubMed] [Google Scholar]
- 17.Pinto A.L. and Steitz,J.A. (1989) Proc. Natl Acad. Sci. USA, 86, 8742–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins C.A. and Guthrie,C. (1999) Genes Dev., 13, 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potashkin J., Li,R. and Frendewey,D. (1989) EMBO J., 8, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abovich N., Liao,X.C. and Rosbash,M. (1994) Genes Dev., 8, 843–854. [DOI] [PubMed] [Google Scholar]
- 21.Zamore P.D. and Green,M.R. (1989) Proc. Natl Acad. Sci. USA, 86, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S., Romfo,C.M., Nilsen,T.W. and Green,M.R. (1999) Nature, 402, 832–835. [DOI] [PubMed] [Google Scholar]
- 23.Zorio D.A. and Blumenthal,T. (1999) Nature, 402, 835–838. [DOI] [PubMed] [Google Scholar]
- 24.Merendino L., Guth,S., Bilbao,D., Martinez,C. and Valcárcel,J. (1999) Nature, 402, 838–841. [DOI] [PubMed] [Google Scholar]
- 25.Prabhala G., Rosenberg,G.H. and Käufer,N.F. (1992) Yeast, 8, 171–182. [DOI] [PubMed] [Google Scholar]
- 26.Käufer N.F., Simanis,V. and Nurse,P. (1985) Nature, 318, 78–80. [DOI] [PubMed] [Google Scholar]
- 27.Yan D., Perriman,R., Igel,H., Howe,K.J., Neville,M. and Ares,M.,Jr (1998) Mol. Cell. Biol., 18, 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q. and Sharp,P.A. (1996) Science, 274, 605–610. [DOI] [PubMed] [Google Scholar]
- 29.Reed R. (1996) Curr. Opin. Genet. Dev., 6, 215–220. [DOI] [PubMed] [Google Scholar]
- 30.Manley J.L. and Tacke,R. (1996) Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 31.Tang Z., Kuo,T., Shen,J. and Lin,R.-J. (2000) Mol. Cell. Biol., 20, 816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Z., Yanagida,M. and Lin,R.-J. (1998) J. Biol. Chem., 273, 5963–5969. [DOI] [PubMed] [Google Scholar]
- 33.Berglund J.A., Chua,K., Abovich,N., Reed,R. and Rosbash,M. (1997) Cell, 89, 781–787. [DOI] [PubMed] [Google Scholar]
- 34.Abovich N. and Rosbash,M. (1997) Cell, 89, 403–412. [DOI] [PubMed] [Google Scholar]
- 35.Grey M., Dusterhoft,A., Henriques,J.A. and Brendel,M. (1996) Nucleic Acids Res., 24, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley J.P. and Guthrie,C. (1999) Mol. Cell, 3, 55–64. [DOI] [PubMed] [Google Scholar]
- 37.Murray M.V., Kobayashi,R., and Krainer,A.R. (1999) Genes Dev., 13, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Chua K. and Reed,R. (1999) Genes Dev., 13, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chua K. and Reed,R. (1999) Nature, 402, 207–210. [DOI] [PubMed] [Google Scholar]
- 41.Forsburg S.L. (1999) Trends Genet. Sci., 15, 340–344. [DOI] [PubMed] [Google Scholar]