Abstract
An increase in the dose of the Su(var)3-7 locus of Drosophila augments heterochromatin-promoted variegated silencing. The deduced protein sequence of Su(var)3-7 reveals seven widely spaced zinc fingers. We found that Su(var)3-7 has affinity for DNA in vitro and that the minimal protein sequence requirement for DNA binding is any module containing two zinc fingers and the interval between them. As Su(var)3-7 is a heterochromatin-associated protein, we tested its affinity for various satellite DNA sequences in vitro. The AATAT and 353-bp elements have the highest affinity. If affinity for satellite DNAs contributes to the presence of Su(var)3-7 in heterochromatin, a general affinity for DNA, or sequences yet to be determined, suggests a function in the genomic silencing of position-effect variegation: expansion of heterochromatin, whether continuous by spreading or discontinuous by pairing with sequence elements scattered through euchromatin, could use the affinity of Su(var)3-7 for DNA.
INTRODUCTION
In 1930, Müller described a number of X-ray-induced mutants characterized by a mosaic phenotype. These mutants are chromosomal rearrangements relocating the variegating genes next to pericentric heterochromatin (Schultz, 1936). Two types of models were put forward to explain the phenomenon of position-effect variegation (PEV). Schultz was the first to propose that inactivation by heterochromatin could spread from the rearrangement breakpoint, and Zuckerkandl (1974) proposed that the spreading depends on the dose of building blocks of heterochromatin. The other school of thought also has ancient roots. Pontecorvo suggested in 1944 that heterochromatin could form from folding of DNA at any region comprising repetitive sequences (Pontecorvo, 1944). Recent observations of discontinuous expansion of silencing and of long-distance effects raise the possibility that sequence elements dispersed in the genome could anchor the formation of heterochromatin by association with pericentric heterochromatin (Dorer and Henikoff, 1994; Talbert and Henikoff, 2000). Dominant modifiers of PEV provide an avenue to identifying the proteins involved in PEV (Reuter and Spierer, 1992). Indeed, three loci with a haplo-suppressor triplo-enhancer phenotype were found to encode heterochromatin-associated proteins. These include Su(var)2-5 (Eissenberg et al., 1990), Su(var)3-7 (Cléard et al., 1997) and Su(var)3-9 (Tschiersch et al., 1994; Aagaard et al., 1999).
Attempts at explaining PEV require that at least one component binds to DNA. Silencing should reflect the amounts of this component, and therefore the haplo-suppressor and triplo-enhancer modifier loci are first-choice candidates. Moreover, as PEV affects different genes in a variety of rearrangements, this component should bind DNA with affinity to a wide range of sequences. Su(var)3-7 is a good candidate, as its amino-acid sequence encodes seven zinc fingers, which are motives known to bind DNA (Harrison, 1991). In this study, we found that Su(var)3-7 associates with DNA in vitro and that each pair of zinc fingers forms a DNA-binding domain.
RESULTS
Su(var)3-7 binds DNA
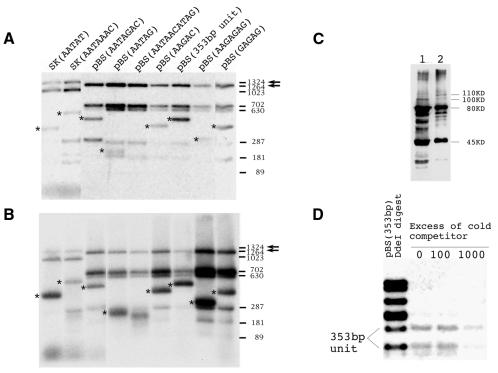
To test the affinity of Su(var)3-7 for DNA, we constructed a glutathione S-transferase (GST) Su(var)3-7 fusion protein. The construct encodes 1047 of the 1169 amino acids of the complete protein. One hundred and twenty-one amino acids from the N-terminus are missing, but they are known to be dispensable for the modifier of variegation function (Reuter et al., 1990; Cléard et al., 1995). The fragment was cloned in an expression vector and introduced in Escherichia coli. The expressed fusion protein is shorter than expected, probably because of premature arrest of translation. From its size on western blot (Figure 1), we estimate that the major protein component is interrupted before the sixth zinc finger. Nonetheless, the protein expressed contains five of the seven zinc fingers, and we decided to use it for DNA-binding assays in vitro. In a preliminary test, we determined by southwestern blot that the GST–Su(var)3-7 protein retains Drosophila genomic DNA (data not shown). As Su(var)3-7 is a heterochromatin-associated protein (Cléard et al., 1997), we tested potential targets, namely satellite DNA sequences. Nine plasmids containing different Drosophila satellite sequence DNA (Lohe and Brutlag, 1986) were digested to separate the insert from plasmid DNA and were labelled. The mixture was incubated with the GST–Su(var)3-7 protein bound to a solid matrix through its GST moiety. Figure 1 shows an autoradiogram of the fragments generated by digestion (A) compared with the same fragments selected by the Su(var)3-7 protein (B). In the absence of a non-specific competitor such as poly(dI–dC), binding is detected with essentially all the DNA fragments, but there is a strong enrichment for some of the satellite DNA. Enrichment was quantified by scanning the autoradiograms to verify the qualitative differences visible in Figure 1 (data not shown). Except for the repeats of the sequence motif GAGAG, the satellite DNA insertions are preferentially selected by the fusion protein. Affinity of Su(var)3-7 is 8-fold stronger for AATAT repeats, and the 353-bp unit, than for AATAAAC repeats. Figure 1 also shows a competition experiment designed to further assess the binding affinity for satellite DNA compared with plasmid DNA. Increasing amounts of cold competitor (the same, but unlabelled digest) were added. The binding to plasmid is readily competed out, but association of Su(var)3-7 to the satellite DNA fragments is still visible with a 1000-fold excess of identical cold fragments.
Fig. 1. Binding of different DNA probes by GST–Su(var)3-7. (A) Each lane of a 2.5% agarose gel corresponds to a digest of a 32P-labelled recombinant plasmid. The satellite DNA insert is marked by a star and identified above each lane. The arrows indicate the plasmid fragments used to estimate the relative affinity. (B) Fragments selected by the GST–Su(var)3-7 fusion protein in the absence of poly(dI–dC). Same reaction and probe as in (A). (C) Western blot of the GST–Su(var)3-7 constructs containing seven (lane 2) or six (lane 1) zinc fingers. Both lanes show a minor band of the correct size (110 and 100 kDa, respectively). The major products are truncated proteins, one of 80 kDa (corresponding to a protein with five zinc fingers) and one of 45 kDa (ending just after the second finger). (D) Competition for binding of labelled plasmid pBS(353 bp) by GST–Su(var)3-7. The binding reaction mixtures contain poly(dI–dC) at a final concentration of 0.1 µg/µl and ∼10 ng of labelled plasmid, and 0, 1.0 or 10.0 µg of the same, but unlabelled, digested plasmid was added. In this experiment, the pBS(353 bp) plasmid was cut with EcoRI, HindIII and DdeI, the latter splitting the insert into two fragments.
A module of two zinc fingers is necessary and sufficient for DNA binding
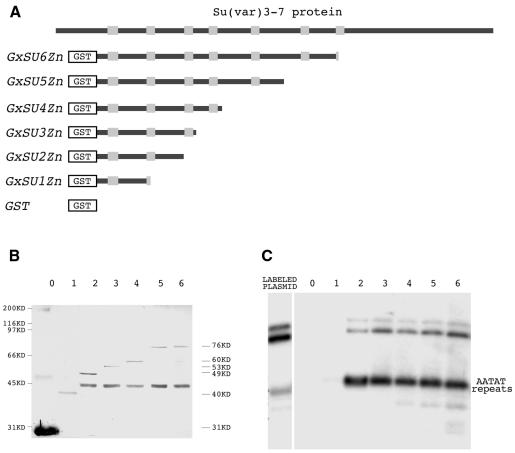
The deduced sequence of Su(var)3-7 reveals a number of repeated motifs (Reuter et al., 1990; Cléard et al., 1995). Among them, the seven zinc fingers separated by large intervals stand out. We have shown above that a segment of the protein comprising five fingers does bind DNA. In order to delineate the protein sequence required for DNA binding, we constructed a panel of proteins with a stepwise reduction in length and consequently in the number of zinc fingers (Figure 2A). The DNA sequence tested was the AATAT repeat. Figure 2C illustrates the results. Stepwise reduction from five down to two fingers does not affect significantly the binding. In contrast, the fragment containing only one complete finger, and the spacer between this one and the next, does not bind the satellite DNA. The same was observed with a segment comprising the last finger, namely the seventh, and the entire C-terminus part of the protein (data not shown).
Fig. 2. Binding of the AATAT satellite by different GST–Su(var)3-7 derivatives. (A) The Su(var)3-7 protein is represented as a black bar within which the zinc fingers appear as grey boxes. (B) Western blot of the different recombinant proteins schematized in (A), bound to glutathione–Sepharose 4-B. Lanes 0–6 correspond to proteins with zero to six zinc fingers revealed with anti-GST antibody. (C) Autoradiogram of the agarose gel after electrophoresis of the 300-bp AATAT repeats fragment retained by the same proteins. The amounts of protein fragments in the binding assay were those deposited on the western blot in (B), and the experiment was performed in the presence of 0.1 µg/µl of the non-specific competitor poly(dI–dC).
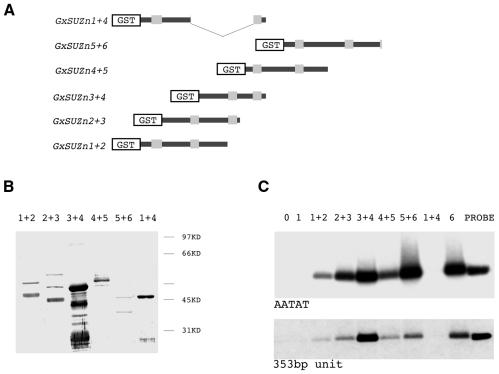
From these results, one could argue that the requirement is not for a pair of fingers, but that the second or the seventh finger plays a specific role. To refine the analysis, we designed five additional constructs representing a collection of different pairs of zinc fingers, including an artificial one, made of the first and the fourth finger (Figure 3A). The segments were tested for interaction with the AATAT and the 353-bp satellite DNAs. The results are shown in Figure 3. Any pair of zinc fingers, together with the spacer sequence, binds efficiently both the AATAT and the 353-bp satellites. Differences in intensity of the bands seem to be parallel to the amounts of protein in the assay (data not shown). There is one notable exception to the rule of the pair of fingers. The construct GxSUZn1+4 comprises two zinc fingers, but, in contrast to all others of its kind, it is unable to retain DNA. In this construct, however, the cystein doublet at the base of the finger has become a cystein triplet because of the cloning strategy. The sequence -C-X-X-C- is mutated to -C-X-X-C-X-X-C-, and this probably renders the finger non-functional. This result reinforces the observation that a DNA-binding module must comprise two functional fingers.
Fig. 3. Binding of the AATAT and 353-bp satellites by different zinc finger pairs derived from Su(var)3-7. (A) Different parts of the fusion proteins are schematized as in Figure 2. (B) Western blot of the proteins corresponding to (A), after purification on glutathione–Sepharose 4-B, revealed with an anti-GST antibody. Lanes are named according to the pair of fingers present in the fusion proteins. (C) Autoradiography of the agarose gel after electrophoresis of the 300-bp AATAT repeats fragment and the 500-bp insert containing the 353-bp unit bound by the GST-tagged protein constructs. The amounts of protein fragments in the binding assay were those deposited on the western blot in (B), and the experiment was carried out in the presence of 0.1 µg/µl of the non-specific competitor poly(dI–dC).
Defining a consensus DNA sequence for binding
We addressed this question using two approaches. First, we performed footprints experiments using the 353-bp unit, which also contains three AATAT repeats. The DNA fragments were protected by an increase in protein concentration, but we could not detect a specific footprint (data not shown). Secondly, we tried to select fragments of Drosophila genomic DNA with a Su(var)3-7 fragment comprising two zinc fingers according to Cuvier et al. (1998). Following five cycles of selection and PCR amplification, 60 fragments were sequenced (data not shown). Although repeated elements were predominant, we were unable to define a consensus sequence.
DISCUSSION
PEV was first characterized as an effect in cis of blocks of pericentric heterochromatin onto euchromatic genes (Weiler and Wakimoto, 1995). In addition to its cytological definition, the main characteristic of heterochromatin is its concentration of satellite DNA and of middle repetitive elements (Lohe and Brutlag, 1986). With the exception of genes naturally embedded in heterochromatin, the phenotypes in PEV result from the silencing of genes normally in euchromatin. The different models invoke linear expansion of heterochromatin, discontinuous expansion allowing for ‘skipping genes’ and sequestration at distance by pairing of sequences within euchromatin with others of their kind in heterochromatin (Weiler and Wakimoto, 1995). These models could be reconciled by assuming that there are sequences scattered throughout the genome that have a potential affinity for association with heterochromatin. When in the proximity of a large block of heterochromatin, possibly because of random movements or higher concentrations of associated ‘nucleating’ factors, these sequences could bind heterochromatin constituents and eventually fuse with the bulk of pericentric heterochromatin. A higher concentration of components would favour these interactions and the resulting expansion of the repressed state. The candidate to play this role is obviously the haplo-suppressor triplo-enhancer of PEV.
The results reported here lead us to propose that Su(var)3-7 is one of the links between these scattered sequences and the pericentric heterochromatin. However, there are other candidates. HP1 seems to have a non-specific affinity for DNA and nucleosomes (Zhao et al., 2000) and interacts with histones in mammals (Nielsen et al., 2001). In Drosophila, HP1 associates with a DNA-binding protein ORC (Shareef et al., 2001) and induces ectopic chromosomal loops (Seum et al., 2001). Su(var)3-9 might also play a central role, as its mutations are epistatic to those in HP1 and Su(var)3-7 (G. Reuter, personal communication). Interpretation of this result is delicate, however, as the authors used Su(var)2-5 and Su(var)3-7 haplo-insufficient mutants. Finally, DDP1, a single-stranded nucleic-acid-binding protein, associates with satellite DNA and co-localizes with HP1 (Cortés et al., 1999), and Prod, a satellite DNA-binding protein, was also proposed to function in heterochromatin compaction (Török et al., 2000).
The preference of Su(var)3-7 in vitro for some satellite DNA sequences, followed by a lesser affinity for others, and an apparent general affinity for DNA, suggests a mechanism whereby the increase in dose might cause the expansion of heterochromatin-induced silencing. At the ‘physiological’ dose, Su(var)3-7 associates only with heterochromatin sequences, but at higher concentrations it binds to other sequences in the genome and to other partners to mediate heterochromatin formation. This nucleation is facilitated by proximity of large blocks of pericentric heterochromatin, thus explaining the spreading effect. This mechanism allows for the discontinuous expansion observed in a number of instances (for cytological evidence, see Belyaeva and Zhimulev, 1991; for genetic evidence, see Talbert and Henikoff, 2000, and references therein). Variegation induced by insertion of a tandem array of repeated elements in euchromatin (Dorer and Henikoff, 1994) is also explained if Su(var)3-7 has a particularly good affinity for them. Future research should now be addressed at identifying targets of Su(var)3-7 in euchromatin. For example, we have shown that Su(var)3-7 associates at the locus of a large insertion of the AAGAG satellite DNA, a sequence we have not tested in vitro, and affects variegation of a neighbouring gene (Delattre et al., 2000).
A second interesting finding in this report is the modular structure of Su(var)3-7. From its deduced sequence, Su(var)3-7 encodes seven zinc fingers rather regularly spaced over 700 amino acids, starting from the N-terminus of the 1169 amino acid long protein (Cléard et al., 1995). These fingers are very similar and are each preceded by a tryptophan motif. By testing a variety of segments of the protein for binding to DNA, we have determined that the minimal requirement is a pair of fingers separated by a spacer sequence. Although we have not tested all the possible combinations, we propose that each finger and its immediate neighbourhood is exchangeable with any other and that a combination of two with any of the spacers provides DNA-binding properties. Obviously, there are a number of other constraints to accommodate. For example, Su(var)3-7 is part of a larger complex containing HP1 (Cléard et al., 1997; Delattre et al., 2000). The affinity of Su(var)3-7 in vivo may differ from that observed in vitro by its participation in a large complex of proteins. Another speculation to assess is an alternative role for Su(var)3-7: the multimodular structure of Su(var)3-7 could be instrumental in the DNA compaction characteristic of silent heterochromatin.
METHODS
The Su(var)3-7 fusion protein. Various fragments of the Su(var)3-7 cDNA (Cléard et al., 1995) were cloned in-frame downstream of the gene encoding the enzyme GST in the vectors pGEX-3X, pGEX-4T-1 and pGEX-4T-2 from Pharmacia. The resulting clones were introduced by transformation in the E. coli BL21 strain. Expression was induced by 0.1 mM IPTG (final concentration). After 4 h of growth, cells were collected by centrifugation and suspended in 5% of the initial volume of ice-cold PBS prior to disruption by mild sonication. Lysis was completed by incubation for 30 min on ice after the addition of Triton X-100 to a final concentration of 1%. Debris were removed by centrifugation at 10 000 r.p.m. for 10 min at 4°C. For binding on glutathione–Sepharose 4-B (Pharmacia) and purification, the lysate was gently mixed for 30 min at room temperature with beads (50 µl per ml of lysate) equilibrated in PBS. Unbound protein was removed by several washes with PBS. All proteins used in binding assays were analysed by SDS–PAGE and western blot with anti-GST and/or anti-Su(var)3-7 antibodies.
DNA probes and DNA binding assays. Plasmids containing satellite DNA sequences (Lohe and Brutlag, 1986) were digested to separate the insert. The digestion mixture was treated with shrimp alkaline phosphatase followed by heat inactivation. When an isolated insert instead of the mixture of fragments was used as a probe, the reaction was submitted to electrophoresis on agarose gel and the fragment of interest purified by Geneclean (Bio 101). For the labelling reaction, 50 ng of fragment or whole digest were incubated for 30 min at 37°C in 50 µl of 1 × PNK buffer, 50 mM DTT, 10 µCi [γ32P]dATP and 10 U of polynucleotide kinase. The probe was purified on a spin column. A typical reaction was performed in 25 µl final volume using 2.5 µl of matrix bound protein and 22.5 µl of a mixture containing 10 000 c.p.m. of radiolabelled fragment (∼0.5 ng) and 2.5 µg poly(dI–dC) in NETN (20 mM Tris–Cl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40) (Kaelin et al., 1991). The reaction was incubated for 30 min at room temperature. After removing the unbound material, beads were washed three times with 1 ml of NETN containing NaCl at a final concentration of 0.2 M. After the last wash and addition of 40 µl of loading buffer (10 mM Tris–Cl pH 7.5, 0.025% Bromophenol Blue, 3% glycerol, 0.1% SDS), beads were heated for 10 min at 80°C, pelleted by centrifugation and the supernatant loaded onto a 2.5% agarose gel. After electrophoresis, the gel was dried and exposed for autoradiography.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rakesh Mishra for his expert advice on binding assays. This work was supported by grants from the Swiss National Science Foundation and by the State of Geneva, Switzerland.
REFERENCES
- Aagaard L. et al. (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva E.S. and Zhimulev, I.F. (1991) Cytogenetic and molecular aspects of position-effect variegation in Drosophila melanogaster. III. Continuous and discontinuous compaction of chromosomal material as a result of position effect variegation. Chromosoma, 100, 453–466. [DOI] [PubMed] [Google Scholar]
- Cléard F., Matsarskaia, M. and Spierer, P. (1995) The modifier of position-effect variegation Suvar(3)7 of Drosophila: there are two alternative transcripts and seven scattered zinc fingers, each preceded by a tryptophan box. Nucleic Acids Res., 23, 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléard F., Delattre, M. and Spierer, P. (1997) SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J., 16, 5280–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A., Huertas, D., Fanti, L., Pimpinelli, S., Marsellach, F.X., Pina, B. and Azorin, F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J., 18, 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O., Hart, C.M. and Laemmli, U.K. (1998) Identification of a class of chromatin boundary elements. Mol. Cell. Biol., 18, 7478–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Tonka, C. and Spierer, P. (2000) The genomic silencing of position-effect variegation in Drosophila melanogaster: interaction between the heterochromatin-associated protein Su(var)3-7 and HP1. J. Cell Sci., 113, 4253–4261. [DOI] [PubMed] [Google Scholar]
- Dorer D.R. and Henikoff, S. (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell, 77, 993–1002. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., James, T.C., Foster-Hartnett, D.M., Hartnett, T., Ngan, V. and Elgin, S.C. (1990) Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 87, 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. (1991) A structural taxonomy of DNA-binding domains. Nature, 353, 715–719. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. Jr, Pallas, D.C., Decaprio, J.A., Kaye, F.J. and Livingston, D.M. (1991) Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell, 64, 521–532. [DOI] [PubMed] [Google Scholar]
- Lohe A.R. and Brutlag, D.L. (1986) Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 83, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.J. (1930) Types of viable variations induced by X-rays in Drosophila. J. Genet., 22, 299–334. [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani, M., Ortiz, J.A., Remboutsika, E., Chambon, P. and Losson, R. (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell, 7, 729–739. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G. (1944) Structure of heterochromatin. Nature, 153, 365–367. [Google Scholar]
- Reuter G. and Spierer, P. (1992) Position effect variegation and chromatin proteins. BioEssays, 14, 605–612. [DOI] [PubMed] [Google Scholar]
- Reuter G., Giarre, M., Farah, J., Gausz, J., Spierer, A. and Spierer, P. (1990) Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature, 344, 219–223. [DOI] [PubMed] [Google Scholar]
- Schultz J. (1936) Variegation in Drosophila and the inert chromosome regions. Proc. Natl Acad. Sci. USA, 22, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C., Delattre, M., Spierer, A. and Spierer, P. (2001) Ectopic HP1 promotes chromosome loops and variegated silencing in Drosophila. EMBO J., 20, 812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef M.M., King, C., Damaj, M., Badagu, R., Huang, D.W. and Kellum, R. (2001) Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol. Biol. Cell, 12, 1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B. and Henikoff, S. (2000) A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics, 154, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török T., Gorjanacz, M., Bryant, P.J. and Kiss, I. (2000) Prod is a novel DNA-binding protein that binds to the 1.686 g/cm3 10 bp satellite repeat of Drosophila melanogaster. Nucleic Acids Res., 28, 3551–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann, A., Krauss, V., Dorn, R., Korge, G. and Reuter, G. (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J., 13, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler K.S. and Wakimoto, B.T. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 577–605. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk, T., Allis, C.D. and Eissenberg, J.C. (2000) Heterochromatin protein 1 (HP1) binds to nucleosomes and DNA in vitro. J. Biol. Chem., 275, 28332–28338. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E. (1974) A possible role of ‘inert’ heterochromatin in cell differentiation. Action of and competition for ‘locking’ molecules. Biochimie, 56, 937–954. [DOI] [PubMed] [Google Scholar]