Abstract
The ubiquitin–proteasome system catalyses the immediate destruction of misfolded or impaired proteins generated in cells, but how this proteolytic machinery recognizes abnormality of cellular proteins for selective elimination remains elusive. Here, we report that the C-terminus of Hsc70-interacting protein (CHIP) with a U-box domain is an E3 ubiquitin-ligase collaborating with molecular chaperones Hsp90 and Hsc70. Thermally denatured firefly luciferase was multiubiquitylated by CHIP in the presence of E1 and E2 (Ubc4 or UbcH5c) in vitro, only when the unfolded substrate was captured by Hsp90 or Hsc70 and Hsp40. No ubiquitylating activity was detected in CHIP lacking the U-box region. CHIP efficiently ubiquitylated denatured luciferase trapped by the C-terminal region of Hsp90, which contains a CHIP binding site. CHIP also showed self-ubiquitylating activity independent of target ubiquitylation. Our results indicate that CHIP can be regarded as ‘a quality-control E3’ that selectively ubiquitylates unfolded protein(s) by collaborating with molecular chaperones.
INTRODUCTION
Ubiquitin (Ub) is covalently attached to substrate proteins via an isopeptide linkage between the C-terminal Gly of Ub and the ɛ-NH2 group of Lys of the acceptor substrate by a cascade system, consisting of activating (E1), conjugating (E2) and/or ligating (E3) enzymes (Hershko et al., 2001). Modification of cellular proteins by Ub targets them for proteolytic attack by the 26S proteasome (a eukaryotic ATP-dependent protease complex) (Voges et al., 1999). Of these ubiquitylating enzymes, E3s are believed to exist as molecules with a large diversity, presumably in hundreds of species, and play a critical role in the selection of substrate for degradation, because each distinct E3 usually binds a protein substrate with a degree of selectivity for ubiquitylation (Pickart, 2001). To date, all known E3s are classified into two types: the HECT-type E3 and the RING-finger type E3 (Weissman, 2001). The former group has a domain capable of binding Ub as a thioester bond, termed ‘HECT’, which harbours an ∼350 amino acid region of homology to the E6-AP (a HECT-type E3 first identified) C-terminus. The RING-type E3 is the general term for Ub ligases with a RING-finger motif(s) containing a Cys-rich motif capable of binding Zn2+. It is plausible that RING-finger type E3s are not covalently bound to Ub and that RING fingers recruit E2s to the vicinity of proteins to be ubiquitylated and thus mediate ubiquitylation by facilitating the direct transfer of Ub from E2-Ub to target the Lys residue.
Intriguingly, Koegl et al. (1999) frecently reported a new class of ubiquitylating enzyme involved in multi-Ub chain assembly (termed E4) that is identical to yeast Ufd2. E4 belongs to a family of proteins with a common domain in ubiquitylation called ‘U-box’, which is a derived version of the RING-finger domain lacking the metal-chelating residues (Aravind and Koonin, 2000). Hatakeyama et al. (2001) reported that some members of the U-box family proteins, including E4 and CHIP (C-terminus of Hsc70-interacting protein), show self-ubiquitylating activity. Of these, CHIP was originally discovered as a co-chaperone with a new tetratricopeptide repeat (TPR)-containing protein that negatively regulates the ATPase and chaperone activities of Hsc70 (Ballinger et al., 1999). Furthermore, it was shown that CHIP is involved in the ubiquitylation of the cystic-fibrosis transmembrane-conductance regulator (CFTR) in the endoplasmic reticulum-associated degradation (ERAD) pathway and functions as a co-chaperone that converts Hsc70 from a protein-folding machine into a degradation factor involved in endoplasmic reticulum (ER) quality control (Meacham et al., 2001). At the same time, CHIP was found to induce ubiquitylation of the glucocorticoid receptor bound to Hsp90 for proteasomal degradation, modulating protein triage decisions that regulate the balance between protein folding and degradation of chaperone substrates (Connell et al., 2001). Based on these findings, CHIP is considered as a new category of E3 enzyme responsible for quality control of cellular proteins linked to the function of molecular chaperones (McClellan and Frydman, 2001). More recently, Jiang et al. (2001) reported that CHIP is a U-box-dependent ubiquitin ligase that targets Hsc70 for ubiquitylation.
However, there is no experimental evidence to show that CHIP indeed acts as an E3 Ub-protein ligase capable of distinguishing non-native states from native states of target proteins for ultimate ubiquitylation. In the present study, we provide direct in vitro evidence that CHIP selectively ubiquitylates thermally denatured substrate captured by molecular chaperones, such as Hsp90 and Hsc70. CHIP appears to function as ‘quality-control E3’ involved in the selective ubiquitylation of target proteins by recognizing the non-native state in a molecular chaperone-assisted manner. Our findings indicate that molecular chaperones assisting protein folding are also capable of linking substrate (unfolded protein) to the Ub pathway (CHIP:E3) for efficient ubiquitylation.
RESULTS
CHIP ubiquitylates thermally denatured luciferase captured by Hsp90
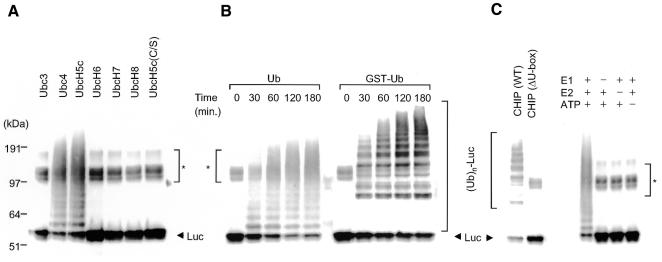
To directly demonstrate the E3 activity in CHIP, we devised a fully reconstituted system for ubiquitylation of thermally denatured firefly luciferase in the presence of Hsp90. We produced all components involved in these systems as recombinant or purified proteins (see Methods). CHIP was incubated at 30°C for 2 h with E1 and various species of E2 in the presence of Ub, ATP and firefly luciferase that has been pretreated with Hsp90 at 45°C for 5 min (called here heat shock). For this heat treatment, we previously reported that Hsp90 captures unfolded luciferase (Minami et al., 2000). As shown in Figure 1A, ubiquitylation of heat-denatured luciferase was detected as high-molecular-mass smear bands by western blotting with an anti-luciferase antibody after separation by SDS–PAGE when Ubc4 and UbcH5c were used as E2 enzymes. No ubiquitylating activity by CHIP was observed when the active site mutant UbcH5c(C/S) (Cys at position 85 was replaced by Ser) was used (Figure 1A, right lane). These results indicate that CHIP has ubiquitylating activity towards unfolded proteins. In contrast, all other E2s examined, such as Ubc3, UbcH6, UbcH7 and UbcH8, did not function as a partner of CHIP.
Fig. 1. CHIP is a U-box ubiquitin E3 ligase collaborating with Ubc4 and UbcH5c. (A) Thermally denatured luciferase with Hsp90 was subjected to in vitro ubiquitylation assay by CHIP, as described in Methods. The ubiquitylation assay of luciferase pretreated at 45°C for 5 min was performed at 30°C for 2 h with CHIP, Ub and various Ub-conjugating enzymes (E2s). Ubiquitylation of luciferase was detected by western blotting with an anti-luciferase antibody. (B) Time course of luciferase ubiquitylation by CHIP. Ubiquitylation of luciferase by CHIP was carried out as a function of time as in (A) using Ub (left panel) or GST–Ub (right panel). (C) The U-box structure is essential for E3 activity of CHIP. The ubiquitylation assay of luciferase was performed for 2 h as in (B, right), except for the use of GST–CHIP lacking the U-box region (ΔU-box) (left panel). For the control experiment, ubiquitylation of luciferase was conducted by omission of E1, E2 or ATP from the reaction mixtures used in (A) (right panel). Unmodified luciferase (designated Luc) is shown by an arrowhead. Ubiquitylated luciferase with a high molecular mass is shown as (Ub)n-Luc. The regions marked by asterisks represent non-specific bands.
The level of multiubiquitylated luciferase increased in a time-dependent manner, coincident with progressive decrease of unmodified luciferase (Figure 1B, left panel). To confirm that these multiple bands are indeed Ub-modified forms of luciferase, we used GST–Ub instead of Ub. As shown in Figure 1B (right panel), anti-luciferase reactive bands shifted to higher molecular mass positions in a time-dependent manner. No ubiquitylating activity was observed when CHIP lacking the C-terminal U-box region (ΔU-box) was used (Figure 1C, left panel), indicating that the U-box region of CHIP plays an essential role in the ubiquitylating activity. In addition, ubiquitylating activity was not detected without E1, E2 or ATP (Figure 1C, right panel).
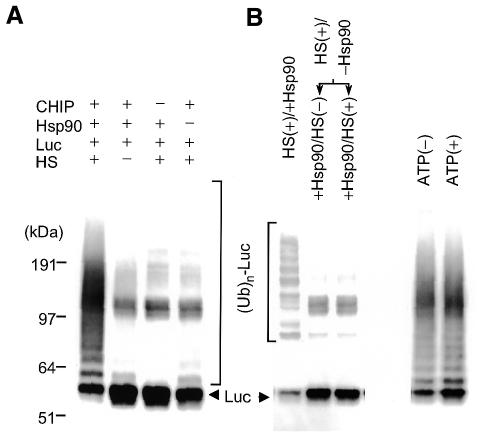
Next, we examined whether Hsp90 is essential for CHIP-dependent ubiquitylation of heat-denatured luciferase. As shown in Figure 2A, no appreciable ubiquitylating activity by CHIP was detected under heat treatment of luciferase without Hsp90 or without its heat treatment even in the presence of Hsp90, indicating that CHIP does not target free luciferase, irrespective of the native or non-native state. CHIP ubiquitylated thermally denatured luciferase only when it was heat denatured in the presence of Hsp90 (Figure 2A). In addition, the ubiquitylating activity evoked by CHIP was negligible or not seen when Hsp90 was added after luciferase was heat denatured in the absence of Hsp90, even though Hsp90 was heat treated prior to addition (Figure 2B, left panel). Addition of ATP during heat shock of luciferase had no effect on the ubiquitylating activity, indicating that ATP is not required for trapping of unfolded luciferase by Hsp90 (Figure 2B, right panel). These results indicate that CHIP does not target severely aggregated luciferase, which was produced by heat shock without Hsp90 and unable to bind to Hsp90, and that unfolded luciferase captured by Hsp90 becomes a substrate for CHIP.
Fig. 2. CHIP ubiquitylates heat-denatured luciferase captured by Hsp90. (A) Effects of Hsp90 and/or heat shock (HS) on CHIP-dependent luciferase (Luc) ubiquitylation activity. The ubiquitylation assay of luciferase pretreated at 45°C for 5 min at indicated combinations was performed at 30°C for 2 h with CHIP, Ub and UbcH5c (left panel) as in Figure 1A. (B) Effects of Hsp90 on the ubiquitylation reaction by CHIP. For luciferase that had been heat denatured in the absence of Hsp90, heat untreated or pretreated Hsp90 was added to the reaction mixtures (middle and right lanes, left panel). Control assay was conducted using luciferase heat denatured with Hsp90 (left lane, left panel). Heat treatment of luciferase with Hsp90 was carried out in the presence or absence of ATP prior to the ubiquitylation assay (right panel). Ubiquitylation assay was performed using GST–Ub. Symbols and abbreviations used are similar to those in Figure 1.
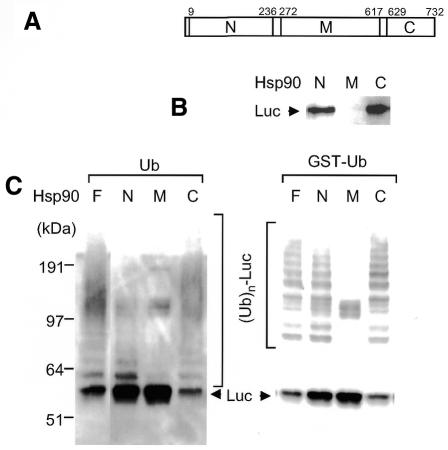
Hsp90 has two independent sites for associating target unfolded proteins: i.e. the N-terminal ATPase domain and the C-terminal domain (Young et al., 1997; Minami et al., 2001). In addition, CHIP is known to be associated with the C-terminal region of Hsp90 via its N-terminal TPR motif (Ballinger et al., 1999). Therefore, to test which domain of Hsp90 co-operates with CHIP activity, we dissected Hsp90 into three parts: i.e. the N-terminal ATPase domain (N), the middle region (M) and the C-terminal region containing the TPR domain (C), as depicted in Figure 3A. These fragments were expressed as GST-fused proteins. Approximately equivalent amounts of luciferase were associated with the N- or C-terminal, but not with the middle region of Hsp90 (Figure 3B), as reported recently (Minami et al., 2001). When the C-terminal region of Hsp90 was used for heat shock, CHIP caused ubiquitylation of unfolded luciferase, as native Hsp90 did (Figure 3C, left panel), indicating that the C-terminal domain is sufficient to trap both denatured protein and CHIP for effective ubiquitylation. The ATPase activity of Hsp90 is not essential for ubiquitylation of unfolded substrates captured by Hsp90, but faint ubiquitylation bands were seen when the N-terminal region of Hsp90 was used. To confirm this conclusion, we repeated the same experiment using GST–Ub instead of Ub. As shown in Figure 3C (right panel), significant ubiquitylating activity was observed even when the N-terminal region of Hsp90 was used. However, this activity was considerably lower compared with that obtained by the C-terminal region of Hsp90 (Figure 3C). This is probably because CHIP does not bind to the N-terminal region of Hsp90, and thus free CHIP may ineffectively target the N-terminal region-associated unfolded luciferase, which encounters CHIP in the solution by chance. No appreciable activity was observed when the middle region of Hsp90 was used, irrespective of the usage of Ub or GST–Ub. These results indicate that association of the substrate with either the N- or C-terminal region of Hsp90 is essential for ubiquitylation of the substrate by CHIP.
Fig. 3. Role of Hsp90 domains on CHIP-dependent ubiquitylating activity. (A) Schematic representation of Hsp90. The N-terminal (N), middle (M) and C-terminal (C) regions were expressed as GST-fused proteins. (B) Association of luciferase with GST-fused N-, M- and C-fragments of Hsp90 after thermal treatment. After GST-fused Hsp90 fragments (11 µM) were heated at 45°C for 5 min with 0.26 µM luciferase in 50 mM Tris–HCl buffer pH 7.5, they were pulled down with GSH beads. The resulting precipitates were subjected to SDS–PAGE followed by western blotting with an anti-luciferase antibody. (C) Ubiquitylation of thermally denatured luciferase by deletion mutants of Hsp90 and CHIP. Eleven micromoles of full-length Hsp90 (F) and GST-fused N-, M- and C-fragments of Hsp90 were heated with luciferase at 45°C for 5 min and used for ubiquitylation assay using Ub or GST–Ub as indicated (left and right panels).
CHIP ubiquitylates heat-denatured luciferase trapped by Hsc70 and Hsp40
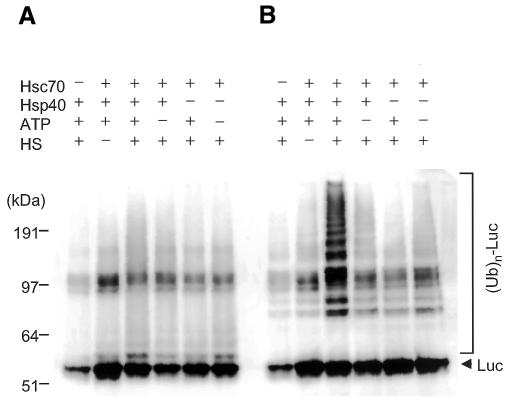
In addition to Hsp90 (Young et al., 2001), Hsc70 also functions as molecular chaperones responsible for protein folding (Frydman, 2001), and CHIP was found to bind Hsc70 through its TPR domain (Ballinger et al., 1999). Accordingly, we examined whether CHIP also teams with Hsc70 for ubiquitylation of unfolded protein, like Hsp90. To clarify the ubiquitylated forms, the assay was conducted using Ub or GST–Ub (Figure 4A and B, respectively). When Hsc70 was pre-incubated with luciferase at 43°C for 10 min, a very low ubiquitylating activity was observed, and the ubiquitylated level of luciferase resembled that without heat shock (Figure 4B). However, when Hsp40 was added with Hsc70 under the same heat-shock conditions, CHIP-dependent ubiquitylating activity was greatly promoted. Unlike Hsp90, addition of ATP during a heat-treatment period markedly influenced the effect of Hsc70 and Hsp40 for ubiquitylation, because obvious CHIP activity was not observed in the absence of ATP, even if it was present for the ubiquitylation assay (Figure 4). These observations are consistent with previous findings that Hsc70 and Hsp40 capture collaboratively unfolded proteins in an ATP-dependent fashion (Minami et al., 1996). Collectively, the results clearly indicate that CHIP exerts its ubiquitylating function for targets captured by Hsc70, whose reaction was assisted by Hsp40.
Fig. 4. Importance of Hsc70 and Hsp40 for CHIP-dependent ubiquitylation of heat-denatured luciferase. Luciferase was heated at 43°C for 10 min with Hsc70 and/or Hsp40 in the presence or absence of ATP, as described in Methods. The routine ubiquitylation assay by CHIP was carried out with UbcH5c and Ub (A) or GST–Ub (B), as described in Figure 1A.
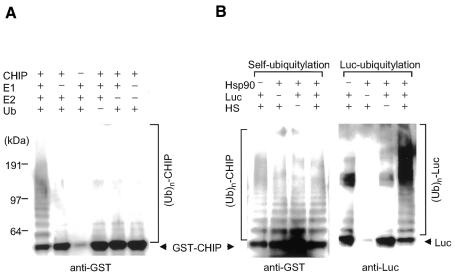
The self-ubiquitylating activity of CHIP
It was reported recently that at least five U-box proteins including CHIP have self-ubiquitylating activity (Hatakeyama et al., 2001). When CHIP was incubated with E1, E2 (Ubc-H5c) and Ub (Figure 5A), western blot analysis of CHIP revealed multiple smear bands, confirming that CHIP is a U-box ligase with auto-ubiquitylating activity. No obvious change in the ladder bands was observed in the presence or absence of Hsp90 and/or luciferase (Figure 5B, right panel), indicating that this self-ubiquitylating activity was independent of ubiquitylation of the target protein.
Fig. 5. Self-ubiquitylating activity of CHIP. (A) Combination of various components. GST–CHIP was incubated at 30°C for 2 h with E1, E2 (UbcH5c), Ub and ATP at indicated combinations. The assay mixture was the same as conducted for ubiquitylation, except that luciferase and Hsp90 were not added. After incubation, the reaction was terminated by the addition of 20 µl sample buffer for SDS–PAGE. The boiled supernatant was subjected to SDS–PAGE followed by western blotting with an anti-GST antibody. (B) The effect of Hsp90 and heat-denatured luciferase. Self-ubiquitylation of GST–CHIP was performed in the presence of luciferase that had been heated at 45°C for 5 min (HS) as in (A) (left panel). Simultaneously, the ubiquitylated assay of luciferase by CHIP was detected by western blotting with an anti-luciferase antibody (right panel).
DISCUSSION
It has been proposed that >30% of newly synthesized proteins are discarded without being properly folded (Yewdell, 2001). In addition, even if proteins are normally formed into tertiary structures, they are continuously impaired by spontaneous denaturation, chemical modification (e.g. oxidation) and environmental stresses, such as increased temperatures, reactive oxygen species (ROS; e.g. free radicals) or heavy metals (especially mercury). In rare cases, dysfunctional proteins with highly abnormal conformations result from genetic mutations (Sherman and Goldberg, 2001). In considering these circumstances, one would expect accumulation of these improperly folded proteins or abnormal proteins in the cells. However, no such accumulation is detected in normal cells, because such cells are fully equipped with a surveillance system that rapidly eliminates such aberrant proteins, which could be conceptually considered as a protein quality control in cells.
The protein quality-control system is well known as unfolded protein response (UPR) and ERAD responsible for stresses in the ER of eukaryotic cells, but a similar adaptive system also operates in the cytoplasmic and nuclear compartments. For this purpose, two mechanistically antagonistic strategies have been acquired during evolution. One is the chaperone system, including heat shock proteins Hsp90 and Hsc70 that can recognize misfolded or unfolded proteins and assist in their conversion to a functional conformation, preventing aggregation and accumulation of such abnormal proteins (Frydman, 2001; Young et al., 2001). The other is called the Ub-proteasome system (Hershko et al., 2001). When severely damaged proteins cannot be refolded into their functional forms, cells hydrolyse the abnormal proteins to small polypeptides by this protein-destroying machinery and ultimately convert back to amino acids by oligo- and exopeptidases (Voges et al., 1999). However, it remains to be determined whether these two apparently antagonistic systems are mechanistically related or how the cells distinguish the balance between chaperone-assisted refolding and proteolytic abrogation of unfolded proteins during recovery from stress. Interestingly, there is accumulating evidence addressing the importance of molecular chaperones, such as GroEL, DnaJ, Ydj1 and Hsc70, for certain short-lived and abnormal proteins in bacterial and mammalian cells (Lee et al., 1996; Bercovich et al., 1997; Kandror et al., 1999; Huang et al., 2001). These observations suggest a functional link between chaperone systems and proteolysis machinery, but the details have remained to be uncovered.
In the present study, we have shown that CHIP is a key molecule involved in protein quality control. CHIP is a Ub-protein ligase that selectively ubiquitylates denatured proteins when the substrate is captured by molecular chaperones such as Hsp90 and Hsc70. These results indicate strongly a new function for molecular chaperones: assisting in aggressive destruction of proteins with aberrant structure, in addition to participating in protein folding. Based on these findings, we hypothesize here the existence of an E3 Ub-protein ligase(s) responsible for ubiquitylation of markedly unfolded proteins that cannot be refolded, termed provisionally ‘quality-control E3’, and that CHIP is a possible candidate for this category of enzymes. In this context, Meacham et al. (2001) recently reported that Hsc70, but not Hsp90, is involved in the CHIP-dependent ubiquitylation of CFTR in vivo. In addition, Connell et al. (2001) reported that ubiquitylation-dependent instability of the Hsp90-trapped glucocorticoid receptor is promoted by CHIP. Our in vitro analyses extended these original observations by demonstrating directly that CHIP is a U-box E3 enzyme belonging to a new category of E3 enzyme responsible for quality control of cellular proteins linked to the function of molecular chaperones.
There is growing interest in the importance of the Ub-proteasome system in maintaining protein homeostasis in cells. Indeed, this proteolytic pathway has been recently highlighted in non-dividing cells of the brain, since neuronal intracellular inclusions are composed of Ub-positive protein aggregates, which have recently been described as a common ultrastructural feature of most neurodegenerative diseases (Lowe et al., 2001). Typical examples of these particles are the cytosolic Lewy bodies in Parkinson’s disease, neurofibrillary tangle containing tau in Alzheimer disease and nuclear polyglutamine aggregates in CAG-repeat diseases. The frequent appearance of Ub-positive intracellular inclusions in many neurodegenerative disorders indicates failure of protein quality control in the cells. Furthermore, Bence et al. (2001) recently reported that accumulation of abnormal proteins causes dysfunction of the Ub-proteasome pathway. In considering these circumstances, our results suggesting CHIP as a quality-control E3 undoubtedly provide new insights into protein homeostasis in cells coupled to pathological aspects induced by the abnormality. However, it is still unclear how molecular chaperones control the balance between protein folding and degradation. Further studies should be designed to elucidate the in vivo role of chaperones in proteolysis.
METHODS
Materials. Firefly luciferase was from Roche (Mannheim, Germany), bovine Ub from Sigma (St Louis, MO), anti-luciferase from Sigma and anti-GST antibodies from Calbiochem (La Jolla, CA). Hsp90, Hsc70 and Hsp40 were prepared as described previously (Minami et al., 1996, 2000).
Preparation of recombinant proteins. Recombinant His-Ubc3, His-Ubc4, His-UbcH5c, His-UbcH6, His-UbcH7, His-UbcH8 and GST–Ub were produced in Escherichia coli. Recombinant E1 (Uba1) was produced from baculovirus-infected HiFive insect cells. These proteins, which are all human species except mouse E1, were affinity purified by His Trap column (Pharmacia Biotech, Buckinghamshire, UK). The mutant UbcH5c(C/S) (Cys at position 85 in the UbcH5c was replaced by Ser) was synthesized by a PCR-assisted method using a site-directed mutagenesis kit (Stratagene, La Jolla, CA). The CHIP cDNA was amplified by RT–PCR using RNA from mouse liver with appropriate primers. CHIP and CHIP (ΔU-box) lacking the C-terminal region (amino acids 199–304) were produced as glutathione S-transferase (GST)-fused proteins. The deletion mutants termed the N-, M- and C-region of Hsp90 kindly provided by F.U. Hartl (Max-Planck-Insitut für Biochemie, Martinsried, Germany) and expressed as fusion proteins with GST, as described previously (Young et al., 1997; Minami et al., 2000).
Ubiquitylation assay. Heat treatment of firefly luciferase was carried out as described previously (Minami et al., 1996, 2000). Briefly, 0.26 µM of luciferase was heated at 45°C for 5 min with or without 11 µM of Hsp90 or at 43°C for 10 min with 4.7 µM of Hsc70 and/or 3.2 µM of Hsp40 in the presence or absence of 4 mM ATP in 50 mM Tris–HCl buffer pH 7.5 containing 2 mM MgCl2. After heating, they were quickly chilled in an ice bath. One microlitre of these heat-denatured or native luciferase was routinely incubated for 2 h at 30°C, unless otherwise specified, in a reaction volume of 50 µl of 50 mM Tris–HCl buffer pH 7.5 containing 50 ng of recombinant mouse E1, 0.5 µg of E2, 2 µg of GST–CHIP, 6 µg of bovine Ub or 12 µg of GST–Ub, 1 mM DTT, 2mM MgCl2 and 4 mM ATP. After terminating the reaction by the addition of 20 µl sample buffer for SDS–PAGE, the boiled supernatant was separated by 4–12% SDS–PAGE and visualized by western blotting with anti-luciferase antibody for detection of ubiquitylating activity of CHIP or anti-GST antibody to assay the self-ubiquitylation of GST–CHIP (chemiluminescence).
REFERENCES
- Aravind L. and Koonin, E.V. (2000) The U box modified RING finger—a common domain in ubiquitination. Curr. Biol., 4, R132–R134. [DOI] [PubMed] [Google Scholar]
- Ballinger C.A., Connell, P., Wu, Y., Hu, Z., Thompson, L.J., Yin, L.Y. and Patterson, C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol., 19, 4535–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N.F., Sampat, R.M. and Kopito, R.R. (2001) Impairment of the ubiquitin–proteasome system by protein aggregation. Science, 292, 1552–1555. [DOI] [PubMed] [Google Scholar]
- Bercovich B., Stancovski, I., Mayer, A., Blumenfeld, N., Laszlo, A., Schwartz, A.L. and Ciechanover, A. (1997) Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem., 272, 9002–9010. [DOI] [PubMed] [Google Scholar]
- Connell P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L.J., Hohfeld, J. and Patterson, C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nature Cell Biol., 3, 93–96. [DOI] [PubMed] [Google Scholar]
- Frydman J. (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem., 70, 603–647. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Yada, M., Matsumoto, M., Ishida, N. and Nakayama, K. (2001) U-Box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem., 276, 33111–33120. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover, A. and Varshavsky, A. (2001) The ubiquitin system. Nature Med., 6, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Huang H.C., Sherman, M.Y., Kandror, O. and Goldberg, A.L. (2001) The molecular chaperone DnaJ is required for the degradation of a soluble abnormal protein in Escherichia coli. J. Biol. Chem., 276, 3920–3928. [DOI] [PubMed] [Google Scholar]
- Jiang J., Ballinger, C.A., Wu, Y., Dai, Q., Cyr, D.M., Hohfeld, J. and Patterson, C. (2001) CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Kandror O., Sherman, M. and Goldberg, A.L. (1999) Rapid degradation of an abnormal protein in Escherichia coli proceeds through repeated cycles of association with GroEL. J. Biol. Chem., 274, 37743–37749. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U. and Jentsch, S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Lee D.H., Sherman, M.Y. and Goldberg, A.L. (1996) Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Mayer, J., Landon, M. and Layfield, R. (2001) Ubiquitin and the molecular pathology of neurodegenerative diseases. Adv. Exp. Med. Biol., 487, 169–186. [DOI] [PubMed] [Google Scholar]
- McClellan A.J. and Frydman, J. (2001) Molecular chaperones and the art of recognizing a lost cause. Nature Cell Biol., 3, E51–E53. [DOI] [PubMed] [Google Scholar]
- Meacham G.C., Patterson, C., Zhang, W., Younger, J.M. and Cyr, D.M. (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nature Cell Biol., 3, 100–105. [DOI] [PubMed] [Google Scholar]
- Minami Y., Höhfeld, J., Ohtsuka, K. and Hartl, F.-U. (1996) Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J. Biol. Chem., 271, 19617–19624. [DOI] [PubMed] [Google Scholar]
- Minami Y., Kawasaki, H., Minami, M., Tanahashi, N., Tanaka, K. and Yahara, I. (2000) A critical role of the proteasome activator PA28 in the Hsp-90-dependent protein folding. J. Biol. Chem., 275, 9055–9061. [DOI] [PubMed] [Google Scholar]
- Minami M., Nakamura, M., Emori, Y. and Minami, Y. (2001) Both the N- and C-terminal chaperone sites of Hsp90 participate in protein folding. Eur. J. Biochem., 268, 2520–2524. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Sherman M.Y. and Goldberg, A.L. (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron, 29, 15–32. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl, P. and Baumeister, W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variation on ubiquitination. Nature Rev. Mol. Cell Biol., 169, 169–178. [DOI] [PubMed] [Google Scholar]
- Yewdell J.W. (2001) Not such a dismal science: the economics of protein synthesis, folding, degradation and antigen processing. Trends Cell Biol., 11, 294–297. [DOI] [PubMed] [Google Scholar]
- Young J.C., Schineider, C. and Hartl, F.U. (1997) In vitro evidence that hsp90 contains two independent chaperone sites. FEBS Lett., 418, 139–143. [DOI] [PubMed] [Google Scholar]
- Young J.C., Moarefi, I. and Hartl, F.U. (2001) Hsp90: a specialized but essential protein-folding tool. J. Cell Biol., 154, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]