SUMMARY
Tumor microbiota can produce active metabolites that affect cancer and immune cell signaling, metabolism, and proliferation. Here, we explore tumor and gut microbiome features that affect chemoradiation response in patients with cervical cancer using a combined approach of deep microbiome sequencing, targeted bacterial culture and in vitro assays. We identify that an obligate L-lactate-producing lactic acid bacterium found in tumors, Lactobacillus iners, is associated with decreased survival in patients, induces chemotherapy and radiation resistance in cervical cancer cells, and leads to metabolic rewiring, or alterations in multiple metabolic pathways, in tumors. Genomically similar L-lactate-producing lactic acid bacteria commensal to other body sites are also significantly associated with survival in colorectal, lung, head and neck, and skin cancers. Our findings demonstrate that lactic acid bacteria in the tumor microenvironment can alter tumor metabolism and lactate signaling pathways, causing therapeutic resistance. Lactic acid bacteria could be promising therapeutic targets across cancer types.
Graphical Abstract
eTOC Blurb
Colbert et al. describe how tumoral Lactobacillus iners strongly predict poor chemoradiation response and survival for patients with cervical cancer. Further, L. iners, and potentially other obligate L-lactate producing Lactic acid bacteria, appear to rewire tumor metabolism and could serve as biomarkers or therapeutic targets in multiple cancer types.
INTRODUCTION
Tumors, even in allegedly sterile organs, have unique microbiomes that can modify treatment response and survival1–8. While the gut microbiome indirectly affects tumor response through systemic mechanisms, including innumerable immune9–12 and metabolism-mediated pathways13–16,17–20, tumor-resident bacteria may directly impact tumor growth, survival, and function4–7.
There are several challenges to understanding the complex mechanisms of tumor-microbiota interactions. In preclinical models, microbiome manipulation of in vivo tumor models does not reliably recapitulate changes in the human microbiome. In clinic, studies of the tumor microbiome are limited by longitudinal tumor biopsy availability, pitfalls of optimized sequencing and analysis protocols for formalin-embedded tumor samples21–26, and sequencing reference libraries. Tumor strains develop in a unique environmental niches and selective pressures, and adapt or acquire additional genes and functions necessary to survive in low nutrient, low oxygen, or low pH environments. In-depth mechanistic study of the tumor microbiome remains a significant challenge in most tumor types.
Exophytic cervical cancers develop in mucosal surfaces and are amenable to repeated tumor microbiome sampling. For this reason, we used cervical cancer and a combined deep sequencing, immune profiling, and targeted bacterial culture platform to explore the potential mechanisms of direct tumor-microbiome interactions during cancer therapy. In this study, we analyzed tumor-resident bacteria for associations with poor treatment responses in 101 patients with cervical cancer undergoing chemoradiation, enrolled in a prospective, serial biomarker collection study across two institutions (MD Anderson Cancer Center, Houston TX; Lyndon B. Johnson Hospital of Harris Health System, Houston TX). We then integrated these data with targeted culture to identify and profile the key tumor microbiota associated with treatment resistance.
RESULTS
A total of 101 patients with newly diagnosed, locally advanced cervical cancer were enrolled; ninety-six patients had pre-treatment samples collected prior to standard of care treatment (CRT; 45Gy of external beam radiation therapy with weekly concurrent cisplatin at 40mg/m2 and brachytherapy) (Fig. 1A–B, Supp. Table 1). Samples were sent for 16S ribosomal RNA gene sequencing (16S), shotgun metagenome sequencing (SMS), T cell repertoire sequencing (TCR), and/or metabolomics (Supp. Table 2). 93 baseline gut microbiome samples were collected for 16S and/or SMS. 244 serial tumor swabs were also collected during and after treatment (1, 3 and 5 weeks of RT and 12-week follow-up).
Figure 1. Tumor-resident Lactobacillus iners is associated with decreased recurrence-free and overall survival in cervical cancer patients.
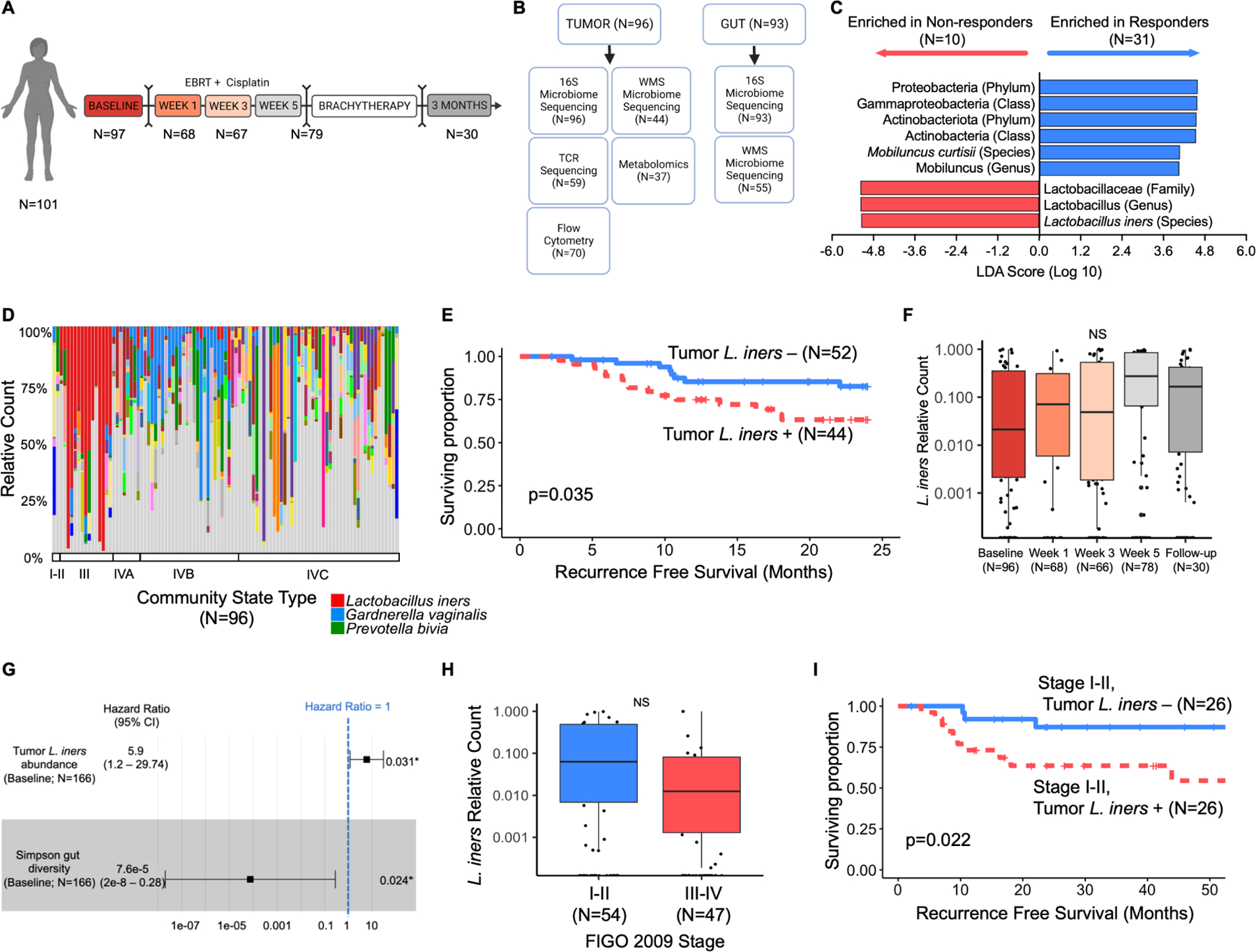
A. Study design and standard of care treatment algorithm for patients on study with number of cervical tumor swabs collected at each timepoint for 16S ribosomal RNA sequencing (16S). Patients received 5 weeks of EBRT with concurrent cisplatin followed by brachytherapy and repeat imaging for disease response at 3 months. Sampling was at baseline, Weeks 1, 3 and 5 of radiation, and at 3 month follow up.
B. Sample types collected and available for each analysis at baseline (pre-treatment). Tumor and gut samples were collected where possible from each patient at each timepoint; however, not all samples were collected or available for sequencing at each timepoint.
C. Linear discriminant analysis effect size (LEfSe) analysis of 16S data from cervical tumor swabs for bacteria enriched in non-responders to radiation in a pilot cohort (N=41). Default parameters were used for LEfSe analysis with an LDA threshold of 4.0 for statistical significance and visualization.
D. 16S compositional stacked bar plots of cervical tumor swabs for all patients at baseline (N=97), sorted by vaginal community state type (CST), including L. iners (red), G. vaginalis (blue), and P. bivia (green).
E. Kaplan-Meier recurrence-free survival (RFS) curves stratified by presence (N=44) or absence (n=52) of tumoral L. iners. Survival curves censored at 24 months. Log-rank test for comparison. Total # of events = 33.
F. 16S relative counts of L. iners in cervical tumor swabs collected during CRT. Week 1 (N=68), week 3 (N=66), week 5 (N=78), and follow-up (N=30) compared to baseline (N=96) using paired t-tests and false discovery rate (FDR) adjusted p-value. Box represents interquartile range (25th to 75th), bar indicates median, whiskers represent minimum and maximum values.
G. Multivariate cox proportional hazard analysis for overall survival (OS), adjusting for gut microbiome diversity (N=90) and tumoral L. iners (N=90). Total # of events = 14. Square represents hazard ratio (HR), bars represent 95% confidence intervals on HR.
H. Baseline relative counts of L. iners stratified by tumor size (FIGO 2009 Stage I-II [N=54] vs Stage III-IV [N=47]). Unpaired t-test. NS=p>0.05. Box represents interquartile range (25th to 75th), bar indicates median, whiskers represent minimum and maximum values.
I. Kaplan-Meier RFS curves for patients with FIGO 2009 Stage I-II tumors, stratified by presence (N=26) or absence (n=26) of tumoral L. iners. Survival curves censored at 54 months. Log-rank test for comparison.
Tumor Lactobacillus iners are associated with non-response to CRT and decreased recurrence-free survival
To identify initial bacteria of interest in the tumor microbiome (tumor-resident bacteria), we first performed linear discriminant analysis (LDA) effect size (LEfSe) in a pilot cohort of 43 patients using 16S ribosomal RNA sequencing data (16S) for associations with chemoradiation response. Lactobacillus iners (L. iners) was significantly associated with non-response to CRT (N=10), while Proteobacteria (phylum), Gammaproteobacteria (class), and Actinobacteriota (phylum) were associated with response to CRT (Fig. 1C; N=31; CDA score >4) in the pilot cohort. Next, we evaluated the relationship of these organisms with recurrence-free survival (RFS). Increased relative counts of tumor-resident L. iners were significantly associated with decreased RFS (RFS; Cox Proportional hazard ratio [Cox HR] 5.29 [95% CI 2.44–8.14]; log-rank p=0.0003), while Proteobacteria (phylum), Gammaproteobacteria (class), and Actinobacteriota (Phylum) were not associated with RFS in the pilot cohort (Supp. Fig. 1A–C; all p>0.05). Cervical tumor microbial diversity (Simpson, Faith’s phylogenetic diversity, Fisher) evenness (Pielou), or richness (total observed features) were not associated with response or RFS (Supp. Fig. 1D–M), using both rarefied and non-rarefied data and MaAsLin2 (FDR q value=0.07, p value=0.0007). To validate the association of L. iners with RFS, we enrolled an additional 58 patients across both institutions. The presence of any tumor-resident L. iners at baseline remained significantly associated with CRT non-response. In all patients, tumoral L. iners was present in 46% of samples (Fig. 1D) and the presence of L. iners remained associated with decreased RFS (log-rank p=0.035; Fig. 1E).
In univariate analysis of all patients with baseline samples (N=96), increased relative counts of L. iners at baseline were also significantly associated with lower RFS (Table 1; Cox HR 3.7 [95% CI 1.0 – 13.3; p=0.04) and lower overall survival ([OS], Table 1; Cox HR 10.4 [95% CI 1.8 – 60.3]; p=0.009). Proteobacteria, Gammaproteobacteria, and Actinobacteriota remained unassociated with RFS or OS, as did tumor microbiome evenness, diversity, and richness (Supp. Table 3). Other clinical and demographic variables associated with shorter RFS on univariate analysis included higher FIGO 2009 stage (III-IV vs. I-II; p=0.01) and lower gut microbiome diversity (p=0.049).
Table 1.
Univariate (UV) and multivariate (MV) Cox proportional hazard models for recurrence-free and overall survival for all patients with baseline samples (N=96).
| Variable | Recurrence-free Survivala | Overall Survival | |||||
|---|---|---|---|---|---|---|---|
| UV | MV | UV | MVl | ||||
| pval |
HR (95%CI)b | pval | pval |
HR (95%CI) |
pval |
||
| Tumor L. Iners c | <0.01 d | 5.79 (1.98–16.95) | <0.01 | 0.02 | 8.60 (1.77–71.77) | <0.01 | |
| Institution | |||||||
| LBJe | – | – | |||||
| MDACCf | 0.29 | 0.07 | |||||
| Age (years) | 0.36 | 0.41 | |||||
| Race | |||||||
| Asian/Black/Other | – | – | |||||
| Hispanic | 0.16 | 0.99 | |||||
| White | 0.52 | 0.20 | |||||
| BMIf (kg/m2) | 0.84 | 0.40 | |||||
| Smoking Status | |||||||
| Never | – | – | |||||
| Former | 0.76 | 0.29 | |||||
| Current | 0.43 | 0.63 | |||||
| Histology | |||||||
| Non-squamousg | – | – | |||||
| Squamous | 0.68 | 0.12 | |||||
| LVSI | |||||||
| No | – | – | |||||
| Yes | 0.91 | 0.86 | |||||
| Unknown | 0.57 | 0.79 | |||||
| FIGO 2009 Stage h | |||||||
| II-II | – | – | – | ||||
| III-IV | 0.01 | 2.49 (1.18–5.25) | 0.02 | 0.27 | 2.82 (0.93–8.49) | 0.07 | |
| Grade | |||||||
| Other/Unknown | – | – | |||||
| 1 | 0.99 | 0.99 | |||||
| 2 | 0.09 | 0.22 | |||||
| 3 | 0.83 | 0.70 | |||||
| HPV Type | |||||||
| HPV 16/18 | – | – | |||||
| Negative/Other | 0.19 | 0.07 | |||||
| Cisplatin Cycles | 0.36 | 0.81 | |||||
| Radiation Dose (Gy) | 0.92 | 0.03 | |||||
| Antibiotic Use i | |||||||
| No | – | – | |||||
| Yes | 0.58 | 0.08 | |||||
| Nodal Status | |||||||
| Negative | – | – | |||||
| Positive | 0.84 | 0.99 | |||||
| Tumor Dimension (cm) | 0.51 | 0.44 | |||||
| Gut Diversity j | <0.05 | 0.03 (5.6E–05 –21.19) | 0.30 | 0.01 | |||
| Gut Evenness k | 0.18 | 0.03 | |||||
N=33 recurrence events.
Hazard Ratio and 95% Confidence interval on HR for cox proportional hazard (Cox PH) models.
Increasing relative counts in baseline (pretreatment) tumor swabs.
Bold font indicates p<0.05 variables included in multivariate Cox PH models.
Lyndon B. Johnson Hospital, Harris Health System, Houston TX.
The University of Texas M.D. Anderson Cancer Center, Houston TX.
Adenocarcinoma and adenosquamous carcinoma.
The International Federation of Gynecology and Obstetrics.
Antibiotic use within 30 days of baseline swab collection extracted from inpatient and outpatient pharmacy and electronic medical record data, antifungals not included.
Simpson gut diversity (continuous).
Pielou’s evenness metric.
Overall survival multivariate model includes only gut diversity due to limited number of events.
Relative counts of L. iners and overall tumor microbiome diversity (Simpson, Faith, Fisher), evenness (Pielou) or overall richness (Observed features) did not change significantly during or after CRT (Fig. 1F; Supp. Fig. 2A–E).
L. iners abundance is an independent predictor of poor recurrence-free and overall survival on multivariate analysis
On multivariate (MV) Cox proportional hazard (PH) analysis for RFS in all patients, adjusting for stage and gut microbiome diversity, only higher L. iners abundance remained associated with decreased RFS (Table 1; Cox HR 5.79 [1.98–16.95]; p=0.001), as did higher FIGO stage (2.49 [1.18–5.25]; Cox PH p=0.016). Gut microbiome diversity was no longer significant (Cox PH p=0.30). Sensitivity analyses for model stability with only gut diversity and L. iners abundance confirmed L. iners was significant for RFS, while gut diversity was not. There was no difference in RFS based on relative counts of L. iners for patients with small tumors (FIGO 2009 stage I-II; N=52) versus large tumors (Fig. 1H, FIGO 2009 stage III-IV; N=47). Even in these patients, the presence of tumoral L. iners (N=26) still predicted significantly shorter RFS (Fig. 1I; 26; log-rank p=0.022).
High L. iners abundance was also significantly associated with worse OS on MV analysis (Table 1; Cox HR 12.7 [2.4–66.5]; p=0.006) when adjusted for stage (p=0.07). Independent models for L. iners adjusted for gut microbiome diversity and evenness were constructed independently due to small OS event numbers, and L. iners remained significant in both models (Fig. 1G, Gut Simpson diversity L. iners Cox HR 5.9 [95% CI 1.2–29.7]; p<0.0031; Supp. Fig. 3A, Gut Pielou evenness L. iners Cox HR 6.5 [95% CI 1.3–32.9]; p=0.024).
L. iners is not a surrogate for another gut or tumor microbe, microbial signature or clinical feature
No clinical, demographic, or gut microbiome metrics were associated with tumoral L. iners (Table 2). L. iners+ tumors had slightly lower overall tumor microbiome alpha diversity; however, diversity was not associated with RFS or OS. No other tumor or gut compositional metrics, besides gut Simpson diversity and Pielou evenness, were significantly associated with L. iners, RFS, or OS (Supp. Table 3), including gut and tumor Faith’s phylogenetic diversity, Fisher’s alpha, observed features, Shannon diversity, or Simpson evenness (all p>0.05).
Table 2.
Pathologic and clinical variables associated with the presence of tumoral L. iners (N=96).
| Tumor L. Iners Status |
|||||
|---|---|---|---|---|---|
| Variable | L. iners − (N=52) | L. iners + (N=44) | |||
| N (%) |
Mean (SD) |
N (%) |
Mean (SD) |
p-valuea |
|
| Institution | 0.07 | ||||
| LBJ Hospitalb | 15 (29) | 6 (14) | |||
| MDACCc | 37 (71) | 38 (86) | |||
| Age (years) | 47.1 (9) | 43.0 (11) | 0.06 | ||
| Race | 0.21 | ||||
| Black/Asian/Other | 6 (12) | 6 (14) | |||
| Hispanic | 27 (52) | 15 (34) | |||
| White | 19 (37) | 23 (52) | |||
| BMId (kg/m2) | 30.2 (6) | 28.9 (7) | 0.34 | ||
| Smoking Status | 0.61 | ||||
| Never | 32 (62) | 26 (59) | |||
| Former | 14 (27) | 15 (34) | |||
| Current | 6 (12) | 3 (7) | |||
| Histology | 0.50 | ||||
| Non-squamouse | 10 (19) | 11 (25) | |||
| Squamous | 42 (81) | 33 (75) | |||
| LVSI f | 0.80 | ||||
| No | 12 (23) | 9 (21) | |||
| Yes | 3 (6) | 4 (9) | |||
| Unknown | 37 (71) | 31 (70) | |||
| FIGO 2009 Stage g | 0.37 | ||||
| I-II | 26 (50) | 26 (59) | |||
| III-IV | 26 (50) | 18 (41) | |||
| Grade | 0.08 | ||||
| Indeterminate/ Unknown | 7 (14) | 11 (25) | |||
| 1 | 6 (12) | 1 (2) | |||
| 2 | 25 (48) | 15 (34) | |||
| 3 | 14 (27) | 17 (39) | |||
| HPV Genotype | 0.11 | ||||
| HPV 16/18 | 35 (71) | 24 (56) | |||
| Negative/Other | 14 (29) | 19 (44) | |||
| Cisplatin Cycles | 5.0 (1) | 5.0 (1) | 0.92 | ||
| Antibiotic Use h | 0.12 | ||||
| No | 36 (69) | 23 (54) | |||
| Yes | 16 (31) | 20 (47) | |||
| Nodal Status | 0.98 | ||||
| Negative/Unknown | 20 (38) | 17 (39) | |||
| Positive | 32 (62) | 27 (61) | |||
| Tumor Dimension (cm) | 5.5 (2) | 5.1 (2) | 0.38 | ||
| Faith's PDi (Gut) | 12.1 (3) | 12.7 (3) | 0.30 | ||
| Fisher's Alpha (Gut) | 20.5 (9) | 23.0 (8) | 0.17 | ||
| Observed Features (Gut) | 150.8 (61) | 166.4 (58) | 0.22 | ||
| Pielou's Evenness (Gut) | 0.7 (0) | 0.7 (0) | 0.23 | ||
| Shannon Diversity (Gut) | 5.0 (1) | 5.2 (1) | 0.15 | ||
| Simpson's Evenness (Gut) | 0.1 (0) | 0.1 (0) | 0.53 | ||
| Simpson Diversity (Gut) | 0.9 (0) | 0.9 (0) | 0.36 | ||
Chi-square or Fisher’s exact test for categorical variables. T-test for continuous variables.
Lyndon B. Johnson Hospital, Harris Health System, Houston TX.
The University of Texas M.D. Anderson Cancer Center, Houston TX.
Body mass index.
Adenocarcinoma and adenosquamous carcinoma.
Lymphovascular space invasion.
The International Federation of Gynecology and Obstetrics.
Antibiotic use within 30 days of baseline swab collection extracted from inpatient and outpatient pharmacy and electronic medical record data, antifungals not included.
Unsupervised clustering revealed two L. iners+ clusters, co-occurring with either Gardnerella vaginalis (G. vaginalis) or Atopobium vaginae (A. vaginae), and an L. iners-cluster with Prevotella bivia (P. bivia); cluster membership did not change significantly during CRT and was not indepentently associated with outcomes (Supp. Fig. 2F–I, Supp. Fig. 2J–N, Supp. Table 3, all p>0.05).
There was also no association of overall viral, HPV-specific viral load or fungal load with presence of L. iners (Supp. Fig. 2N–P). Antibiotic use was not associated with L. iners (Table 2) or outcomes (Table 1).
Presence and abundance of L. iners in the gut was significantly associated with initial CRT response on LEfSe, in addition to Escherichia/shigella (E/shigella) (Supp. Fig. 3B), but not with RFS or OS (Supp. Fig. 3C–D; Supp. Table 3; all Cox PH and KM p>0.05). Patients with gut L. iners also had tumor L. iners: the only bacterial species in the gut enriched (LDA score ≥4) in patients with L. iners+ tumors was L. iners with no direct correlation between abundances (Supp. Fig. 3E–FG).
L. iners does not affect baseline or dynamic overall or antigen-specific T cell repertoire
In other cancers, bacteria prime immune response to standard cancer therapy and immunotherapy. To explore whether there were differences in T cell repertoire or clonal expansion in response to cancer therapy, we performed tumoral T cell repertoire at serial timepoints27 (N=199). L. iners− tumors had overall higher counts of TCR templates (8,175 vs. 14,600 t-test p=0.03; Supp. Fig. 4A), CD4+ T cells and CD8+ T cells (Supp. Fig. 4B, C) at baseline, suggesting higher T cell infiltration. Both L. iners+ and L. iners- tumors had a decrease during CRT in productive clonality and overall templates (Supp. Fig. 4D–I), L. iners – tumors rebounded slightly earlier at the end of treatment and by week 12; We identified no differences in serial clonal TCR repertoire or motifs (Supp. Fig. 4), or in clonal expansion of HPV-specific TCRs (Supp. Fig. 4U). T cell recognition and expansion did not appear to be the primary mechanism for poor survival.
L. iners induces chemoradiation resistance in cervical cancer cell lines
To test whether L. iners from cervical cancers could directly cause radiation resistance independent of an immune effect, we cultured, isolated and characterized L. iners strains from cervical tumors, then filtered bacterial supernatant for cell-free supernatant (CFS). To identify the optimal supplement ratio of CFS in the cell culture medium and the culture condition, we performed serial dilution assays using a HPV16+ cervical squamous cell carcinoma (CSCC) cell line, CaSki (Supp. Fig. 5A). We generated a cervical cancer patient-derived organoid line (PDO, B1188) which develops a dense 3D morphology, exhibits immunofluorescence profiles consistent with CSCC (Fig. 2A, B), and is sensitive to both IR and cisplatin treatments (Supp. Fig. 5B, C). A line of genomically stable primary cells was generated after several passages (B1188 primary cells).
Figure 2. L. iners induces treatment resistance in vitro.
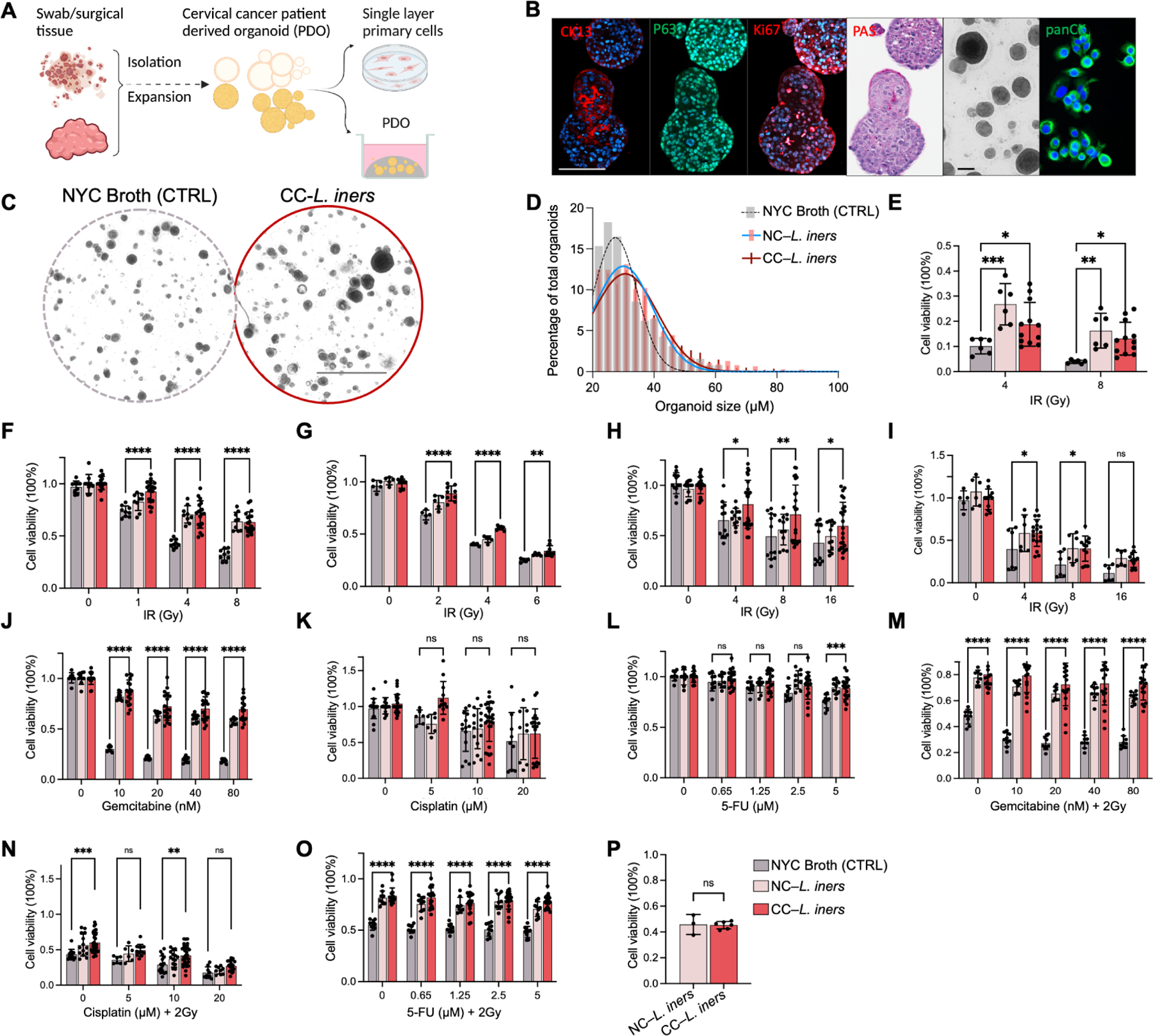
A) Workflow for the establishment and maintenance of patient-derived organoids (PDO) and B188 primary cells.
B) Positive staining of PDOs B1188 with antibodies for anti-P63 and anti-Ki67, together with decreased expression of the differentiation marker staining of anti-CK13 antibody and PAS, confirming squamous carcinoma origin. Positive staining of anti-panCK marker demonstrates primary cancer cell origin. Scale bars,100 μm
C) Bright view of PDO B1188 pretreated with cancer-derived L. iners (CC-L. iners) cell-free supernatant (CFS) vs. control (NYC Broth) followed by 4Gy irradiation. Scale bars,1 mm
D) Histogram of organoid size and count (percentage of total counted) for organoids from PDO B1188 pretreated with CC-L. iners CFS (red) vs. non-cancer derived L. iners (NC-L. iners; pink) vs. control (NYC Broth; grey) followed by 4Gy irradiation.
E) Cell viability (measured by CellTiter Glo) of irradiated organoids from PDO B1188 pretreated with CC-L. iners cell-free supernatant (CFS) vs. control (NYC Broth) followed by 4Gy and 8Gy irradiation. 2 experiments, 3 replicates. One way ANOVA (CFS vs. control).
F) B1188 cell viability after irradiation.
G) HeLa cell viability after irradiation.
H) SiHa cell viability after irradiation.
I) CaSki cell viability after irradiation.
J) B1188 cell viability after gemcitabine (GEM) treatment.
K) B1188 cell viability after cisplatin (CIS) and 2Gy irradiation.
L) B1188 cell viability after 5-fluorouracil (5-FU).
M) B1188 cell viability after GEM and 2Gy irradiation.
N) B1188 cell viability after CIS + 2Gy irradiation.
O) B1188 cell viability after = 5-FU and 2 Gy.
P) B1188 cell viability with UV-killed bacterial fragments and IR.
1 experiment, 3 replicates. Cell viability (measured by CellTiter Glo) following pretreatment of cells with control (NYC Broth), NC-L. iners CFS or CC-L. iners CFS (F-O); 3 experiments, 3 replicates each (F-O); 2 patient-derived CC-L. iners strains pooled (E-P); one way ANOVA between CC-L. iners with Mean and SEM are presented (E-P).
B1188 PDOs were incubated for two weeks with 20% CFS harvested from two cancer-derived L. iners strains (CC-L. iners), one commercial non-cancer-associated L. iners strains (NC-L. iners), or Control (CTRL, 20% NYC Broth), prior to treatment with ionizing radiation (IR), cisplatin (CIS), gemcitabine (GEM) or 5-fluorouracil (5FU), or their combination. PDOs cultured with CC-L. iners CFS displayed more aggressive growth and radiation resistance than CTRL, with higher organoid count, larger organoid size (Fig 2C, D; Supp. Fig 5D, E), and increased cell viability after IR (Fig 2E). CC-L. iners and NC-L. iners CFS also caused increased cell viability in B1188 primary cells (Fig. 2F). HeLa, SiHa, and CaSki cells all exhibited significantly increased cell viability with L. iners treatment at all doses of irradiation (Fig. 2G–I). CC-L. iners treated B1188 cells were resistant to GEM (Fig. 2J), but not cisplatin (CIS; Fig. 2K) or 5-fluorouracil (5FU; Fig. 2L) alone. With the addition of IR, they exhibited resistance to GEM-IR, CIS-IR, and 5FU-IR (Fig. 2M–O). L. iners similarly induced GEM resistance in CaSki cells (Supp. Fig. 5F–H), but to no chemotherapeutics without IR in SiHa or HeLa cells (Supp. Fig. 5I–M). We also evaluated cell viability after IR in HeLa cells using cancer-derived and non-cancer derived L. crispatus. L. crispatus did not induce treatment resistance (Supp. Fig. 5N). We observed no radiation sensitization with UV-killed, PBS-washed L. iners (Fig. 2P), suggesting the factors mediating radiation resistance were secreted by L. iners rather than an effect of bacterial cell wall components.
L. iners alter gene expression in lactate signaling pathways
Next, to explore how L. iners CFS could alter cancer cell sensitivity to IR, we performed RNA sequencing of pretreated B1188 cells. Our proposed mechanism for the effect of L. iners on cancer cell metabolism is given in Fig. 3A. We found that cells treated with L. iners CFS versus NYC broth control had significantly altered gene expression (Fig 3B). Metacore pathway analysis revealed enrichment in several pathways closely linked to lactate signaling and lactate dehydrogenase (LDH) activity, including reactive oxygen species (ROS)-induced cellular signaling and hypoxia-inducible-factor 1 (HIF-1) transcription targets, FGFR signal transduction28–30, Her2/ERBB2 signaling31–33, and p53/p73 dependent apoptosis34 (Fig. 3C); Gene Set Enrichment Analysis (GSEA) Hallmark Pathway analysis confirmed enrichment in oxidative stress/ ROS-induced cellular signaling and HIF-1a transcription targets, along with the GSEA pathway for skeletal muscle genes, which is also highly regulated by lactate (Supp. Fig. 6A).
Figure 3. L. iners causes treatment resistance through increased L-lactate production in the tumor microenvironment.
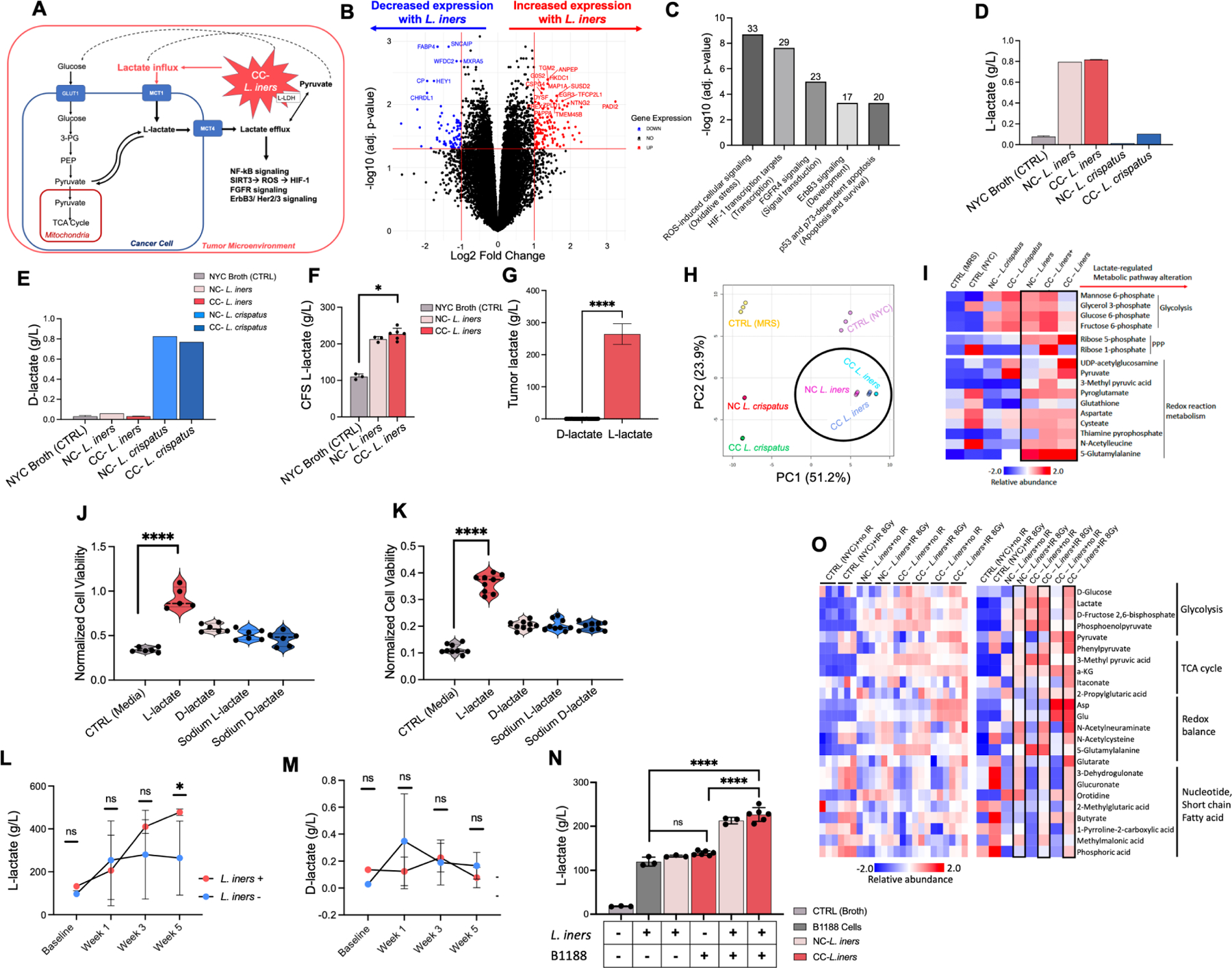
A. Hypothetical schematic of L. iners production of L-lactate in the tumor microenvironment “priming” cervical cancer cells for lactate addiction, driving the feedback loop of lactate utilization via upregulation of GLUT1, MCT1 and MCT4, and lactate-regulated induction of reactive oxygen species signaling, HIF-1, NFkB, FGFR, ErbB3/HER 2/3, and p53 dependent pathways.
B. B1188 cells pre-treated with L. iners (1 NC-L. iners strain, 2 CC-L. iners strains) vs. control (NYC broth) CFS prior to RNA sequencing. Fold change in gene expression from control (right) to L. iners (left) is shown. Log2 (Fold Change) threshold of −1, 1. –Log10 (FDR-adjusted p-value) threshold is 1.2.
C. Metacore pathway analysis of significantly altered genes. Top 5 most significantly altered pathways are shown ranked by -Log10 (FDR-adjusted p-value). Number above bar represents the proportion of genes altered in each pathway.
D. L-lactate production in bacterial culture of cancer-derived CC-L. crispatus, CC-L. iners, NC-L. crispatus, NC-L. iners, and control (NYC broth).
E. D-lactate production in bacterial culture of cancer-derived CC-L. crispatus, CC-L. iners, NC-L. crispatus, NC-L. iners, and control (NYC broth).
F. L-lactate levels in bacterial culture for control (NYC Broth), NC-L. iners or CC-L. iners.
G. L-lactate and D-lactate relative levels (g/L) for cervical tumor Cytobrush samples (log scale). N=29.
H. Principal component analysis (PCA) of metabolites.
I. Unsupervised hierarchical clustering of most differentially abundant metabolites, grouped by metabolic process.
J. Cell viability (CellTiter Glo) of pretreated B1188 cells with 20mM lactate isoforms (L-lactate, D-lactate, Sodium L-lactate, Sodium D-lactate, media control) after irradiation (4Gy).
K. Cell viability (CellTiter Glo) of pretreated B1188 cells with 20mM lactate isoforms after GEM.
L. L-lactate levels in cervical tumor Cytobrush samples before, during (Week 1, Week 3) and after EBRT (Week 5) for L. iners + patients (BL N=1; Wk1 N=6; Wk3 N=2; Wk5 N=4) and L. iners− patients (BL N=3; Wk1 N=4; Wk3 N=6; Wk 5 N=4).
M. D-lactate levels in cervical tumor Cytobrush samples.
N. L-lactate levels for media control (−/−), L. iners in culture alone (+/−), B1188 cells in culture alone(−/+), vs. B1188 cells treated with L. iners CFS (+/+) for NC-L. iners and CC-L. iners.
O. Differentially abundant metabolites present in primary cells B1188 treated with NYC Broth (control), NC-L. iners (N=1) CFS, and CC-L. iners (N=2) CFS, either nonirradiated (0Gy) or irradiated (8Gy), grouped by metabolic process.
Unsupervised hierarchical clustering of most differentially abundant metabolites, grouped by metabolic process (I,O); Analyzed by Megazyme Kit (D,E), TC-MS (L-N) or HR-MS/IC-MS (I,O); Wilcoxon rank-sum test (J-M), unpaired t-test with mean and SEM (G) or 2-way ANOVA (F) with NS P > 0.05, *P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001; 1 (B,F,J),N, 2 (H-J) or 3 (K) experiments, 3 replicates each, 2 CC-L. iners strains pooled (B,F,J); 1 experiment, 1 culture plate, no statistical comparisons (D-E). Wilcoxon rank-sum test vs. CTRL unadjusted (J-L) and adjusted (N); Comparisons for cells treated with MRS Broth (L. crispatus control), NYC Broth (L. iners control), and cancer and non-cancer derived L. iners (N=3) and L. crispatus strains (N=2) (H-I). Normalized to unirradiated media control (J-K).
L. iners are obligate L-lactate producers and CC-L. iners+ tumors are L-lactate enriched
All lactobacilli produce lactate as the final product of fermentation via lactate dehydrogenase (LDH) activity after carbohydrate utilization35–37. One of the distinguishing characteristics of L. iners, compared to beneficial lactobacilli38, is that its smaller genome uniformly does not contain the D-LDH gene, rendering it an obligate L-lactate producer. L-lactate is also the predominant enantiomer (97–99%) in mammalian cells and tumors39. Although cancer cells can produce D-lactate by the methylglyoxal pathway, this likely negligibly affects metabolism.
CC-L. iners produced only L-lactate in vitro (Fig. 3D), while cancer-derived and non-cancer derived L. crispatus produced primarily D-lactate (Fig. 3E); All cancer-derived L. iners genomes did not contain D-LDH. We validated this using quantitative L and D-lactate ion chromatography-mass spectrometry (IC-MS) assays. CC and NC-L. iners had significantly higher L-lactate levels than broth controls (Fig. 3F). In cervical tumor samples, we confirmed that L-lactate levels were >1,000 fold greater than D-lactate (Fig. 3G).
Non-targeted metabolic profiling of CFS from NC and CC-L. iners and L. crispatus strains revealed distinct metabolic network alterations in glycolysis, the pentose phosphate pathway (PPP), and the regulation of redox balance for CC-L. iners, all of which are linked to oncogenic lactate metabolism (Fig. 3H, I).
L-lactate recapitulates treatment resistance in cervical cancer cell lines
To determine whether L-lactate alone could induce chemotherapy and IR resistance similar to L. iners CFS, we pretreated cervical cancer cell lines with four isoforms of lactate: L-lactate, D-lactate, sodium L-lactate and sodium D-lactate for two weeks prior to chemotherapy or IR treatment. The lactate concentrations and pH were maintained until harvest. L-lactate, but not other isoforms, in the culture medium consistently recapitulated the treatment resistance of cervical cancer cells to IR and GEM in all cell lines, with varying effects observed for CIS and 5-FU (Fig. 3J–K; Supp. Fig. 6B–E), which was consistent with the effect of L. iners CFS.
L. iners increases tumor metabolic activity in response to radiation-induced stress
Although at baseline, there was no difference in measured L-lactate levels between L. iners+ and L. iners – tumors; interestingly, L-lactate levels (but not D-lactate levels) in L. iners+ tumors increased steeply from baseline to the end of CRT, nearly doubling by week 5 (Fig. 3L–M). Irradiated L. iners treated B1188 cells also significantly increased L-lactate production versus broth (CTRL) or L. iners alone, and demonstrated remarkable upregulation in glycolysis, TCA cycle, redox balance and nucleotide, short chain fatty acid assembly, particularly after irradiation (Fig. 3N–O).
These data demonstrate that L. iners can potentiate lactate utilization, production and metabolic activity in response to metabolic stress, including from IR.
L. iners+ tumors have upregulated glycolysis compared to L. iners− tumors
L. iners is a facultative anaerobe, and can make ATP via aerobic respiration or switch to fermentation under anaerobic conditions, efficiently producing lactate. Thus, we hypothesized that the lactate production and metabolic rewiring might be magnified in the hypoxic tumor microenvironment in patients. Principal component analysis (PCA) of non-targeted metabolomics confirmed distinct metabolite profiles for L. iners+ (N=30) and L. iners- tumors (N=36; Fig 4A; DSC p<0.005), and significant enrichment of unique metabolites in L. iners+ tumors (Fig. 4B), overall indicative of higher metabolic activity. Supervised clustering confirmed clusters of metabolites differentially enriched in L. iners+ vs. L. iners− tumors (Fig. 4C). Metabolites significantly enriched in L. iners+ tumors were pyruvate, indicative of increased glycolysis, adenosine triphosphate (ATP) and NADH, both indicative of upregulated TCA cycling and downstream electron transport chain activity, and deoxyguanosine triphosphate (dGTP) and deoxyuridine monophosphate (dUMP), both precursors for increased DNA synthesis, which can be driven by excess ATP. Galactaric acid was also significantly enriched in L. iners+ tumors; galactaric acid is an indicator of increased fermentation and lactate production in lactobacilli. The metabolic pathways most upregulated in L. iners + tumors were the Warburg effect, glycolysis, glutamate metabolism, and galactose metabolism (Fig. 4D, Supp. Fig. 6F–H).
Figure 4. L. iners positive tumors have metabolic alterations compared to L. iners negative tumors.
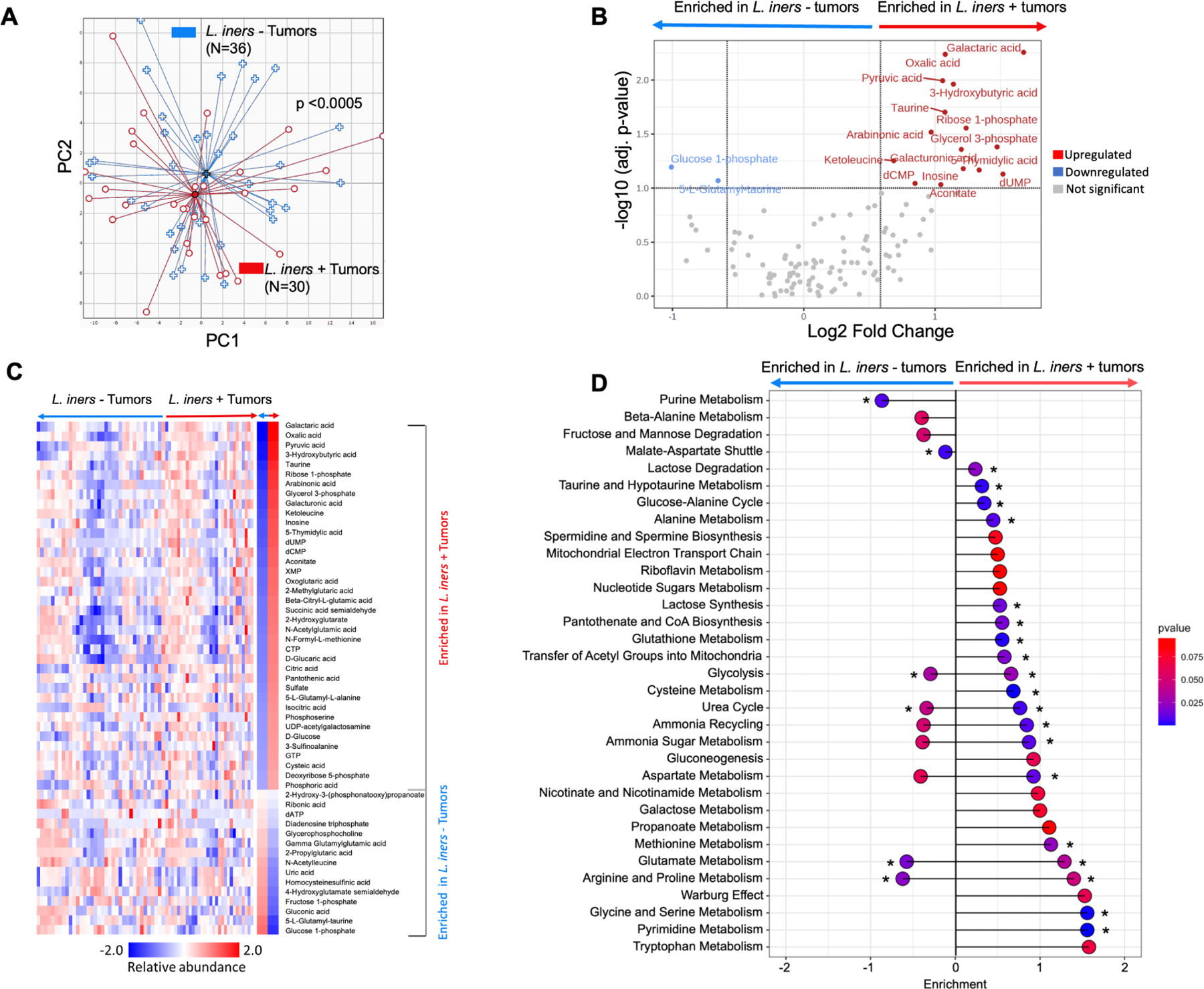
A. Principal coordinate analysis of relative abundances of tumor metabolites for L. iners+ (N=36) and L. iners− tumors (N=30). Dispersion Separability Criterion, p<0.005.
B. Volcano plot of differentially abundant metabolites. –Log10 (adj. p-value) threshold=1.0; log2 (Fold Change) threshold −1 to 1.
C. Supervised hierarchical clustering of differentially abundant metabolites.
D. Lollipop plot of pathway assignments for differentially enriched metabolites. Sorted by effect size on a log10 scale. * FDR-adj p<0.05.
Analyzed by HR-MS and IC-MS (A-D).
The most significantly enriched metabolite in L. iners- tumors was 5-Aminoimidazole-4-carboxamide ribonucleotide (AICAR). AICAR is an analog of adenosine monophosphate (AMP) and activates the AMP-kinase cascade in response to ATP deprivation, such as in downregulated glycolysis. AICAR is pro-apoptotic in this setting, and strongly suppresses cancer cell proliferation in response to ATP deprivation. It is used as a cancer therapy sensitizer in various cancers27–32 and can reverse Warburg metabolism33. These findings strongly suggest that L. iners plays a significant role in contributing to energy production and DNA synthesis within the tumor microenvironment.
Cancer-derived L. iners acquires additional genes for lactose utilization during carcinogenesis
L. iners is a common organism in a healthy cervicovaginal microbiome and regularly undergoes horizontal gene transfer to evade antibiotics and adapt to changing nutrient, pH and oxygenation conditions40. We hypothesized that CC-L. iners in patients with cervical cancer acquired additional genes during carcinogenesis, which may contribute to the metabolic affects observed.
To identify potential functions unique to CC-L. iners, we sequenced the complete genomes of two CC-L. iners isolates and two NC-L. iners isolates. We also performed SMS on baseline tumor samples (N=44) and assembled Lactobacillus genomes. We compared these to complete genomes of L. iners KY41, isolated from a healthy individual. All contigs annotated by L. iners in the metagenomes were combined and contigs that were annotated by any Lactobacillus (99% L. iners) to represent a cervical cancer Lactobacillus “pan-genome” (Supp. Fig. 7A). 593 genes (81%) were shared between CC-L. iners and NC-L. iners. 120 (16%) genes were unique to CC-L. iners, while NC-L. iners contained only 18 unique genes (2%; Fig. 5A). There were also more shared KOs (N=100; 14%) between dysplasia- associated L. iners (N=14) and CC-L. iners than between NC-L. iners and CC-L. iners (Fig. 5B). Overall, this higher genetic commonality between dysplasia-associated L. iners and CC-L. iners suggests these genes were acquired prior to or during carcinogenesis, rather than after cancer development.
Figure 5. Cancer-derived L. iners acquires additional genes for lactate production over NC-L. iners and alters cancer cell gene expression in pathways involved in intrinsic radiation sensitivity.
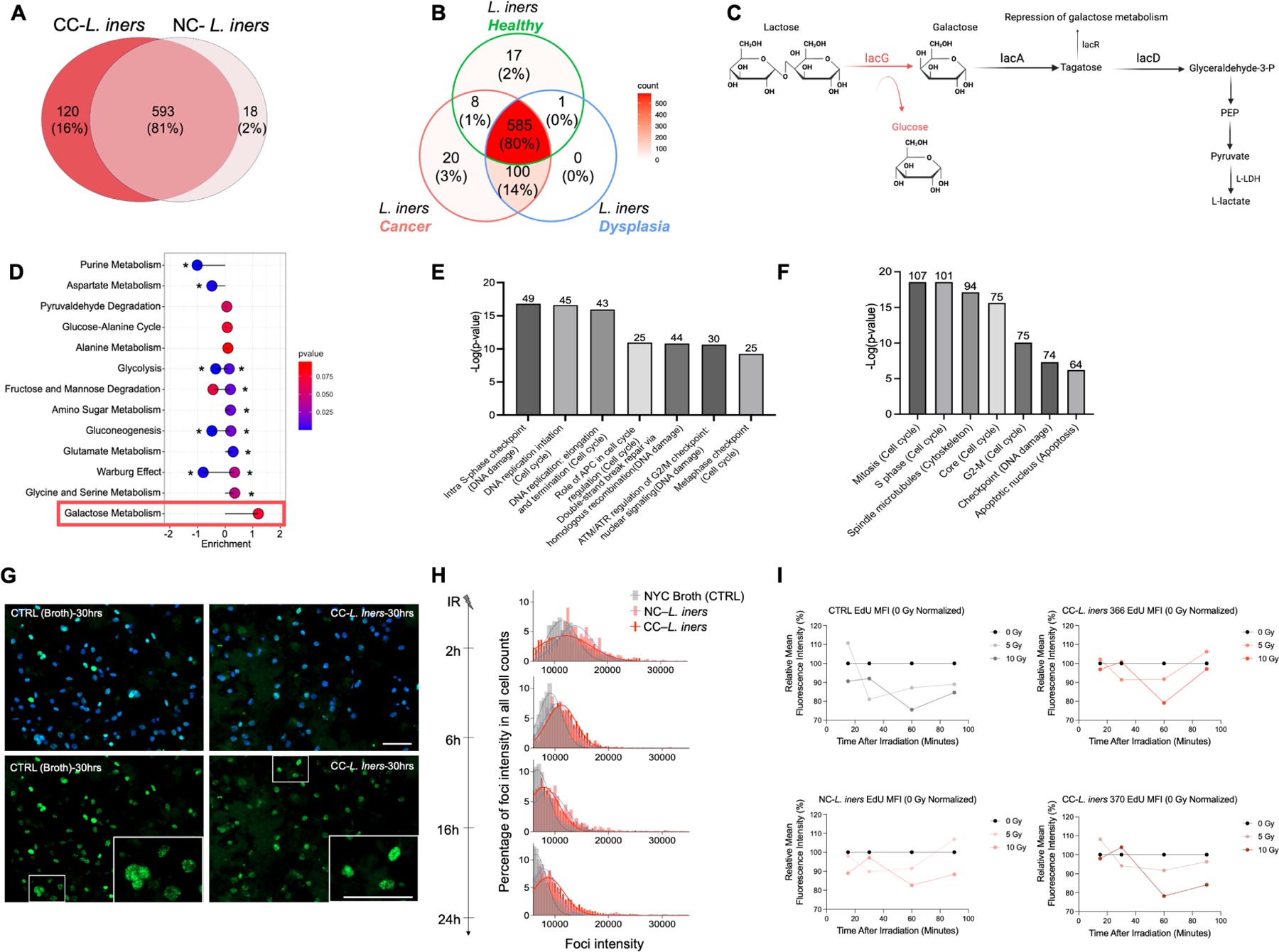
A. Overlapping and unique Genes on comparative genomic analysis for CC- L. iners (16%) vs NC-L. iners (2%) vs. shared (81%).
B. Overlapping genes on comparative genomic analysis for Healthy L. iners vs. dyplasia L. iners vs. CC- L. iners.
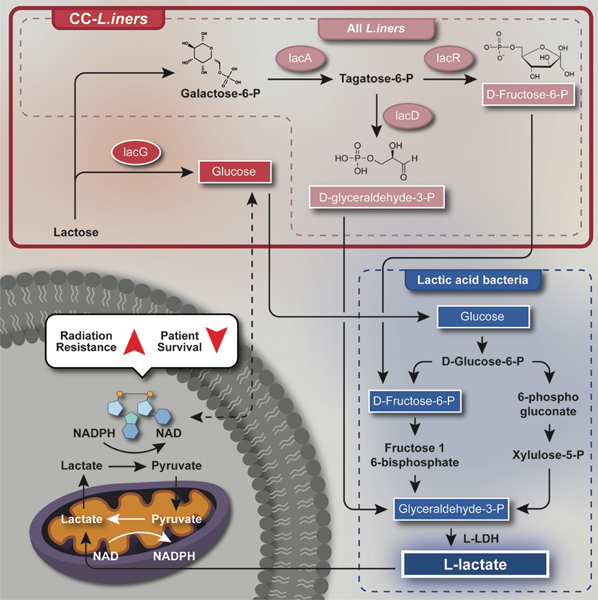
C. Sequential genes in pathways common to CC- L. iners and CC- L. iners (black) and unique to CC- L. iners (red) on comparative genomic analysis. LacG in CC- L. iners encodes the reversible enzyme, 6-phospho-beta-galactosidase, which converts lactose to galactose, while CC- L. iners ers utilizes only lactose in the lacDRA pathway. LacR is a repressor switch to turn off lactose metabolism to lactate.
D. Lollipop plot of metabolite pathways assignments for metabolites enriched in CC- L. iners isolates vs. NC-L. iners isolates. Galactose metabolism is the only upregulated pathway, consistent with lacG gene acquisition and 6-phospho-beta-galactosidase activity.
E. Top 7 differentially expressed metacore pathways for B1188 cells.
F. Top 7 differentially expressed metacore processes for B1188 cells.
G. Gamma H2AX and DAPI fluorescent staining of pretreated B1188 cells 30 hours after irradiation (8 Gy). Scale bars,100 μm
H. Gamma H2AX dynamics for primary cells B1188 treated with 8Gy in each CFS condition.
I. Radio-resistant EdU DNA synthesis assays for pretreated B1188 primary cells. Normalized to 0 Gy (black).
Log2 (Fold Change) cutoff of 2.0 (E,F); 1 independent experiment, 1 (I) or 3 (E) replicates, 2 CC-L. iners strains pooled (E) or separate (I); No statistical comparisons made (H,I).
We also performed gene, function, and pathway analysis (KO, KEGG, BRITE) to determine the function of genes acquired by CC-L. iners that were not present in NC-L. iners. We compared the genomes of CC-L. iners isolates and a near complete assembly of L. iners obtained from a patient sample with NC-L. iners. A complete comparison of genes, functions, and pathways unique to and shared by CC-L. iners and NC-L. iners (Supp. Fig. 7B–C, Supp. Table 4) demonstrated that, while all L. iners strains had the lacA, lacD, and lacR genes involved in conversion of galactose to D-glyceraldehyde-3-P and Fructose-6-P, only CC-L. iners also contained the additional lacG gene, which encodes 6-phospho-beta-galactosidase, which converts lactose to Galactose-6-P and Glucose (Fig. 5C, Supp. Fig 7C–D, Supp. Table 4). Lactobacilli can easily convert lactose to lactate, and galactose to lactate in the reverse direction, via the Leloir pathway and the Tagatose-6-phosphate pathway36,42,43. All L. iners+ patient samples and L. iners isolates contained only L-LDH, while none contained the gene for D-LDH. Non-targeted metabolomics of CC-L. iners and NC-L.iners isolates revealed the only upregulated pathway in CC-L. iners vs. NC-L. iners was galactose metabolism, consistent with the genomic findings (Fig. 5D).
DNA damage and response in cervical cancer cells after CFS treatment
We also investigated intrinsic DNA sensitivity and repair in CC-L. iners and NC-L. iners treated cells. While gene expression with any L. iners versus control involved lactate signaling pathways (Fig. 3, Supp. Fig. 7E), there were also notable differences in gene expression specifically for unirradiated CC-L. iners treated cells versus NC-L. iners treated cells (Fig. 5E, F; Supp. Fig. 7F–G), with significant downregulation of several DNA damage response pathways, DNA replication and initiation, G2/M and intra-S phase checkpoints and E2F transcription targets. We also noted a general, slightly higher resistance to IR for CC-L. iners vs NC-L. iners treated cells (Fig 2). After irradiation, CC-L. iners versus NC-L. iners treated cells demonstrated significant gene expression alterations, with upregulation of epithelial-mesenchymal-transition (EMT), NFKb signaling, and KRAS signaling, and downregulation of hypoxia response signaling (Supp. Fig. 7H Supp. Table 5). The gene expression data for treatment with L. iners demonstrated increased expression of oxidative stress/ ROS-induced cellular signaling and HIF-1 transcriptional targets in the cells cultured with L. iners CFS compared to cells cultured with broth both before (Fig. 3C and Supp. Fig. 7C) and after irradiation (Supp. Fig. 7D), suggestive of increased ROS and hypoxia in the microenvironment. ROS elevation correlates with increased γ-H2AX and causes induction of double-strand breaks and assurance of DNA damage response activation44–47. We evaluated γ-H2AX foci kinetics at different time points (2 hours [2h] to 24 hours [24h]) after 8Gy irradiation. All cells expressed the strongest foci intensity 2h after IR, with decreasing intensity over time, indicating DNA damage repair. CC-L. iners exhibited less initial foci generation as compared to NC-L. iners, while both exhibited more initial foci formation versus CTRL (Fig. 5G, H). CC-L. iners exhibited the most rapid return to normal levels, indicating efficient repair overall.
We also noted altered expression in S phase and G2/M checkpoint regulation genes (Fig. 4E-F), and a trend toward increased incorporation of EdU by cells treated with L. iners (Fig. 5I) suggestive of a dysfunctional intra-S phase checkpoint, and slight distinctions in redox reaction metabolism for CC-L. iners vs. NC-L. iners (Fig. 3H, 3O). Lactose degradation via the lacG gene in CC-L. iners produces glucose as a byproduct (Fig. 5D), which can also induce G2/M checkpoint arrest by the Cdk1/ CyclinB complex, and enhance tumor survival post-irradiation despite DNA damage52–55, consistent with the gene expression data. Further investigation is needed.
L. iners and similar lactic acid bacteria (LAB) are relevant in other cancer types
We also analyzed microbiome composition of tumor samples from anal, (N=70), vaginal and vulvar cancer (N=44) at MDACC and reprocessed raw reads from whole genome sequencing (WGS) and RNA sequencing (RNAseq) in The Cancer Genome Atlas (TCGA) for head and neck squamous cell carcinoma (HNSCC; N= 171), colorectal adenocarcinoma (COAD; N=502), non-small cell lung cancer (NSCLC; N=1047), and melanoma (SKIN; N= 106). L. iners was identified in anal, vaginal, vulvar, colorectal and lung cancers but, not unexpectedly, extremely rarely, since L. iners is a commensal vaginal microbe. Still, L. iners in NSCLC (N=57; 5.4%) was strongly associated with decreased RFS (Fig 6A; 21 months vs. Not reached [NR]; log-rank p=0.0077).
Figure 6. L. iners and genomically similar, commensal, L-lactic acid producing bacteria (LAB) portend poor prognosis across cancer types.
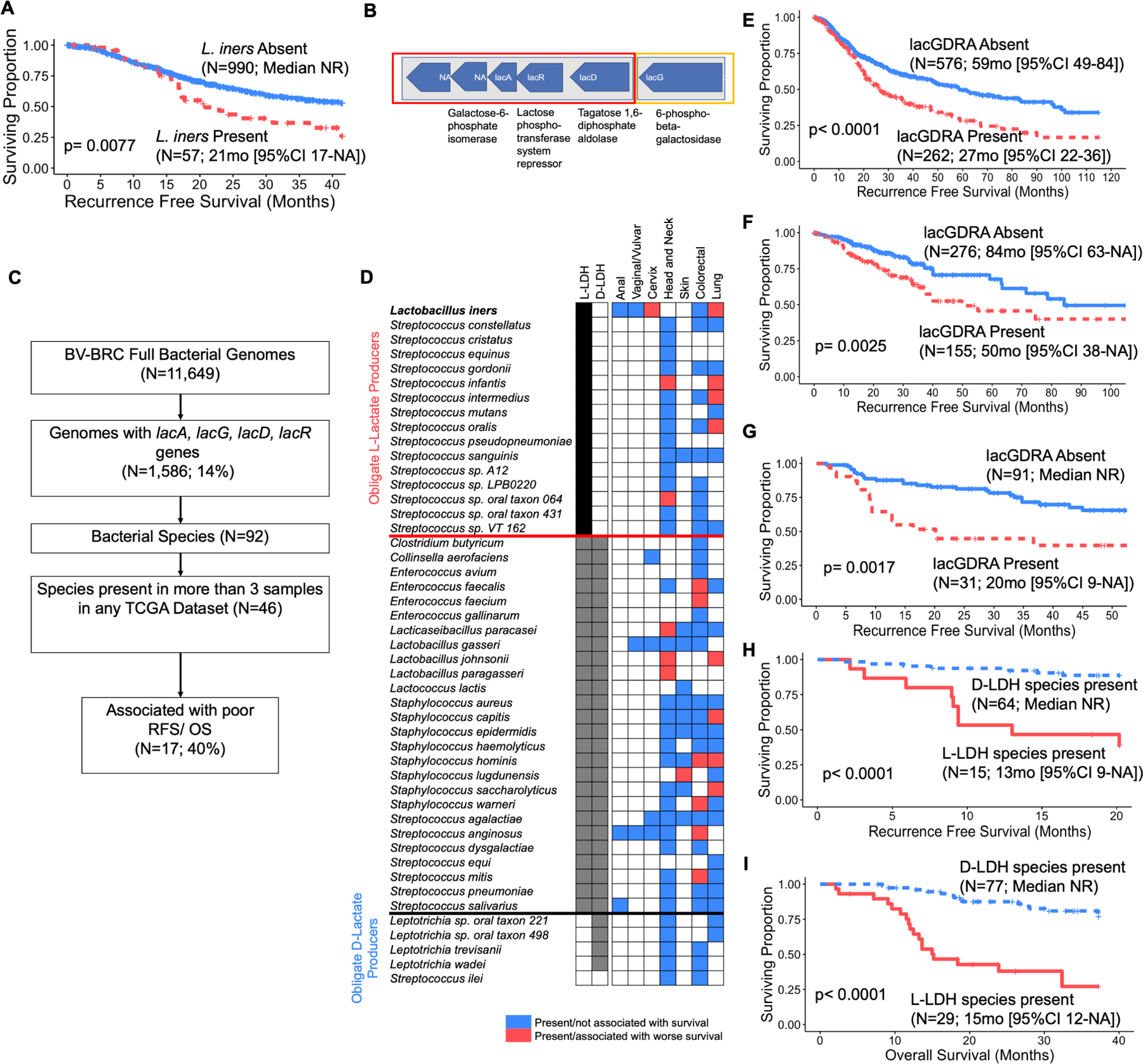
A. Recurrence-free survival (RFS) for L. iners presence (N=57) or absence (N=990) in primary tumor samples from the non-small cell lung carcinoma (NSCLC) TCGA dataset.
B. Consecutive operons found in L. iners isolates and deposited genomes (CC-L. iners = 2, NC-L. iners = 2) on comparative genomic analysis. Orange denotes lacG gene found only in CC-L. iners isolates, vs. red which denotes lac genes found in all L. iners isolates.
C. Flowchart for identification of genetically similar LAB species in TCGA datasets.
D. Frequency of lacGDRA bacteria genomically similar to L. iners (N=46) across MDACC (anal, vaginal/vulvar, cervix) and TCGA datasets (head and neck, skin, colorectal, lung). Red box denotes species is present and associated with decreased RFS and/or overall survival (OS) in individual dataset. Blue box denotes species is present in dataset, but not associated with RFS or OS.
E. RFS for patients with NSCLC stratified by presence (N=262) or absence (N=576) of any lacGDRA species from Bacterial and Viral Bioinformatics Resource Center (BV-BRC).
F. RFS for patients with Colorectal adenocarcinoma stratified by presence (N=155) or absence (N=276) of any lacGDRA species.
G. RFS for patients with Head and Neck Squamous Cell Carcinoma (HNSCC) stratified by presence (N=31) or absence (N=91) of any lacGDRA species.
H. RFS for patients with HNSCC stratified by presence of at least one obligate D-lactate producing lacGDRA bacterial species (Leptotrichia trevisanii or Leptotrichia wadei) and no obligate L-lactate producing lacGDRA bacterial species (N=64) vs. at least one L-lactate producing species (Lactobacillus paragasseri, Streptococcus infantis, Lactobacillus johnsonii, Streptococcus sp. oral taxon 064, or Lacticaseibacillus paracasei; N=15).
I. OS for patients with HNSCC stratified by presence of at least one obligate D-lactate producing lacGDRA bacterial species (Leptotrichia trevisanii or Leptotrichia wadei) and no obligate L-lactate producing lacGDRA bacterial species (N=77) vs. at least one L-lactate producing species (Lactobacillus paragasseri, Streptococcus infantis, Lactobacillus johnsonii, Streptococcus sp. oral taxon 064, or Lacticaseibacillus paracasei; N=29).
Kaplan-meier survival curves with log-rank test for comparison. (A, E-I).
Although L. iners is almost exclusively a cervicovaginal microbe, LAB are ubiquitous across body sites. We hypothesized that functionally similar LAB could impact tumors in their respective niches. We identified 92 bacterial species whose genomes (N=1,586) contained the lacA, lacG, lacD, and lacR genes found in CC-L. iners; 46 in more than 3 patients (Fig. 6B–D). 40% of these species (17/46) were associated with decreased RFS and/ or OS in other cancers (Fig. 6D, Supp. Table 5).
The presence of any tumoral lacGDRA bacteria was significantly associated with lower RFS in all four cancers, including NSCLC (Fig. 6E; 27 months vs. 59 months; p<0.0001), COAD (Fig. 6F; 50 months vs. 84 months; log-rank p=0.0025), HNSCC (Fig. 6G; 20 months vs. NR; p=0.0017), and SKIN (Supp. Table 5; 6 months vs. NR; log-rank p=0.0041). Significant species included: Enterococcus (E.) faecalis (COAD), E. faecium (COAD), L. paracaseii (HNSCC), L. johnsonii (HNSCC, NSCLC), L. paragaseii (HNSCC). Staphylococcus (S.) capitis (NSCLC), S. hominis (COAD, NSCLC), S. lugdunensis (SKIN), S. saccharolyticus (NSCLC), S. warneri (COAD), S. anginosus (COAD), and Streptococcus mitis (COAD) (Supp. Table 5).
Obligate L or D-lactate production by genetically similar LAB is associated with survival.
Thirty percent (5/16) of the obligate L-lactate producers (with L-LDH gene but no D-LDH) were associated with poor patient survival (L. iners, S. infantis, S. intermedius, S. oralis, and S. sp. Oral taxon 064), while none of the five obligate D-lactate producers (Leptotrichia sp. oral taxons 221 and 498, Leptrotrichia trevisanii [L. trevisanii], Leptrotrichia wadeii [L. wadeii], S. ilei) were (Fig. 6D); 40% (2/5) of the D-lactate producers (L. trevisanii and L. wadeii) were associated with increased rather than decreased RFS and/or OS, consistent with previous reports that D-lactate producing LAB in the gut microbiome are protective56–59,59–61. No other obligate L-lactate or mixed L/D-lactate producers were associated with decreased RFS.
In HNSCC, there was nearly 100% RFS (Fig. 6H; Median NR vs. 13 months; p<0.0001) and OS (Fig. 6I; Median NR vs. 15 months; p<0.0001) in patients with only obligate D-lactate producers versus a median of 13 months in patients with at least one detrimental L-lactate producing LAB, suggesting that D-lactate could outcompete L-lactate for monocarboxylate transporters (MCTs) in tumor cells. While L. iners is a commensal cervicovaginal microbe, its functions are likely relevant in other cancers.
DISCUSSION
Here, we demonstrate that cervical cancer-associated Lactobacillus iners, an obligate L-lactate producing facultative anaerobic bacterium, induces treatment resistance through efficient L-lactate production and metabolic rewiring in tumors. This is supported by strong clinical associations, the induction of chemotherapy and radiation resistance by L. iners or L-lactate, and its high lactate production in vitro and in tumors after irradiation. This finding is not limited to cervical tumor bacteria or cervical tumors; instead, we argue that tumor-associated LAB participate in lactate-mediated metabolic rewiring in a similar fashion to described tumor-stroma or tumor-immune cell interactions, with broad implications in many cancer types.
Lactate is a powerful exchangeable metabolic coupling molecule between cancer cells, rather than simply a metabolic waste product, and cross-talks with many known treatment resistance mechanisms. Metabolic coupling occurs via exploitation of the reversible LDH enzyme to convert lactate to pyruvate or vice versa, and in turn, recycling NAD to NADH+. Tumor lactate and LDH expression are correlated with aggressive tumor biology and poor survival across cancer types62–65, including cervical cancer66, perhaps because a higher lactate level in tumors is representative of higher glucose-to-lactate metabolic flux. It can serve as a metabolic fuel, a ‘hormone’ sensed by membrane receptors, and an epigenetic modifier through histone lactylation, all of which can directly affect DNA synthesis and lead to radiation and chemotherapy resistance.
Lactate is a “lactormone,” acting in a hormonal positive feedback loop and serving as an exchangeable molecule and a key regulator of interactions between tumor cells and surrounding cells,67 including tumor-stroma, cancer cell-cancer cell, cancer cell- astrocyte, and cancer cell-fibroblast interactions. Cancer-associated fibroblasts (CAFs)68–70 can be activated by SIRTUIN3 release as a result of tumor-stroma contact, which in turn causes mitochondrial oxidation and upregulates MCT4 expression, lactate biosynthesis, glucose transporter 1 (GLUT1), and glycolysis. SIRTUIN3 is a key molecule in cervical cancer71. Lactate is also a strong promoter of dendritic cells and tumor-associated macrophage (TAM) recruitment in the tumor microenvironment. We propose that L. iners in the tumor microenvironment has a symbiotic relationship with cancer cells to fuel growth via these pathways, similar to what has been demonstrated for fibroblasts or immune cells.
Lung cancers, which share many key metabolic genes with cervical cancers, including PI3K/AKT, STK11, and TP53, exhibit a striking preference for lactate utilization over glucose to fuel the citric acid cycle63. This activity is particularly profound when cancer cells adapt to oxidative stress, wherein excess lactate leads to upregulation of MCT1 and MCT4 for overall lactate influx and efflux. This phenomenon of “lactate addiction” results in efficient use of lactate to fuel the TCA cycle and other metabolic pathways, and an even higher preference for lactate over glucose. L. iners in the tumor microenvironment may similarly “prime” cervical cancer cells and magnify the lactate feedback loop that results from oxidative stress after radiation or chemotherapy.
Many therapeutic opportunities exist to target LAB involved in therapeutic resistance. Modification of the vaginal microbiome is feasible, inexpensive, and relatively low risk. For example, in bacterial vaginosis (BV), the application of topical metronidazole followed by reconstitution of a beneficial strain of L. crispatus, known as LACTIN-V, resulted in vaginal colonization by L. crispatus in nearly 80% of patients72. Other potential therapies to eliminate L. iners, such as topical application of bacteriocins, lytic phages, bioengineered bacteria, or clinically proven probiotics73,74 could be repurposed for cancer therapy. Lactobacilli themselves are promising candidates for bioengineered microbial cancer therapeutics, due to the simplicity of genome editing and general lack of pathogenicity36,75–79. L. iners itself could be exploited to deliver anti-cancer drugs with tumor specificity, given its strong commensality to cervical tumors and the cervicovaginal niche.
Outside of bacterial targeting, systemic use of targeted therapies related to lactate uptake and conversion could also be useful in patients with tumoral LAB. Several clinically available LDH inhibitors exist, which could prevent conversion of lactate to pyruvate in these tumors. There is known strong synergy between gemcitabine and LDH inhibitors, so our data demonstrating extreme treatment resistance with L-lactate provides rationale for adjuvant or concurrent use of an LDH inhibitor in these patients80. Further, there are several promising MCT inhibitors in various stages of clinical and pre-clinical development, including first in class single inhibitors of MCT1 and MCT2, and MCT1/MCT4 and MCT2/MCT4 dual inhibitors81. MCT1 inhibitors with tumor specificity and a very high affinity for MCT182 or LDH inhibitors could be used as a targeted therapy in patients with lactate-producing bacteria.
Overall, these data provide strong evidence of an opportunity for targeted interventions in the tumor microenvironment of lactic-acid bacteria populated tumors for future clinical translation.
Limitations of the Study
Our study is limited by the absence of an in vivo L. iners colonization model to distinguish tumor and gut microbiome effects, assess tumor metabolism, or explore reversibility by eliminating L. iners, as such models arenť currently available for mice or non-human primates. Future models are needed for these investigations. Our analysis of L. iners in the tumor microenvironment only considered CD4+ CD8+ T cell composition and clonal expansion. Lactate in the tumor microenvironment affects various unexamined immune processes, including macrophage polarization and dendritic cell recruitment. Additionally, iťs unclear if L. iners, especially cancer-associated strains, confer intrinsic resistance to double-stranded DNA breaks or repair mechanisms beyond lactate's effects on radiation resistance and DNA damage. Further mechanistic studies are required and it is possible that there are other mechanisms of DNA damage and repair, which are outside the scope of this study.
STAR METHODS
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lauren Colbert (lcolbert@mdanderson.org)
Materials availability
Patient-derived Lactobacilli are grown at MD Anderson and available for sharing per request under an MTA, where applicable. The sequences of these strains have been uploaded to NCBI. The cervical cancer patient-derived organoid line, B1188, is also available at MD Anderson and available for sharing upon request under an MTA. They have been characterized, and sequencing data uploaded to SRA.
Data and code availability
16S and WGS have been deposited in SRA and will be made publicly available upon publication of the manuscript (BioProject accession numbers: PRJNA989630, PRJNA702617, PRJNA685389). L. iners strain assemblies using WMS have been deposited in NCBI and will be made publicly available upon publication (BioSample accession numbers: SAMN27176861, SAMN27176862, SAMN27176863, SAMN27176864). TCR Sequencing data has been deposited in immuneACCESS and will be made public upon acceptance. Processed microbiome, TCR sequencing, RNA Sequencing and TCGA data are available in supplementary material, along with sample metadata and clinical data.
All original code used to generate figures or tables is available in this paper’s supplemental information.
Any additional information required to reanalyze the data or code reported in this paper is available from the lead contact upon request.
Experimental Model and Study Participant Details
Study design, patient inclusion criteria and chemoradiation treatment
Patients with cervical cancer were enrolled on a University of Texas MD Anderson Cancer Center (MDACC) institutional review board (IRB) approved prospective biomarker collection study (MDACC 2014–0543, 2019–1059). We upheld all required ethical standards and approvals during the study, including, but not limited to, those set forth by the Declaration of Helsinki, Institutional Review Board (IRB) or Ethics Committees, and relevant national and international guidelines. Patients with locally advanced, non-metastatic cervical cancer planned for standard-of-care chemoradiation with curative intent were consented to the study. Patients were required to have visible cervical tumor amenable to sampling. 101 patients were enrolled from MDACC main campus and 23 from Harris Health System, Lyndon B. Johnson General Hospital Oncology Clinic (LBJ) (Supp. Table 1) between September 2015 and March 2022. Patients underwent initial staging including PET/CT and MRI prior to enrollment. The majority of tumors were International Federation of Gynecologic Oncology (FIGO) 2009 stage IIB (37%). Patients with stage IB1 disease were treated with CRT due to gross nodal disease. Patients received a minimum of 45Gy of external beam radiation therapy (EBRT) using intensity modulated- radiation therapy (IMRT) in 25 fractions over five weeks concurrently with weekly cisplatin 40 mg/m2 in 2Gy fractions, followed by two pulsed- dose rate brachytherapy treatments at approximately week five and week seven with EBRT between brachytherapy treatments for gross nodal disease or persistent disease in the parametria to a minimum dose to gross disease of 60Gy including brachytherapy contributions. Fused PET/CT and MRI scans were used for treatment planning. Brachytherapy was planned using 3-D volumetric planning, generally with MRI guidance in addition to CT. The week five sample was taken in the operating room during the first brachytherapy treatment. Response to radiation was monitored using clinical exams through the course of treatment, MRI at week five, and surveillance PET/CT +/− MRI at approximately three months post-treatment. Non- response to radiation was defined as residual FDG-avidity at the time of first follow-up PET/CT. For survival analysis, any biopsy-proven recurrence identified on physical exam or follow-up imaging was coded as a recurrence event, and any recurrence or death due to any cause was coded as an overall survival event. Recurrence-free survival (RFS) time was calculated from diagnosis to first recurrence event or last known MDACC/ LBJ clinic visit with physical exam and/or imaging. Overall survival (OS) time was calculated from diagnosis to last known contact with patient or tumor board vital status record.
Cell Lines
HeLa cells were a generous gift of the Sam Mok lab. Cells were cultured in 1X MEM with 10% FBS and 1% Penicillin/Streptomycin at 37°C and 5% CO2. These cells have not been authenticated. SiHa cells were ordered from ATCC (HTB-35). SiHa cells were cultured in 1XMEM with 10% FBS and 1% Penicillin/Streptomycin at 37°C and 5% CO2. CaSki cells were ordered from ATCC (CRL-1550). CaSki cells were cultured in RPMI1640 supplemented with 10% FBS and 1% Penicillin/Streptomycin at 37°C and 5% CO2. Both SiHa and CaSki cells were authenticated through ATCC. HeLa, SiHa, and CaSki cell lines are female.
Bacterial Strains
Lactobacillus iners isolates (ATCC, Pt-1 (IN366), Pt-2 (IN370)) were cultured in NYC III broth at 37°C in anaerobic conditions (10% CO2, 5% H2, nitrogen balance). Lactobacillus crispatus isolates (ATCC and Pt-3 (I012T4)) were cultured in MRS broth at 37°C in anaerobic conditions (10% CO2, 5% H2, nitrogen balance).
Method Details
Sample Collection
Physicians collected swabs and cytobrush samples of the cervical tumor at five time points: baseline, week one (after five radiotherapy fractions), week three (10–15 fractions), week five (20–25 fractions; first brachytherapy treatment), and first follow-up (12 weeks post-treatment). Tumor swabs for microbiome sequencing were collected with an Isohelix Buccal Swab (Isohelix, DSK-50). Swabs were placed into individual collection tubes and transported at room temperature to the lab within 4 hrs. 400 μLs of stabilization buffer (Isohelix) was added to each tube, or prefilled tubes were used when available (BFX S1/05/50). Tubes were vortexed for 15 secs and stored at −80°C until DNA extraction. DNA for TCR sequencing was extracted from tumor swabs using the Isohelix Xtreme DNA lysis kit per the manufacturer’s instructions (Isohelix, cat. #XME-50). Bacterial genomic DNA was extracted using MoBIO PowerSoil DNA Isolation Kit (Qiagen). DNA samples were stored at −20°C.
Tumor cells and supernatant for metabolomics and flow cytometry were collected using Cytobrush Plus Endocervical Samplers (Cooper Surgical) from the tumor and immediate region using previously validated techniques. Brushes were placed into individual conical tubes and immediately transported at room temperature to the lab. In the lab, 10 mLs of sterile complete RPMI-1640 media, containing 1% penicillin-streptomycin and gentamicin antibiotics (HyClone, Corning, Lonza, respectively) and 10% fetal bovine serum (FBS, Corning), were added to each tube, which were then vortexed for 1 min to dislodge and suspend cells. When large amounts of mucus were present, 5 mLs of dithiothreitol solution (1X Hank’s balanced salt solution, 4% bovine serum albumin, 2 mM dithiothreitol; Invitrogen) were added to the cell suspensions and passed through a 70-μm cell strainer into new conical tubes. Cells were pelleted by centrifugation and resuspended in sterile complete RPMI-1640 media for counting. For flow cytometry, pellets were immediately used for flow cytometry. For metabolomics, cells were pelleted again and resuspended in sterile freeze media, composed of 90% FBS (Corning) and 10% DMSO (Sigma-Aldrich), and stored at −80°C.
Two BBL CultureSwabs (BD Biosciences) were swabbed in the tumor region by a physician and transported to the lab within 30 minutes for downstream culturing to isolate patient Lactobacillus strains.
For blood samples, 1 mL of blood was collected into 3 6-mL serum clot activator-containing vacutainers (BD Biosciences) and transported at room temperature to the lab within 4 hrs. Upon arrival in the lab the blood was immediately stored at −80°C until metabolomics processing.
For human tissue used for organoid culture, fresh cancer tissue was obtained from a surgical resection specimen of a cervical cancer patient under the designated ethical protocol. She participated in this study under an IRB-approved protocol (2019–1059) and signed informed consent form approved by the responsible authority.
16S rRNA Sequencing
16S rRNA gene sequencing was performed at the Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine. 16S rRNA was sequenced using methods adapted from the methods used for the Human Microbiome Project and Earth Microbiome Project83,84 Briefly, the 16S rDNA V4 region was amplified by PCR using a 515F-806R primer pair and sequenced on the MiSeq platform (Illumina) using the 2×250 bp paired-end protocol. The primers used for amplification contain adapters for MiSeq sequencing and single-end barcodes allowing pooling and direct sequencing of PCR products83.
Shotgun Metagenomic Sequencing (SMS)
For clinical isolates, DNA was isolated from Lactobacillus strains with Gentra Puregene Yeast/Bact. Kit (Qiagen) following manufacturer’s instructions. For patient tumor samples, after 16Sv4 sequencing, DNA isolates from tumor swabs were sequenced with Illumina sequencers. SMS was performed by personnel of the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine. Whole Genome Shotgun sequencing was performed on genomic bacterial DNA (gDNA), which was extracted to maximize bacterial DNA yield from specimens while keeping background amplification to a minimum83–85. Libraries were constructed from each sample using the KAPA Hyper Prep Kit (Kapa Biosystems, Wilmington, MA, USA) and sequenced using the Illumina HiSeqX platform with the 2 × 150 bp paired-end read protocol. Sequencing reads were derived from raw BCL files which were retrieved from the sequencer and called into fastqs by Casava v1.8.3 (Illumina). Appropriate read preparation steps, such as quality control, trimming and filtering and host DNA removal prior to further analysis were performed using an in-house pipeline (Statistical analysis section).
T cell Receptor Repertoire Sequencing (TCR)
DNA isolated from tumor swabs was submitted for TCR sequencing to the Cancer Genomics Laboratory at the University of Texas MD Anderson Cancer Center (Houston, TX). Survey depth sequencing was performed using the Adaptive Biotechnologies immunoSEQ human T cell receptor beta (hsTCRB) Kit, Version 3 (Adaptive Biotechnologies, ISK10101). Two replicates of 200 ng DNA per sample were prepared for qPCR with the V- and J-gene specific primers provided in the immunoSEQ hsTCRB kit and the QIAGEN Multiplex PCR Kit (Qiagen, catalog no. 206145). First, 31 cycles of qPCR were performed on all replicates, then, sample manifest barcodes generated with immuSEQ Analyzer and Illumina adapters were added to each PCR replicate for eight additional qPCR cycles. The libraries were purified using a bead-based system to remove residual primers, pooled at equal volume, and checked for quality control with Agilent D1000 screen tapes to determine the size-adjusted concentration. The libraries were quantified with the Applied Biosystems QuantStudio 6 and the KAPA Biosystems library quantification kit, using manufacturer's instructions.
On the basis of the qPCR results, approximately 15 pmol/L of the pooled libraries were loaded onto the Miseq Sequencing System for a single end read which includes a 156-cycle Read 1 and a 15-cycle Index 1 read run. Raw sequences output from the Miseq were transferred to Adaptive's immunoSEQ Data Assistant, where the data were processed to report the normalized and annotated TCRB repertoire profile for each sample.
Isolation of L. iners strains from patient tumors
Culture swabs were collected from tumors and immediately placed in an anaerobic transport bag (BD 260683). Within 30 minutes of collection, tumor swabs were plated onto a TSA plate (BD Biosciences, Cat# 221239) and a MRS plate (Moltox, Cat# 51–40S020.140) and incubated at 37°C in anaerobic conditions for 3–5 days. Bacterial growth from plates was sub-cultured until single colonies could be isolated. MALDI-TOF mass spectroscopy was used to identify the bacterial isolate species and performed according to the standard protocol described in the Bruker MALDI-TOF user manual.
Generation of patient-derived organoids
Patient-derived organoids were generated as described by Lõhmussaar et al86, with several modifications. Cervical cancer tissue was mechanically minced using scalpels, followed by digestion in a dissociation mixture (1 mg/ml collagenase (Sigma) and 0.4 mg/ml Hyaluronidase (Sigma) in complete RPMI medium, supplemented with 10% FBS and 1% penicillin-streptomycin, for 1.5 hours at 37°C in a water bath shaker. The resulting cells were centrifuged down at 350g for 5 min, followed by adding 1–5ml of Trypsin-EDTA (Gibco) supplemented with 100ug/ml DNase I (Sigma) and digesting the cell clumps for 8–10 min at 37°C waterbath shaker. The resulting cell suspensions were washed three times with AdDF+++ (Advanced DMEM/F12 supplemented with 1x Glutamax, 10 mM HEPES, and penicillin-streptomycin), and erythrocytes were lysed in Red Blood Cell Lysis Buffer (Roche). The cells and small cell clumps were embedded into Basement Membrane Extract (BME, Cultrex BME RGF type 2, Trevigen) and plated in 10 ul-volume droplets on a pre-warmed 24-well suspension culture plates and allowed to solidify at 37°C for 30 min before addition of full growth medium (AdDF+++ supplemented with 1% Noggin conditioned medium (made in-house), 10% of RSPO1 conditioned medium (made in-house), 1x B27 supplement (GIBCO), 2.5 mM nicotinamide (Sigma), 1.25 mM n-Acetylcystein (Sigma), 10 mM ROCK inhibitor (Abmole), 500 nM A83–01 (Tocris), 10 mM forskolin (Bio-Techne), 25 ng/ml FGF7 (Peprotech), 100 ng/ml FGF10 (Peprotech) and 1 mM p38 inhibitor SB202190 (Sigma), 50 ng/ml EGF (Peprotech) and 100 mg/ml Primocin (InvivoGen)). For splitting, organoids were pipetted up and down 300 times using an electronic pipettor through a small-bore pipette tip to break up the BME and separate organoids from the BME. After centrifugation, organoids were dissociated with TrypLE (Gibco) for 5 min in a cell culture incubator. The cells were pipetted up and down by 100 to break up the remaining cell clumps and organoids into single cells. The approximate splitting ratio was 1:4 every two weeks.
Immunostaining and imaging of organoids
Organoids were fixed 20 min in 4% paraformaldehyde (PFA) at room tempreture followed by dehydration and paraffin embedding. Serial sections were cut as 5 μm and hydrated before staining. Sections were subjected to PAS staining following the manufacturer’s instructions or immunohistochemical staining by using overnight incubation with antibodies raised against P63 (Abcam), KI67 (Abcam), and CK13 (Abcam). The antigen retrieval was performed in citric acid solution (pH 6.0). Fluorescent images were acquired on Axio Observer microscope (Zeiss). The bright view images were taken on Cytation5 Cell Imaging Multimode Reader (Agilent). For organoids size counting, the diameter of each PDO in the image larger than 20 μm was taken in count using GEN 5 software.
Lactobacilli Treatments and cell viability assay
Bacterial strains were cultured under anaerobic conditions at 37°C and 190 rpm for 3–4 days until the cells reached a density of approximately 1 × 109 cells/mL87. The cultures were centrifuged at 4000 rpm for 40 min at 20°C. The supernatants were sterile-filtered to prepare the CFS. After centrifugation, the bacterial pellets were resuspended in PBS to a final concentration of 108/ml and killed using under UV-light for 1h.
For CFS treatment, HeLa, SiHa, and CaSki cells and cervical cancer-derived primary cells were seeded into 6-well plates at 106 cells/well and left to adhere overnight. Once adhered, the cell media was changed to 20% v/v CFS and cultured at 37°C and 5% CO2 for two weeks; passages occurred once a week. After incubation, cells were seeded in 96 well plates at 1000 cells/well and allowed to adhere overnight. The cells were then treated with or without ionizing radiation and cisplatin (Sigma-Aldrich, Cat# P4394), 5-fluorouracil (Sigma-Aldrich, Cat# F6627), gemcitabine (Selleck Chemical LLC S1149100MG), or a combination, as indicated in the figures. After all treatments, cells were allowed to grow for 4–6 days in 20% CFS v/v culture medium until the negative control wells reached approximately 75% confluence before cell viability was assessed. For PDOs, single cells were resuspended in ice cold BME (Trevigen, Cat#3533-005-02) and irradiated as indicated, and then the cells were seeded as 5 µL of BME in a U-bottom 96-well microplate. After 2–3 days, when small organoids emerged from single cells, 20% CFS diluted in full growth medium was added to the wells and incubated overnight for 16h, followed by cisplatin treatments for 1h. After all treatments, the medium was replaced with a full growth medium containing 20% v/v CFS and incubated for 14 days. The PDO in BME were then subjected to a cell viability assay. For the dead bacteria test, cervical cancer-derived primary cells were seeded into 96-well microplates and treated with irradiation or cisplatin. After washing with full growth medium, the dead bacterial pellets were diluted and added to the cells at a density of 160 CFU/cell and co-cultured with the cells for five days, followed by a cell viability assay.
For HeLa, CaSki, SiHa, and cervical cancer-derived primary cells (B1188), a cell viability assay was performed using CellTiter-Glo according to the manufacturer’s instructions. In brief, CellTiter-Glo Reagent (Promega) was applied to the culture medium at a 1:1 ratio. The CellTiter-Glo 3D Reagent (Promega) was applied to the PDOs. The plates were shaken every 5 minutes and incubated for 30 minutes. The luminescence was measured using a Perkin Elmer Victor X3 plate reader. Percent cell viability for each treatment group was calculated by normalizing luminescence readings to those of the non-treated IR or chemo group.
Immunofluorescence staining
For γH2AX and panCK staining, cells were fixed in ice cold methanol for 5 min at room temperature and washed once with phosphate buffered saline (PBS). After fixation, the cells were incubated with antibody overnight in cold room. After primary antibody incubation, the cells were washed three times with PBS and incubated with Alexa Fluor 488-labeled anti-mouse IgG antibody (Invitrogen) containing DAPI for 1 hr at room temperature. Finally, cells were mounted in mounting solution ProLong Gold (Invitrogen). Fluorescent images of panCK were acquired on Axio Observer microscope (Zeiss). Image acquisition of γH2AX was performed with a Cytation 5 Cell Imaging Multimode Reader. Microscopy image analyses were performed using the GEN 5 software. Nuclei were segmented using the DAPI channel and the resulting regions of interest transferred to the fluorescence channel of interest (488 for gammaH2AX). Prism 9 (GraphPad Software) was used to calculate P values based on T-test analyses. Data were considered statistically significant for P values < 0.05.
Bulk RNA sequencing
RNA samples were submitted to Cancer Genomics Center at The University of Texas Health Science Center at Houston. The RNA sample quality was assessed by RNA ScreenTape on a Tapestation (Agilent Technologies Inc., California, USA) and quantified by Qubit 2.0 RNA HS assay (ThermoFisher, Massachusetts, USA). Paramagnetic beads coupled with oligo d(T)25 are combined with total RNA to isolate poly(A)+ transcripts based on NEBNext® Poly(A) mRNA Magnetic Isolation Module manual (New England BioLabs Inc., Massachusetts, USA). Prior to first strand synthesis, samples are randomly primed (5´ d(N6) 3´ [N=A,C,G,T]) and fragmented based on manufacturer’s recommendations. The first strand is synthesized with the Protoscript II Reverse Transcriptase with a longer extension period, approximately 30 minutes at 42°C. All remaining steps for library construction were used according to the NEBNext® Ultra™ II Library Prep Kit for Illumina® (New England BioLabs Inc., Massachusetts, USA). Final libraries quantity was assessed by Qubit 2.0 (ThermoFisher, Massachusetts, USA) and quality was assessed by TapeStation HSD1000 ScreenTape (Agilent Technologies Inc., California, USA). Final library size was about 500bp with an insert size of about 350bp. Illumina® 8-nt dual-indices were used. Equimolar pooling of libraries was performed based on QC values and sequenced on an Illumina® [NovaSeq S4] (Illumina, California, USA) with a read length configuration of 150 PE for [40] M PE reads per sample (20M in each direction).
Lactate isoform treatment of cell lines
Stock solutions of L-lactic acid, D-lactic acid, sodium L-lactate and sodium D-lactate were made at the same concentration, 4M. We used 20 mM lactic acid or lactate to culture the cells for two weeks. And the cells were seeded 1000 cells/well in a 96-well plate for chemoradiation treatments. After treatments, the cells were cultured in medium containing 20 mM lactic acid or lactate for 3–5 days, followed by CellTiterGlo assay.
DNA Synthesis Assay
Cancer derived primary cells were cultured in either broth (control), NC-L. Iners CFS, or CC-L. Iners CFS supplemented culture medium for 2 weeks, and then re-seeded and cultured for 1 day, followed by IR and serial timepoints of recovery. The cells were subjected to the DNA synthesis analysis using the Click-iT™ EdU Alexa Fluor™ 488 Flow Cytometry Assay Kit (Invitrogen, cat. No. C10425). EdU at a final concentration of 40 µM was applied for 15 minutes, then cells were recovered for 60, 90, 120, 150 min and fixed and stained following the manufacturer’s instructions with the PI staining step included. Cells were analyzed on a Thermo Fisher Attune Nxt Flow Cytometer using the 488 nm BL3 filter for EdU and 638 RL2 filter for PI.
Tumor and Clinical isolate Metabolomics
To determine the relative abundance of polar metabolites in samples, extracts were prepared and analyzed by ultra-high resolution mass spectrometry (HRMS). Metabolites were extracted using ice-cold 0.1% Ammonium hydroxide in 80/20 (v/v) methanol/water. Extracts were centrifuged at 17,000 g for 5 min at 4°C, and supernatants were transferred to clean tubes, followed by evaporation to dryness under nitrogen. Dried extracts were reconstituted in deionized water, and 5 μL was injected for analysis by ion chromatography (IC)-MS. IC mobile phase A (MPA; weak) was water, and mobile phase B (MPB; strong) was water containing 100 mM KOH. A Thermo Scientific Dionex ICS-5000+ system included a Thermo IonPac AS11 column (4 µm particle size, 250 × 2 mm) with column compartment kept at 30°C. The autosampler tray was chilled to 4°C. The mobile phase flow rate was 350 µL/min and gradient from 1mM to 100mM KOH was used. The total run time was 60 min. To assist the desolvation for better sensitivity, methanol was delivered by an external pump and combined with the eluent via a low dead volume mixing tee. Data were acquired using a Thermo Orbitrap Fusion Tribrid Mass Spectrometer under ESI negative ionization mode at a resolution of 240,000.
Analysis of D/L-Lactate levels
For rapid, quantitative analysis of D/L lactate levels in bacterial culture, the Megazyme (Neogen) D-/L-Lactic Acid (D-/L-Lactate) (Rapid) Assay Kit was used according to manufacturer instructions with spectrophotometry. To quantitate the relative abundance of D/L-Lactate in primary cell culture media, bacterial culture and tumor cytobrush samples, extracts were prepared and analyzed by Thermo Scientific TSQ Quantiva triple quadruple mass spectrometer coupled with a Dionex UltiMate 3000 HPLC system. Samples were stored in −80 °C freezer and thawed on ice before analysis. 100 μL of samples were aliquoted and metabolites were extracted using cold 80/20 (v/v) methanol/water. Samples were then vortexed, centrifuged at 17,000 g for 10 minutes at 4°C, and organic layers were transferred to clean tubes, followed by evaporation to dryness under nitrogen. Dried extracts were reconstituted in 100 μL of 85/15 (v/v) acetonitrile/50mM ammonium acetate in water, and 5 μL was injected for analysis by liquid chromatography (LC)-MS. The mobile phase A(MPA) is acetonitrile and mobile phase B(MPB) is 33.3 mM ammonium acetate in water. Separation of D/L-Lactate was achieved on an Astec Chirobiotic R, 5um 2.1×150mm column with 15% MPB isocratic condition. The flow rate was 400 µL/min at column temperate was 30 °C. The total run time was 10 minutes. The mass spectrometer was operated in the MRM negative ion electrospray mode with the transitions: m/z 89.1 -> 43.0 for D-Lactate, m/z 89.1 -> 43.1 for L-Lactate, m/z 92.0 -> 45.1 for D-Lactate-13C3 m/z 92.0 -> 45.0 for L-Lactate-13C3. Raw data files were imported to Thermo Trace Finder software for final analysis. The relative abundance of D/L-Lactate was normalized by their stable isotope labeled internal standards.
Comparative genomic analysis of patient-derived isolates and assembled lactobacillus metagenomes
Two patient-derived L. iners strains were also completely sequenced (Pt1, Pt2; Supp. table 4). Near-complete assemblies (99.6% and 90.5% completeness) were produced for these strains and then compared with sequenced strains from patients without cancer: L. iners KY (complete genome; healthy patient) and ATCC 55195 (draft genome, 98.7% completeness; bacterial vaginosis patient) (Supp. Fig. 4E). The comparison of two cancer-derived strains with ATCC 55195 confirmed the similarity of 2 patient-derived L. iners with each other and notable differences from ATCC 55195 (Supp. Fig. 4E). The two cancer-derived strains demonstrated more significant enrichment of functions involved in bacterial immunity and virulence as compared to non-cancer derived strains (Supp. Fig. 5A).
We further used the Pathway Tools software88 to generate PGDBs (Pathway Genome Databases) of the cancer-derived and non- cancer-derived L. iners strains, in addition to a near complete assembly of a strain derived from a third patient’s WMS data, referred as I012T4 (97% completeness).
Quantification and Statistical Analysis
Postprocessing of 16S ribosomal RNA sequencing
The 16S rRNA gene data analysis incorporated phylogenetic-based and alignment-based approaches to maximize data resolution. Sequence read pairs were demultiplexed using unique molecular barcodes, and reads were merged using USEARCH version 7.0.1090. 16S analysis was performed using custom analytic packages and pipelines developed at the Alkek Center for Metagenomics and Microbiome Research at Baylor University to create summary statistics and quality control measurements for each sequencing run, as well as multi-run reports and data-merging capabilities for validating built-in controls and characterizing microbial communities across large numbers of samples or sample groups.
16S rRNA sequence reads were processed and analyzed using the QIIME2 microbiome bioinformatics platform (version 2020.11)89. Rarefaction sampling depth was not utilized and thus the data was not rarefied for the analysis presented in this study. Raw FASTQ paired-end sequences were imported and demultiplexed as QIIME2 artifacts. Amplicon Sequence Variant (ASV) feature tables were constructed using DADA2 denoising90. Phylogenetic reference tree construction was performed using a pre-trained Naïve Bayes classifier and the q2-feature-classifier plugin. For rectal samples, the taxonomic classifier used was trained on the SILVA 138 515F/806R region of sequences91 .For cervical tumor samples we used a custom classifier trained on a cervicovaginal specific database92. Phyla-level eukaryotic, organellar, and unclassified taxa were inspected for removal prior to diversity and compositional analysis for quality control purposes, but rare or low abundant features were not targeted for removal or filtering. Alpha diversity and evenness metrics were calculated through QIIME2. The indices used in this study are Shannon Diversity Index (SDI), Simpson Diversity, Faith’s Phylogenetic Diversity (PD), Fisher’s index, Pielou’s evenness, Simpson’s evenness, and Observed ASV’s (Observed Features).
Postprocessing of tumor SMS
Postprocessing of SMS data was implemented using a set of software tools summarized in the key resources table. The paired-end raw sequence reads in fastq format were filtered and trimmed using BBMap93. To remove contamination by human DNA, which was abundant in the cervical swabs, the Bowtie 294 was used to align filtered and trimmed sequencing reads to a hg38 reference genome (GCA_000001405.28). After removing the contaminated human reads, the remaining reads were assembled into contigs by both MEGAHIT [doi: 10.1093/bioinformatics/btv033] and metaSPAdes95. Only contigs that were larger than 1000 bp were used for binning by MetaBAT296. Genes were predicted in each assembled contigs by Prodigal97 and then annotated by KofamKOALA98, which assign KOs ID, gene name, function, and EC number. Taxonomic classification of each contig was implemented by CAT and BAT99. All the software tools were run with default parameters. Versions and sources of the software tools or packages used in the pipeline are listed in the key resources table. Output of the assembly was a set of assembled contigs for each sample, their taxonomic annotation, predicted genes, their location in the contig, strand, size, KO IDs, genes names, gene functions (description), and EC numbers. Some genes were annotated by 2 or more KOs or/and EC numbers.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD33 antibody | BD Biosciences | Cat# 740293; RRID:AB_2740032 |
| CD86 (B7-2) antibody | BD Biosciences | Cat# 564428; RRID:AB_2738804) |
| CD141 antibody | BD Biosciences | Cat# 565321; RRID:AB_2739180) |
| Brilliant Violet 605(TM) anti-human CD11b antibody | BioLegend | Cat# 301332; RRID:AB_2562021) |
| Brilliant Violet 711(TM) anti-human Ki-67 antibody | BioLegend | Cat# 350516; RRID:AB_2563861) |
| CD152 (CTLA-4) antibody | BD Biosciences | Cat# 563931; RRID:AB_2738491) |
| CD3 antibody | BD Biosciences | Cat# 560835; RRID:AB_2033956) |
| PE anti-human CD45 antibody | BioLegend | Cat# 368510; RRID:AB_2566370) |
| Granzyme B antibody | BD Biosciences | Cat# 562462; RRID:AB_2737618) |
| CD11c antibody | BD Biosciences | Cat# 551077; RRID:AB_394034) |
| CD4 Monoclonal Antibody (S3.5), PE-Alexa Fluor 700 | Thermo Fisher Scientific | Cat# MHCD0424; RRID:AB_10372802) |
| PKR Polyclonal Antibody | Thermo Fisher Scientific | Cat# PA1-990; RRID:AB_560651) |
| Mouse Anti-CD69 Monoclonal Antibody | BD Biosciences | Cat# 555533; RRID:AB_398602) |
| Alexa Fluor(R) 700 anti-human CD8a antibody | BioLegend | Cat# 300920; RRID:AB_528885) |
| APC/Cyanine7 anti-human CD279 (PD-1) antibody | BioLegend | Cat# 329922; RRID:AB_10933429) |
| Brilliant Violet 650 anti-IFN-gamma | BD Biosciences | Cat# 563416 |
| Alexa Fluor 488 anti-Foxp3 | Fisher Scientific | Cat# 53477642 |
| PE-Cy7 anti-HLA-DR | BD Biosciences | Cat# 560651 |
| Anti-GammaH2AX antibody | EMD Millipore | Cat# 05-636 |
| Anti-panCK antibody | Novus Biological | Cat# NBP2-29429 |
| Alexa Fluor 488-labeled anti-mouse IgG antibody containing DAPI | Invitrogen | Cat# A21202 |
| Anti-P63 antibody | Abcam | Cat# AB735 |
| Anti-KI67 antibody | Abcam | Cat# AB16667 |
| Anti-CK13 antibody | Abcam | Cat# AB92551 |
| Biological Samples | ||
| Human Tumor Sampling - Cytobrush | The University of Texas MD Anderson Cancer Center, Houston, TX | Protocol 2014-0543 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BBL CultureSwabs | BD Biosciences | Cat# 220135 |
| Anaerobic Transport Medium Surgery Pack (ATMSP) | Anaerobe Systems | Cat# AS-914 |
| Cytobrush Plus Endocervical Samplers | Cooper Surgical | Cat# C0012 |
| BuccalFix Tubes ™ | Isohelix | Cat# BFX/S1-05-50 |
| Isohelix Swab Pack | Isohelix | Cat# SK-3S |
| Stabilization Buffer | Isohelix | Cat# BFX-25 |
| Serum Blood Collection Tubes | Becton Dickinson | Cat# 367815 |
| Golgiplug | BD Biosciences | Cat# 555029 |
| MRS Agar Plates | Moltox | Cat# 51-40S020.140 |
| TSA Agar Plates (BD BBL™ Trypticase™ Soy Agar with 5% Sheep Blood (BD Trypticase™ Soy Agar II™)) | BD Biosciences | Cat# 221239 |
| Lactobacillus MRS Dehydrated Culture | BD Difco | Cat# 288210 |
| HEPES | Fisher Bioreagents | Cat# BP310100 |
| Proteose Peptone No. 3 | Bacto | Cat# C838M71 |
| NaCl | Fisher Chemical | Cat# S271500 |
| D-Glucose | Avantor Macron | Cat# 491212 |
| Fresh yeast extract | Gibco | Cat# 18180059 |
| Heat Inactivated Horse Serum | Remel | Cat# R55075 |
| Corning™ Minimum Essential Medium Eagle (Mod.) 1X (MEM) with Glutamine | Corning | Cat# 10010CV |
| Corning® Fetal Bovine Serum (FBS), 500 mL, Regular, USDA Approved Origin | Corning | Cat# 35010CV |
| Corning® 100 mL Penicillin-Streptomycin Solution, 100x | Corning | Cat# 30002CI |
| Gentra Puregene Yeast/Bact. Kit | Qiagen | Cat# 158567 |
| Cisplatin (cis-Diammineplatinum(II) dichloride) | Sigma-Aldrich | Cat# P4394 |
| 5-fluoruracil | Sigma-Aldrich | Cat# F6627 |
| Gemcitabine | Selleck Chemical LLC | Cat# S1149100MG |
| CellTiter-Glo and CellTiter-Glo 3D Luminescent Cell Viability Assay | Promega | Cat# G7572 Cat# G9683 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Thermo Fisher | Cat# L34957 |
| Phytohemagglutinin (PHA) | Sigma-Aldrich | Cat# 11249738001 |
| Corning® 500 mL RPMI 1640 | Corning | Cat# 10-040-CV |
| gentamicin antibiotics | Lonza | Cat# 17518Z |
| collagenase | Sigma-Aldrich | Cat# C9407 |
| Hyaluronidase | Sigma-Aldrich | Cat# H3506 |
| Trypsin-EDTA | Gibco | cat# 25200056 |
| DNase I | Sigma-Aldrich | cat# DN25 |
| Red Blood Cell Lysis Buffer | Roche | Cat# 11814389001 |
| Cultrex BME RGF type 2 | Trevigen | Cat# 3533-005-02 |
| B27 supplement | GIBCO | Cat# 175044 |
| nicotinamide | Sigma-Aldrich | Cat# N0636 |
| n-Acetylcystein | Sigma-Aldrich | Cat# A9165 |
| ROCK inhibitor | Abmole | Cat# Y27632 |
| A83-01 | Tocris | Cat# 2939 |
| Forskolin | Bio-Techne | Cat# 1099 |
| FGF7 | Peprotech | Cat# 100-19 |
| FGF10 | Peprotech | Cat# 100-26 |
| p38 inhibitor SB202190 | Sigma-Aldrich | Cat# 7067 |
| EGF | Peprotech | Cat# AF-100-15 |
| Primocin | InvivoGen | Cat# Ant-pm-1 |
| TrypLE™ Express Enzyme (1X), phenol red | Gibco | Cat# 12605036 |
| EDTA-containing vacutainers | BD Biosciences | Cat# 366643 |
| Lymphoprep | Stemcell Technologies or Cosmo Bio Usa | Cat# 07851 or Cat# AXS1114545 |
| dithiothreitol | Invitrogen | Cat# P2325 |
| Corning® Phosphate-Buffered Saline, 1X without calcium and magnesium, PH 7.4 ± 0.1 | Corning | Cat# 21-040-CV |
| Sodium L-Lactate | Sigma-Aldrich | Cat# L7022-5g |
| Sodium D-Lactate | Sigma-Aldrich | Cat# 71716-5g |
| D-Lactic Acid | TCI America | Cat# L02665G |
| L-Lactic Acid | Sigma-Aldrich | Cat# L6402-5g |
| ProLong Gold Antifade Mountant | Invitrogen | Cat# P36930 |
| Kits | ||
| D-/L-Lactic Acid (D-/L-Lactate) (Rapid) Assay Kit | Megazyme/Neogen | Cat# K-DLATE |
| ImmunoSEQ human T-cell receptor beta (hsTCRB) Kit, Version 3 | Adaptive Biotechnologies | Cat# ISK10101 |
| Qiagen Multiplex PCR Kit | Qiagen | Cat# 206145 |
| NEBNext Ultra II Library Prep Kit for Illumina | New England BioLabs | Cat# E7645L |
| Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit | Invitrogen | Cat# C10425 |
| Megazyme (Neogen) D-/L- Lactic Acid (D-/L-Lactate) (Rapid) Assay Kit | Neogen, Megazyme | Cat# K-DLATE |
| Xtreme DNA Isolation Kit | Isohelix | Cat# XME-50 |
| Deposited Data | ||
| 16S | This paper; SRA | BioProject accession numbers: PRJNA989630, PRJNA702617, PRJNA685389 |
| SMS | This paper; SRA | BioProject accession numbers: PRJNA989630, PRJNA702617, PRJNA685389 |
| L. iners strain assemblies | Genome | BioSample accession numbers: SAMN27176861, SAMN27176862, SAMN27176863, SAMN27176864 |
| RNA sequencing | This paper; NCBI/SRA | PRJNA1013527 |
| TCR sequencing | This paper; immuneACCESS | immuneACCESS: https://doi.org/10.21417/LC2022CIR. |
| Code for processing TCGA data | This paper | https://github.com/mda-primetr/Colbert_CancerCell_2023 |
| Experimental Models: Cell Lines | ||
| HeLa | Sam Mok lab, MD Anderson Cancer Center | RRID:CVCL_0030 |
| SiHa | ATCC | Cat# HTB-35 |
| Ca Ski | ATCC | Cat# CRM-CRL-1550 |
| Experimental Models: Organisms/Strains | ||
| ATCC Lactobacillus iners | ATCC | ATCC 55195; NCBITaxon:888801 |
| ATCC Lactobacillus crispatus SJ-3C-US | ATCC | ATCC SJ-3C (PTA-10138); NCBITaxon:575598 |
| Patient-derived Lactobacillus iners I012T4 | This paper, sequences in NCBI | Accession # SAMN27176861 |
| Patient-derived Lactobacillus iners IN370 | This paper, sequences in NCBI | SAMN27176863 |
| Patient-derived Lactobacillus iners IN366 | This paper, sequences in NCBI | SAMN27176862 |
| Software and Algorithms | ||
| ATIMA (Agile Toolkit for Incisive Microbial Analyses) | R Studio | http://atima.jplab.net/ |
| Galaxy: LEfSe | Segata et. al 2010 | https://huttenhower.sph.harvard.edu/galaxy/ |
| Other (Resources) | ||
| FlowJo | FlowJo Software | www.flowjo.com |
| GraphPad Prism | GraphPad Software | www.graphpad.com |
| JMP | SAS | www.jmp.com |
| Living Image Software | IVIS Spectrum - Perkin Elmer | Part #128113 |
| FastQC (version 0.11.8) | https://www.bioinformatics.babraham.ac.uk/projects/fastqc | |
| FastQ_Screen (version 0.14.0) | (Wingett and Andrews 2018) | https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen |
| BBDuk/BBTools (version 20190109) | https://jgi.doe.gov/data-and-tools/bbtools | |
| BBMap/BBTools (version 20190109) | https://jgi.doe.gov/data-and-tools/bbtools | |
| Bowtie2 (version 2.3.5) | (Langmead and Salzberg 2012) | https://github.com/BenLangmead/bowtie2 |
| Samtools (version 1.9) | (H. Li et al. 2009) | https://github.com/samtools |
| IGGsearch (version 20190921) | (Nayfach et al. 2019) | https://github.com/snayfach/IGGsearch |
| MetaPhlAn2 (version 2.0) (mpa_v295_CHOCOPhlAn_201901) | (Truong et al. 2015) | https://bitbucket.org/biobakery/metaphlan2 |
| MEGAHIT (version 1.2.8) | (D. Li et al. 2015) | https://github.com/voutcn/megahit |
| metaSPAdes (version 3.13.1) | (Nurk et al. 2017) | https://github.com/ablab/spades/releases |
| MetaBAT2 (version 20191004) | (Kang et al. 2019) | https://bitbucket.org/berkeleylab/metabat |
| Prodigal (version 2.6.3) | (Hyatt et al. 2010) | https://github.com/hyattpd/Prodigal |
| Diamond (version 0.9.24) | (Buchfink, Xie, and Huson 2015) | https://github.com/bbuchfink/diamond |
| KofamScan (version 1.1.0) | (Aramaki et al. 2020) | https://github.com/takaram/kofam_scan |
| Subread (version 1.6.3) | (Liao, Smyth, and Shi 2014) | http://subread.sourceforge.net |
| DFAST (version 2021.7.12) | https://doi.org/10.1093/bioinformatics/btx713 | https://dfast.ddbj.nig.ac.jp |
| MetaCyc (version 25.5) | https://doi.org/10.1093/nar/gkx935 | https://biocyc.org/download-bundle.shtml |
| Pathway Tools (version 25.5) | https://doi.org/10.1093/bib/bbv079 | https://biocyc.org/download-bundle.shtml |
| FastANI | DOI: 10.1038/s41467-018-07641-9 | https://github.com/ParBLiSS/FastANI |
| QIIME2 (version 2020.11) | https://www.nature.com/articles/s41587-019-0209-9 | |
| DADA2 | doi:10.7717/peerj.5364 | |
| STAR (v. 2.3.0) | DOI: 10.1093/bioinformatics/bts635 | http://code.google.com/p/rna-star/ |
| HTSeq (v. 2.0) | DOI: 10.1093/bioinformatics/btu638 | https://pypi.org/project/HTSeq/ |
| ANNOVAR (v. 20211019) | DOI: 10.1093/nar/gkq603 | http://www.openbioinformatics.org/annovar/. |
| RStudio (v. 2023.03.0+386) | Posit Team | https://posit.co/download/rstudio-desktop/ |
| tidyverse | Rstudio | doi:10.21105/joss.01686 |
| ggplot2 | Rstudio | https://ggplot2.tidyverse.org |
| ggpubr | Rstudio | https://rpkgs.datanovia.com/ggpubr/ |
| dplyr | Rstudio | https://CRAN.R-project.org/package=dplyr |
| ggsignif | Rstudio | doi:10.31234/osf.io/7awm6 |
| plyr | Rstudio | https://www.jstatsoft.org/v40/i01/ |
| rstatix | Rstudio | https://CRAN.R-project.org/package=rstatix |
| survival | Rstudio | https://CRAN.R-project.org/package=survival |
| survminer | Rstudio | https://CRAN.R-project.org/package=survminer |
| broom | Rstudio | https://CRAN.R-project.org/package=broom |
| lmerTest | Rstudio | https://doi.org/10.18637/jss.v082.i13 |
| sjPlot | Rstudio | https://CRAN.R-project.org/package=sjPlot |
| sjmisc | Rstudio | https://doi.org/10.21105/joss.00754 |
| tidyr | Rstudio | https://CRAN.R-project.org/package=tidyr |
| purrr | Rstudio | https://CRAN.R-project.org/package=purrr |
| mclust | Rstudio | https://doi.org/10.32614/RJ-2016-021 |
| circlize | Rstudio | https://cran.r-project.org/web/packages/circlize/index.html |
| ComplexHeatmap | Rstudio | doi:10.1093/bioinformatics/btw313 |
Postprocessing of RNA sequencing data
Processing of RNA-seq reads was implemented by the Genomic Medicine computational pipeline at MDACC. The processed reads were mapped to the hg19 reference by RNA-seq aligner STAR100 and then quantified as counts by HTSeq101,102 and annotated by ANNOVAR. Normalization of the gene counts and identification of differentially expressed genes (DEGs) were implemented by R library ‘edgeR’ and ‘limma’ with ‘voom’ as described103 using R version 4.2.2 (2022-10-31). Differentially active pathways were determined by Quantitative Set Analysis of Gene Expression (QuSAGE) as described104 with default parameters. Hallmark pathways gene set was downloaded from MSigDb v.7.5.1105.
Postprocessing of Lactobacillus isolates shotgun metagenome sequences
Postprocessing of the obtained fastq files was implemented by the same computational workflow used for postprocessing of SMS data. The workflow, which is described above, generated assembled contigs for each strain that were further curated to avoid duplications. The assembled contigs for each strain were annotated using a prokaryotic genome annotation pipeline DFAST106. The software implements gene predictions for protein coding sequences, rRNA, tRNA, and CRISPR, and infers protein functions. Completeness check was calculated by DFAST at the genus level (Lactobacillus, 14 genome, 238 marker sets) and revealed completeness values 98.7% for ATCC 55195 strain and 99.6% and 90.5% for Pt1 and Pt2 respectively. The level of contamination calculated by DFAST was 0.6–0.67%. Identity check evaluated taxonomy identity by calculating average nucleotide identity and by comparison the value with 13000 reference genomes using FastANI software107, HMM scan against TIGRFAM and RPSBLAST against COG were enabled as advanced options of the annotation. Gene predictions were done using Prodigal97 as an option provided by DFAST. Characteristics of the assemblies are provided in Supp. table 4. The PathoLogic component108 of the Pathway Tools software 25.5 and MetaCyc v.25.5109 were used to generate PGDBs for draft genomes of 2 L. iners strains (pt1 and pt2) isolated from cervical swabs of 2 CC patients, ATCC 55195 strain isolated form a patient with bacterial vaginosis, I012T4 strain (draft genome assembled from WMS data of CC patient 12) and for the complete genome of L. iners isolated from a healthy individual. The default parameters were used if not specified to run the tools. The Pathway tools were also used to predict transcription units, generate contigs overviews, pathway diagrams, and to compare the generated PGDBs. Characteristics of the generated PGDBs are provided in Supp. table 4.
Detection and postprocessing of reads of bacterial origin in TCGA datasets
Primary tumor samples from all publicly available datasets on cBioPortal for NSCLC, HNSCC, COAD and SKIN were included. Single and paired-end reads were quality trimmed and quality filtered using BBduk. Reads of human origin were filtered using the bloom filter from the BB tools suite93,110 using a cutoff of >15 31mers matching the human genome (GRChg38)111. Paired-end sequencing reads were merged using BBMerge from the BB tools suite1 under “maxstrict” parameters. Combined merged and single-end reads were dereplicated using VSEARCH112. Dereplicated reads were normalized using normalized-by-median.py from the khmer suite113. The resulting readset was mapped against all bacterial reference entries in GenBank (gbbct) using BLASTn114 with a e-value cutoff of 0.00001, a word length of 29, and a 95% identity cutoff. In parallel, DIAMOND115 was used to perform a translated alignment of the normalized reads against all bacterial proteins in gbbct. All aligned reads (nucleotide and translated alignments) were extracted and aligned to GenBank primate (gbpri), vertebrate (gbvrt), and SILVA116 16S database v138. Reads with alignment bit scores higher for non-bacterial entries were eliminated. Candidate references genomes were selected from the combined nucleotide and translated nucleotide alignments using the minimum set cluster function (clusterReferences.pl) from VirMAP117. Selected references were used as scaffolds to reconstruct putative genomes. Genomes counts were calculated by remapping the original set of reads (post quality trimming and filtering) to the reference genomes selected in the prior step. Taxonomies with a significant skew between total input kmers and kmer diversity were filtered as suspected contaminants.
Statistical Analysis
All code and standardized datasets used for the analysis are publicly available in the supplementary material (Data S1, S2). All raw and processed 16S and SGS sequences, including assembled isolate bacterial genomes are uploaded to SRA and dbGap. All analyses presented were performed on the initial cohort, the validation cohort, and the full cohort independently. We performed strict evaluation for batch effects, including sensitivity analyses for endpoints of interest by batch for each timepoint, institution, and patient demographics. We also analyzed batches separately by institution, with no differences in the outcomes. We also noted that batch 4 had a higher number of features than the other batches (Supp. Fig. 7). Therefore, we performed sensitivity analyses with and without it to ensure that it did not affect our results.
Linear discriminant analysis effect size (LEfSe) was used to identify taxa that were enriched in baseline samples, with the clinical response set as “class”118. A linear discriminant analysis (LDA) score of 4.0 was used with an 0.05 alpha value for the Kruskal-Wallis all-against-all test. To validate LEfSe, we performed MaAsLin2119 with default parameters and the same input as LEfSe. We generated Kaplan-Meier (KM) curves and performed Cox proportional hazards (Cox PH) modeling for the association of species with an LDA score ≥4 for RFS and OS in the pilot cohort. We evaluated the associations between the different tumor alpha diversity, evenness, and richness metrics at baseline with response to CRT and survival in the pilot cohort using Wilcoxon Rank-sum tests and a univariate Cox PH model, respectively. We classified baseline tumor samples into community state types (CST) based on the composition of the vaginal microbial community using VALENCIA38 and visualized the tumor microbiome using a stacked taxonomy bar plot.
Univariate Cox PH models were built for RFS and OS. The models included the tumor and gut baseline alpha diversity metrics, as well as the relative counts of L. iners and species identified on LEfSe with an LDA score ≥4, along with relevant clinical and demographic characteristics. Covariates that were significant at the univariate level (p<0.05) were fitted using a multivariate Cox PH model.
We tested for associations between gut alpha diversity, evenness, and richness, as well as clinical and demographic characteristics, and baseline L. iners status using a Chi-square or Fisher’s exact test for categorical variables and an independent t-test for continuous variables. We quantified changes in tumor alpha diversity metrics and relative counts of L. iners over time using Bonferroni false discovery rate (FDR)-adjusted paired t-tests.
Unsupervised hierarchical clustering of the top 25 species in the baseline samples was conducted using the mclust machine learning algorithm in the full cohort120. We then performed survival analysis on the cluster groups using Cox PH modeling for RFS and OS with G. vaginalis, P. bivia, and A. vaginae. We also analyzed changes in these three species over time using a paired t-test.
For viral annotation, virMAP was used as previously described121. The resulting sequencing reads were trimmed and filtered using the BBtools suite. Reads with at least 50 bp and an entropy value of at least 0.7 were retained. The resulting reads were checked for kmer (k = 31) collisions against all Papillomaviridae (GenBank taxa ID = 151340) using a bloom filter. Reads with no collisions to Papillomaviridae were filtered if a kmer collision against the human genome was detected using a similar methodology. The remaining reads, including the subset that collided with HPV genomes, were used as input for VirMAP. VirMAP was run using default settings. Raw VirMAP viral abundances represent the number of reads assigned to each viral taxa ID. Although these abundances can be interpreted as being proportional to other viral IDs or input reads, they are not normalized against any host-derived metric. Therefore, we normalized VirMAP HPV read coverage against single-copy human gene coverage. First, single-copy gene coverage values were calculated on a per-sample basis. Single-copy genes were identified by aligning all human coding sequences (CDS) to the human genome (hg38) using BLAST. Genes were considered single-copy genes if the alignment span of the CDS was >10,000 base pairs; all CDS were uniquely aligned (genes with overlapping alignment ranges were removed); and the CDS alignment was >99%. In total, 17,737 genes satisfied these constraints.
To calculate coverage per gene, any read that overlapped with the full alignment length per CDS (both exonic and intronic) were included. Mean CDS region coverage was calculated by dividing the total length of aligned reads by the length of the aligned region per CDS. Overall coverage per sample was calculated by taking the mean of all CDS coverage values. HPV coverage values were obtained by dividing the sum of all aligned HPV read lengths by the average complete genome length of the corresponding HPV subtypes. Finally, HPV normalization values were calculated by dividing the HPV coverage values by the per-sample mean gene coverage, thus approximating HPV copies per human genome copy. Total Fungal reads were annotated from NCBI and log transformed to reduce variability and skewness and normalized to the total library size resulting in log normalized fungal reads per million (RPM).
LEfSe was used to identify baseline gut bacteria associated with CRT response. E. shigella was found to be associated with response (LDA ≥4) and so we generated KM survival curves with log-rank tests for comparison and performed Cox PH modeling for RFS and OS. We also used LEfSe to identify bacteria enriched in the baseline gut microbiome of patients with L. iners+ tumors. We correlated baseline gut and tumor L. iners relative counts using the Kendall’s G coefficient.
We used an independent Wilcoxon test to compare T cell repertoire characteristics between L. iners+ and L. iners− tumors at baseline. The T cell repertoire metrics used were the maximum frequency, maximum productive frequency, out-of-frame rearrangements, out-of-frame templates, productive clonality, productive entropy, productive rearrangements, productive templates, sample entropy, total rearrangements, and total templates. Additionally, we tested for differences in the presence of specific motifs and clone groups previously identified among L. iners+ and L. iners− tumors at baseline using chi-square tests.
Clarivate Metacore pathway, network and process enrichment analyses and GSEA Hallmark pathway analysis were used to identify pathways, processes and networks enriched in treatment groups with default parameters.
For metabolomics, no normalization was performed. NGCHM software was used for heatmap clustering analysis. Normalized data was median transformed before generating NGCHM. NGCHM plots are interactive and can be used to search a specific compound, zoom-in and to highlight the value of a specific data point. PCA-plus plots were generated using Batcheffect package. ANOVA was used to find the differentially expressed compounds in two different biological covariates. Differential analysis was performed in several steps using following contrasts. One-Way ANOVA was performed separately for all three time points to identify compounds significantly differing between treatment groups at that specific time point. Two-way ANOVA was performed to estimate the effects of treatment group and/or time on each compound. Interaction P-value indicates the combined P-value of above mentioned both covariates. Tukey’s test was used for post-hoc analysis of pair-wise comparison.
For survival analysis of TCGA data, WGS and RNASeq BacMap feature tables for each cancer type were queried for presence of any of the 93 LAB species in 3 or more samples. We then generated KM survival analyses for dichotomized presence of each bacteria for RFS and OS. We then pooled the species that were found to be significantly associated with poor survival (p <0.05) and generated KM curves for RFS and OS. We used all high quality, reference genomes in the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) to classify the bacteria as obligate L-lactate producers, obligate D-lactate producers, or both.
Statistical significance was set at an α of 5% for a two-sided p-value. All available samples were used for analyses. Analyses were conducted using RStudio 2023.03.0+386 Cherry Blossom122.
Supplementary Material
Highlights.
L. iners is associated with poor tumor response to chemoradiation.
L. iners primes tumor metabolism for increased L-lactate production.
Other lactic acid bacteria are associated with poor survival in other cancer types.
Acknowledgments
Presented in part at the American Society for Therapeutic Radiation Oncology (ASTRO) Annual Meeting, September 24th-27th, 2017 and at the American Society of Clinical Oncology Clinical Immuno-Oncology Symposium (ASCO-SITC), January 25th-27th, 2018. We would like to acknowledge Matthew G. Landry for assistance with illustrations, Joseph Munch for manuscript editing, the High Performance Computing for research facility (HPCC) and Platform for Innovative Microbiome & Translational Research (PRIME-TR), Moon Shots™ Program, The University of Texas MD Anderson Cancer Center.
Funding
This study was partially funded by institutional funding from The University of Texas MD Anderson Cancer Center (HPV-related Cancers Moon Shots™ Program, Institutional Startup Funding, Adverse Incident Grant, Institutional Research Grant), Radiological Sciences North America (RSNA), and NIH (UPR/MDACC: Partnership for Excellence in Cancer Research [U54CA096300–19], NCI Cancer Center Support Grant [P30CA16672]. The Cancer Genomics Center at The University of Texas Health Science Center at Houston is supported by the Cancer Prevention Research Institute of Texas (CPRIT RP180734).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
L.E. Colbert reports grants from American Society for Clinical Oncology, Radiology Society of North America, National Institutes of Health, and MD Anderson Cancer Center during the conduct of the study. A. Jhingran reports personal fees from Genentech during the conduct of the study, as well as personal fees from Genentech outside the submitted work. L. Lin reports other support from AstraZeneca and Pfizer, and grants from NCI outside the submitted work. J.A. Wargo reports other support from Micronoma during the conduct of the study, as well as other support from Imedex, Dava Oncology, Illumina, and PeerView outside the submitted work. P. Okhuysen reports Faculty grant/research support from Merck Sharp and Dohme Corp, Deinove Pharmaceuticals, Summit Pharmaceuticals, Melinta Pharmaceuticals and Napo Pharmaceutical, in addition to consultant work with Napo Pharmaceutical, Ferring Pharmaceutical, Summit Pharmaceutical, and SNIPR Biome Company, all outside of the submitted work.
Inclusion and Diversity
Gender balance in the recruitment of human subjects and sex balance in cell lines were not applicable to this cervical cancer study. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects and through the selection of the cell lines.
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority, a gender minority, a member of the LGBTQIA+ community, or as living with a disability in their field of research or within their geographical location. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. We avoided “helicopter science” practices by including the participating local contributors from the region where we conducted the research as authors on the paper.
SUPPLEMENTAL MATERIAL
Supplemental Table 4. Comparative analysis of complete genomes for all L. iners strains in study, related to Figure 5.
Supplemental Table 5. Differential gene and pathway expression for all samples, related to Figures 4 and 6.
Supplemental Table 6. Kaplan-meier survival analysis for LAB in TCGA data sets, related to Figure 6.
Data S1: Clinical Characteristics, sample metadata and processed microbiome and T cell repertoire sequencing data required to reproduce all analyses, related to Figures 1 and 6; Supplemental Figures 1, 2, 3, and 4; Tables 1 and 2; Supplemental Tables 3 and 6.
Data S2: All original code used in analysis, related to Figures 1 and 6; Supplemental Figures 1, 2, 3, and 4; Tables 1 and 2; Supplemental Tables 3 and 6.
REFERENCES
- 1.Ghaddar B, Biswas A, Harris C, Omary MB, Carpizo DR, Blaser MJ, and De S (2022). Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell 40, 1240–1253.e5. 10.1016/j.ccell.2022.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. (2019). Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 178, 795–806.e12. 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, et al. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574. 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. (2020). The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 368, 973–980. 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, and Wargo JA (2022). Targeting the gut and tumor microbiota in cancer. Nat Med 28, 690–703. 10.1038/s41591-022-01779-2. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, and Zhang Y (2022). Intratumor microbiome in cancer progression: current developments, challenges and future trends. Biomark Res 10, 37. 10.1186/s40364-022-00381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heymann CJF, Bard J-M, Heymann M-F, Heymann D, and Bobin-Dubigeon C (2021). The intratumoral microbiome: Characterization methods and functional impact. Cancer Letters 522, 63–79. 10.1016/j.canlet.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Zhong G, Wei W, Liao W, Wang R, Peng Y, Zhou Y, Huang X, Xian S, Peng S, Zhang Z, et al. (2022). Tumor Microbiome in Nasopharyngeal Carcinoma and Its Association With Prognosis. Front Oncol 12, 859721. 10.3389/fonc.2022.859721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen T, Li Q, Wu J, Wu Y, Peng W, Li H, Wang J, Tang X, Peng Y, and Fu X (2018). Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol Immunother 67, 1635–1646. 10.1007/s00262-018-2233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Y, Yang J, Qian J, Liu R, Huang E, Wang Y, Luo F, and Chu Y (2019). TLR1/TLR2 signaling blocks the suppression of monocytic myeloid-derived suppressor cell by promoting its differentiation into M1-type macrophage. Mol Immunol 112, 266–273. 10.1016/j.molimm.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Kim J-H, Kordahi MC, Chac D, and DePaolo RW (2020). Toll-like Receptor-6 Signaling Prevents Inflammation and Impacts Composition of the Microbiota During Inflammation-Induced Colorectal Cancer. Cancer Prev Res (Phila) 13, 25–40. 10.1158/1940-6207.CAPR-19-0286. [DOI] [PubMed] [Google Scholar]
- 12.He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, Dong X, Huang J, Wang Q, Mackay CR, et al. (2021). Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab 33, 988–1000.e7. 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Bindels LB, Porporato P, Dewulf EM, Verrax J, Neyrinck AM, Martin JC, Scott KP, Buc Calderon P, Feron O, Muccioli GG, et al. (2012). Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br J Cancer 107, 1337–1344. 10.1038/bjc.2012.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931. 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, Kwon O, Ryu TY, Jung C-R, Kim J, Min J-K, Kim D-S, Son M-Y, and Cho H-S (2019). Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer. Mol Med Rep 20, 1569–1574. 10.3892/mmr.2019.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Surendra A, Kumar S, et al. (2014). Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299. 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 17.Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, Khan MAW, Zhang X, White MG, Peterson CB, et al. (2021). Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 374, 1632–1640. 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 20.Zitvogel L, Ayyoub M, Routy B, and Kroemer G (2016). Microbiome and Anticancer Immunosurveillance. Cell 165, 276–287. 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, and Weyrich LS (2019). Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol 27, 105–117. 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Glassing A, Dowd SE, Galandiuk S, Davis B, and Chiodini RJ (2016). Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathogens 8, 24. 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker SP, Tangney M, and Claesson MJ (2020). Sequence-Based Characterization of Intratumoral Bacteria—A Guide to Best Practice. Frontiers in Oncology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz-Flores R, López-Carvallo JA, Cáceres-Martínez J, and Dhar AK (2022). Microbiome analysis from formalin-fixed paraffin-embedded tissues: Current challenges and future perspectives. J Microbiol Methods 196, 106476. 10.1016/j.mimet.2022.106476. [DOI] [PubMed] [Google Scholar]
- 25.Hockney R, Orr CH, Waring GJ, Christiaens I, Taylor G, Cummings SP, Robson SC, and Nelson A (2022). Formalin-Fixed Paraffin-Embedded (FFPE) samples are not a beneficial replacement for frozen tissues in fetal membrane microbiota research. PLoS One 17, e0265441. 10.1371/journal.pone.0265441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam SY, Ioannou A, Konstanti P, Visseren T, Doukas M, Peppelenbosch MP, Belzer C, and Fuhler GM (2021). Technical challenges regarding the use of formalin-fixed paraffin embedded (FFPE) tissue specimens for the detection of bacterial alterations in colorectal cancer. BMC Microbiol 21, 297. 10.1186/s12866-021-02359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colbert LE, El MB, Lynn EJ, Bronk J, Karpinets TV, Wu X, Chapman BV, Sims TT, Lin D, Kouzy R, et al. (2022). Expansion of Candidate HPV-Specific T Cells in the Tumor Microenvironment during Chemoradiotherapy Is Prognostic in HPV16+ Cancers. Cancer Immunol Res 10, 259–271. 10.1158/2326-6066.CIR-21-0119. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Chen G, Liu Z, Liu S, Cai Z, You P, Ke Y, Lai L, Huang Y, Gao H, et al. (2018). Aberrant FGFR Tyrosine Kinase Signaling Enhances the Warburg Effect by Reprogramming LDH Isoform Expression and Activity in Prostate Cancer. Cancer Res 78, 4459–4470. 10.1158/0008-5472.CAN-17-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kantarci H, Gou Y, and Riley BB (2020). The Warburg Effect and lactate signaling augment Fgf-MAPK to promote sensory-neural development in the otic vesicle. eLife 9, e56301. 10.7554/eLife.56301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu J-W, Yeh S-C, Tsai F-Y, Chen H-W, and Tsou T-C (2019). Fibroblast growth factor 21 secretion enhances glucose uptake in mono(2-ethylhexyl)phthalate-treated adipocytes. Toxicology in Vitro 59, 246–254. 10.1016/j.tiv.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Flores AE, Sollome JJ, Thavathiru E, Bower JL, and Vaillancourt RR (2019). HER2/HER3 regulates lactate secretion and expression of lactate receptor mRNA through the MAP3K4 associated protein GIT1. Sci Rep 9, 10823. 10.1038/s41598-019-46954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castagnoli L, Iorio E, Dugo M, Koschorke A, Faraci S, Canese R, Casalini P, Nanni P, Vernieri C, Di Nicola M, et al. (2019). Intratumor lactate levels reflect HER2 addiction status in HER2‐positive breast cancer. J Cell Physiol 234, 1768–1779. 10.1002/jcp.27049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin L, Chun J, Pan C, Alesi GN, Li D, Magliocca KR, Kang Y, Chen ZG, Shin DM, Khuri FR, et al. (2017). Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene 36, 3797–3806. 10.1038/onc.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozenberg JM, Zvereva S, Dalina A, Blatov I, Zubarev I, Luppov D, Bessmertnyi A, Romanishin A, Alsoulaiman L, Kumeiko V, et al. (2021). The p53 family member p73 in the regulation of cell stress response. Biol Direct 16, 23. 10.1186/s13062-021-00307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadimitriou K, Alegría Á, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, et al. (2016). Stress Physiology of Lactic Acid Bacteria. Microbiology and Molecular Biology Reviews 80, 837–890. 10.1128/mmbr.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Wu J, Lv M, Shao Z, Hungwe M, Wang J, Bai X, Xie J, Wang Y, and Geng W (2021). Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Frontiers in Bioengineering and Biotechnology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abedi E, and Hashemi SMB (2020). Lactic acid production – producing microorganisms and substrates sources-state of art. Heliyon 6, e04974. 10.1016/j.heliyon.2020.e04974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.France MT, Mendes-Soares H, and Forney LJ (2016). Genomic Comparisons of Lactobacillus crispatus and Lactobacillus iners Reveal Potential Ecological Drivers of Community Composition in the Vagina. Appl Environ Microbiol 82, 7063–7073. 10.1128/AEM.02385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y-J, Mahieu NG, Huang X, Singh M, Crawford PA, Johnson SL, Gross RW, Schaefer J, and Patti GJ (2016). Lactate metabolism is associated with mammalian mitochondria. Nat Chem Biol 12, 937–943. 10.1038/nchembio.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macklaim JM, Gloor GB, Anukam KC, Cribby S, and Reid G (2011). At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc. Natl. Acad. Sci. U.S.A 108 Suppl 1, 4688–4695. 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwak W, Han Y-H, Seol D, Kim H, Ahn H, Jeong M, Kang J, Kim H, and Kim TH (2020). Complete Genome of Lactobacillus iners KY Using Flongle Provides Insight Into the Genetic Background of Optimal Adaption to Vaginal Econiche. Front Microbiol 11, 1048. 10.3389/fmicb.2020.01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iskandar CF, Cailliez-Grimal C, Borges F, and Revol-Junelles A-M (2019). Review of lactose and galactose metabolism in Lactic Acid Bacteria dedicated to expert genomic annotation. Trends in Food Science & Technology 88, 121–132. 10.1016/j.tifs.2019.03.020. [DOI] [Google Scholar]
- 43.Frontiers | Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows 10.3389/fcimb.2012.00086/full. [DOI] [PMC free article] [PubMed]
- 44.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, and Bonner WM (2010). Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res 704, 152–159. 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziech D, Franco R, Pappa A, and Panayiotidis MI (2011). Reactive oxygen species (ROS)--induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res 711, 167–173. 10.1016/j.mrfmmm.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, Wang Y, Ying M, Jin C, Li J, and Hu X (2021). Lactate dehydrogenases amplify reactive oxygen species in cancer cells in response to oxidative stimuli. Sig Transduct Target Ther 6, 1–13. 10.1038/s41392-021-00595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartesaghi S, Graziano V, Galavotti S, Henriquez NV, Betts J, Saxena J, Minieri V, A, D., Karlsson A, Martins LM, et al. (2015). Inhibition of oxidative metabolism leads to p53 genetic inactivation and transformation in neural stem cells. Proc Natl Acad Sci U S A 112, 1059–1064. 10.1073/pnas.1413165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Z, Kozlov S, Lavin MF, Person MD, and Paull TT (2010). ATM activation by oxidative stress. Science 330, 517–521. 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 49.Cosentino C, Grieco D, and Costanzo V (2011). ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J 30, 546–555. 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soni A, Duan X, Stuschke M, and Iliakis G (2022). ATR Contributes More Than ATM in Intra-S-Phase Checkpoint Activation after IR, and DNA-PKcs Facilitates Recovery: Evidence for Modular Integration of ATM/ATR/DNA-PKcs Functions. Int J Mol Sci 23, 7506. 10.3390/ijms23147506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falck J, Petrini JHJ, Williams BR, Lukas J, and Bartek J (2002). The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet 30, 290–294. 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 52.Qin L, Fan M, Candas D, Jiang G, Papadopoulos S, Tian L, Woloschak G, Grdina DJ, and Li JJ (2015). CDK1 Enhances Mitochondrial Bioenergetics for Radiation-Induced DNA Repair. Cell Rep 13, 2056–2063. 10.1016/j.celrep.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakkenist CJ, and Kastan MB (2004). Initiating cellular stress responses. Cell 118, 9–17. 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 54.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, and Tainer JA (2000). Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800. 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 55.Mittal A, Nenwani M, Sarangi I, Achreja A, Lawrence TS, and Nagrath D (2022). Radiotherapy-induced metabolic hallmarks in the tumor microenvironment. Trends in Cancer 8, 855–869. 10.1016/j.trecan.2022.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Yang Y, and Gu J (2020). Clinical Implications of the Associations Between Intestinal Microbiome and Colorectal Cancer Progression. Cancer Manag Res 12, 4117–4128. 10.2147/CMAR.S240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liotti F, Marotta M, Sorriento D, Pagliuca C, Caturano V, Mantova G, Scaglione E, Salvatore P, Melillo RM, and Prevete N (2022). The probiotic Lactobacillus rhamnosus GG (LGG) restrains the angiogenic potential of colorectal carcinoma cells by activating a pro-resolving program via formyl peptide receptor 1. Mol Oncol 10.1002/1878-0261.13280. [DOI] [PMC free article] [PubMed]
- 58.Liu H, Xu X, Liang J, Wang J, and Li Y (2022). The relationship between Clostridium butyricum and colorectal cancer. Journal of Cancer Research and Therapeutics 18, 1855. 10.4103/jcrt.jcrt_1565_21. [DOI] [PubMed] [Google Scholar]
- 59.Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, Sato S, Yoshida Y, Tsuji S, Uemura K, et al. (2021). Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun 12, 5674. 10.1038/s41467-021-25965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rebersek M (2021). Gut microbiome and its role in colorectal cancer. BMC Cancer 21, 1325. 10.1186/s12885-021-09054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi F, Liu G, Lin Y, Guo CL, Han J, Chu ESH, Shi C, Li Y, Zhang H, Hu C, et al. (2023). Altered gut microbiome composition by appendectomy contributes to colorectal cancer. Oncogene 42, 530–540. 10.1038/s41388-022-02569-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walenta S, Schroeder T, and Mueller-Klieser W (2004). Lactate in solid malignant tumors: potential basis of a metabolic classification in clinical oncology. Curr Med Chem 11, 2195–2204. 10.2174/0929867043364711. [DOI] [PubMed] [Google Scholar]
- 63.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. (2017). Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371.e9. 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu S, and Yang C (2023). Serum lactate dehydrogenase level predicts the prognosis in bladder cancer patients. BMC Urol 23, 65. 10.1186/s12894-023-01239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu C, Han J, Han D, Huang W, and Li B (2023). A new risk score model based on lactate dehydrogenase for predicting prognosis in esophageal squamous cell carcinoma treated with chemoradiotherapy. J Thorac Dis 15, 2116–2128. 10.21037/jtd-23-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolaños-Suárez V, Alfaro A, Espinosa AM, Medina-Martínez I, Juárez E, Villegas-Sepúlveda N, Gudiño-Zayas M, Gutiérrez-Castro A, Román-Bassaure E, Salinas-Nieves ME, et al. The mRNA and protein levels of the glycolytic enzymes lactate dehydrogenase A (LDHA) and phosphofructokinase platelet (PFKP) are good predictors of survival time, recurrence, and risk of death in cervical cancer patients. Cancer Medicine n/a 10.1002/cam4.6123. [DOI] [PMC free article] [PubMed]
- 67.Tauffenberger A, Fiumelli H, Almustafa S, and Magistretti PJ (2019). Lactate and pyruvate promote oxidative stress resistance through hormetic ROS signaling. Cell Death Dis 10, 1–16. 10.1038/s41419-019-1877-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitaker-Menezes D, Martinez-Outschoorn UE, Lin Z, Ertel A, Flomenberg N, Witkiewicz AK, Birbe RC, Howell A, Pavlides S, Gandara R, et al. (2011). Evidence for a stromal-epithelial “lactate shuttle” in human tumors: MCT4 is a marker of oxidative stress in cancer-associated fibroblasts. Cell Cycle 10, 1772–1783. 10.4161/cc.10.11.15659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, and Lisanti MP (2011). Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol 43, 1045–1051. 10.1016/j.biocel.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ippolito L, Comito G, Parri M, Iozzo M, Duatti A, Virgilio F, Lorito N, Bacci M, Pardella E, Sandrini G, et al. (2022). Lactate Rewires Lipid Metabolism and Sustains a Metabolic-Epigenetic Axis in Prostate Cancer. Cancer Res 82, 1267–1282. 10.1158/0008-5472.CAN-21-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L, Shi V, Wang S, Sun L, Freeman RN, Yang J, Inkman MJ, Ghosh S, Ruiz F, Jayachandran K, et al. (2023). SCCA1/SERPINB3 suppresses anti-tumor immunity and blunts therapy-induced T cell responses via STAT-dependent chemokine production. J Clin Invest 10.1172/JCI163841. [DOI] [PMC free article] [PubMed]
- 72.Cohen CR, Wierzbicki MR, French AL, Morris S, Newmann S, Reno H, Green L, Miller S, Powell J, Parks T, et al. (2020). Randomized Trial of Lactin-V to Prevent Recurrence of Bacterial Vaginosis. N Engl J Med 382, 1906–1915. 10.1056/NEJMoa1915254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nataraj BH, and Mallappa RH (2021). Antibiotic Resistance Crisis: An Update on Antagonistic Interactions between Probiotics and Methicillin-Resistant Staphylococcus aureus (MRSA). Curr Microbiol 78, 2194–2211. 10.1007/s00284-021-02442-8. [DOI] [PubMed] [Google Scholar]
- 74.Ariyoshi T, Hagihara M, Takahashi M, and Mikamo H (2022). Effect of Clostridium butyricum on Gastrointestinal Infections. Biomedicines 10, 483. 10.3390/biomedicines10020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuo F, and Marcotte H (2021). Advancing mechanistic understanding and bioengineering of probiotic lactobacilli and bifidobacteria by genome editing. Current Opinion in Biotechnology 70, 75–82. 10.1016/j.copbio.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 76.Fuochi V, Volti GL, and Furneri PM (2017). Probiotic Properties of Lactobacillus fermentum Strains Isolated from Human Oral Samples and Description of their Antibacterial Activity. Curr Pharm Biotechnol 18, 138–149. 10.2174/1389201017666161229153530. [DOI] [PubMed] [Google Scholar]
- 77.Górska A, Przystupski D, Niemczura MJ, and Kulbacka J (2019). Probiotic Bacteria: A Promising Tool in Cancer Prevention and Therapy. Curr Microbiol 76, 939–949. 10.1007/s00284-019-01679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jahanshahi M, Maleki Dana P, Badehnoosh B, Asemi Z, Hallajzadeh J, Mansournia MA, Yousefi B, Moazzami B, and Chaichian S (2020). Anti-tumor activities of probiotics in cervical cancer. J Ovarian Res 13, 68. 10.1186/s13048-020-00668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lebeer S, Vanderleyden J, and De Keersmaecker SCJ (2008). Genes and Molecules of Lactobacilli Supporting Probiotic Action. Microbiol Mol Biol Rev 72, 728–764. 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giovannetti E, Leon LG, Gómez VE, Zucali PA, Minutolo F, and Peters GJ (2016). A specific inhibitor of lactate dehydrogenase overcame the resistance toward gemcitabine in hypoxic mesothelioma cells, and modulated the expression of the human equilibrative transporter-1. Nucleosides Nucleotides Nucleic Acids 35, 643–651. 10.1080/15257770.2016.1149193. [DOI] [PubMed] [Google Scholar]
- 81.Halford S, Veal GJ, Wedge SR, Payne GS, Bacon CM, Sloan P, Dragoni I, Heinzmann K, Potter S, Salisbury BM, et al. (2023). A Phase I Dose-escalation Study of AZD3965, an Oral Monocarboxylate Transporter 1 Inhibitor, in Patients with Advanced Cancer. Clinical Cancer Research 29, 1429–1439. 10.1158/1078-0432.CCR-22-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Floch R, Chiche J, Marchiq I, Naiken T, Ilc K, Murray CM, Critchlow SE, Roux D, Simon M-P, and Pouysségur J (2011). CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc Natl Acad Sci U S A 108, 16663–16668. 10.1073/pnas.1106123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, and Gordon JI (2007). The Human Microbiome Project. Nature 449, 804–810. 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamady M, and Knight R (2009). Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res 19, 1141–1152. 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lõhmussaar K, Oka R, Espejo Valle-Inclan J, Smits MHH, Wardak H, Korving J, Begthel H, Proost N, van de Ven M, Kranenburg OW, et al. (2021). Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell 28, 1380–1396.e6. 10.1016/j.stem.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 87.Tsuda M, Hosono A, Yanagibashi T, Hachimura S, Hirayama K, Itoh K, Takahashi K, and Kaminogawa S (2007). Prior stimulation of antigen-presenting cells with Lactobacillus regulates excessive antigen-specific cytokine responses in vitro when compared with Bacteroides. Cytotechnology 55, 89–101. 10.1007/s10616-007-9104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karp PD, Paley SM, Midford PE, Krummenacker M, Billington R, Kothari A, Ong WK, Subhraveti P, Keseler IM, and Caspi R (2020). Pathway Tools version 24.0: Integrated Software for Pathway/Genome Informatics and Systems Biology. arXiv:1510.03964 [q-bio]
- 89.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37, 852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nearing JT, Douglas GM, Comeau AM, and Langille MGI (2018). Denoising the Denoisers: an independent evaluation of microbiome sequence error-correction approaches. PeerJ 6, e5364. 10.7717/peerj.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, and Glöckner FO (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35, 7188–7196. 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Usyk M, Zolnik CP, Castle PE, Porras C, Herrero R, Gradissimo A, Gonzalez P, Safaeian M, Schiffman M, Burk RD, et al. (2020). Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLOS Pathogens 16, e1008376. 10.1371/journal.ppat.1008376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bushnell B (2014). BBMap: A Fast, Accurate, Splice-Aware Aligner (Lawrence Berkeley National Lab (LBNL), Berkeley, CA (United States)). [Google Scholar]
- 94.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nurk S, Meleshko D, Korobeynikov A, and Pevzner PA (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res 27, 824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, and Wang Z (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359. 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, and Hauser LJ (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, and Ogata H (2020). KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Meijenfeldt FAB, Arkhipova K, Cambuy DD, Coutinho FH, and Dutilh BE (2019). Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol 20, 217. 10.1186/s13059-019-1817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang K, Li M, and Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164. 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, and Ritchie ME (2018). RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR 10.12688/f1000research.9005.3. [DOI] [PMC free article] [PubMed]
- 104.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems 1, 417–425. 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eriksson P, Marzouka N-A-D, Sjödahl G, Bernardo C, Liedberg F, and Höglund M (2021). A comparison of rule-based and centroid single-sample multiclass predictors for transcriptomic classification. Bioinformatics, btab763. 10.1093/bioinformatics/btab763. [DOI] [PMC free article] [PubMed]
- 107.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, and Aluru S (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9, 5114. 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karp PD, Latendresse M, and Caspi R (2011). The pathway tools pathway prediction algorithm. Stand Genomic Sci 5, 424–429. 10.4056/sigs.1794338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.MetaCyc database of metabolic pathways and enzymes | Nucleic Acids Research | Oxford Academic. https://academic.oup.com/nar/article/46/D1/D633/4559117. [DOI] [PMC free article] [PubMed]
- 110.Bushnell B, Rood J, and Singer E (2017). BBMerge – Accurate paired shotgun read merging via overlap. PLOS ONE 12, e0185056. 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. (2017). Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27, 849–864. 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rognes T, Flouri T, Nichols B, Quince C, and Mahé F (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S, et al. (2015). The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res 4, 900. 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Farrell CM, Feldgarden M, Fine AM, Funk K, et al. (2023). Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Research 51, D29–D38. 10.1093/nar/gkac1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Buchfink B, Xie C, and Huson DH (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 116.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glöckner FO (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ajami NJ, Wong MC, Ross MC, Lloyd RE, and Petrosino JF (2018). Maximal viral information recovery from sequence data using VirMAP. Nat Commun 9, 3205. 10.1038/s41467-018-05658-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12, R60. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren B, Schwager EH, et al. (2021). Multivariable association discovery in population-scale meta-omics studies. PLOS Computational Biology 17, e1009442. 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scrucca L, Fop M, Murphy TB, and Raftery AE (2016). mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. The R Journal 8, 289–317. [PMC free article] [PubMed] [Google Scholar]
- 121.Ajami NJ, Wong MC, Ross MC, Lloyd RE, and Petrosino JF (2018). Maximal viral information recovery from sequence data using VirMAP. Nat Commun 9, 3205. 10.1038/s41467-018-05658-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.RStudio Team (2020). RStudio: Integrated Development for R
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S and WGS have been deposited in SRA and will be made publicly available upon publication of the manuscript (BioProject accession numbers: PRJNA989630, PRJNA702617, PRJNA685389). L. iners strain assemblies using WMS have been deposited in NCBI and will be made publicly available upon publication (BioSample accession numbers: SAMN27176861, SAMN27176862, SAMN27176863, SAMN27176864). TCR Sequencing data has been deposited in immuneACCESS and will be made public upon acceptance. Processed microbiome, TCR sequencing, RNA Sequencing and TCGA data are available in supplementary material, along with sample metadata and clinical data.
All original code used to generate figures or tables is available in this paper’s supplemental information.
Any additional information required to reanalyze the data or code reported in this paper is available from the lead contact upon request.


