Abstract
DNA methylation is usually associated with transcriptional silencing, but in the imprinted mouse Igf2 gene, the paternally expressed copy is methylated in two discrete differentially methylated regions (DMRs). DMR1 is located upstream of the fetal promoters and has been shown to be a methylation sensitive silencer. Here we examine the role of the intragenic DMR2 by gene targeting. In contrast to DMR1, deletion of DMR2 on the maternal allele did not lead to activation of the silent Igf2 gene. Deletion of a 54 bp methylated core region in DMR2 on the paternal allele, however, reduced Igf2 mRNA levels and was associated with fetal growth retardation. Nuclear run-on assays showed that the core region influenced transcription initiation, and luciferase reporter assays suggested that its methylation increases transcription. These results reveal a novel mechanism of gene expression whereby intragenic methylation can increase levels of transcription.
INTRODUCTION
Imprinted genes in mammals are expressed from only one of the parental alleles. DNA methylation is an important epigenetic mechanism involved in imprinting. The majority of imprinted genes have differentially methylated regions (DMRs) and a number of elements have been identified in or near DMRs whose epigenetic regulation contributes to the control of imprinted gene expression. These include promoters whose methylation leads to silencing, promoters of antisense transcripts, chromatin boundaries separating promoters and enhancers, and silencers (Brannan and Bartolomei, 1999; Sleutels et al., 2000; Ferguson-Smith and Surani, 2001; Reik and Walter, 2001).
The paternally expressed mouse Igf2 gene has three DMRs, two of which are paternally methylated (Sasaki et al., 1992; Brandeis et al., 1993; Feil et al., 1994; Moore et al., 1997). These observations gave rise to the proposal that DMRs could have silencer functions that might be suppressible by DNA methylation. Alternatively, methylation on the paternal allele might be required as a positive regulator of gene expression. Deletion of the maternal DMR1 region, which is located upstream of the fetal promoters, results in tissue-specific release of silencing of Igf2 (Constância et al., 2000). A silencer element within DMR1 has been identified whose methylation in vitro abrogates silencer function (Eden et al., 2001).
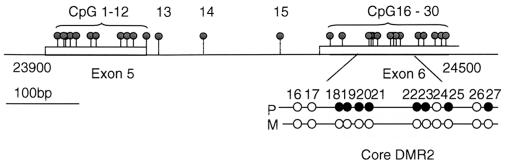
The paternally methylated DMR2 is located within the Igf2 gene and was originally defined as a 2.4 kb BamHI fragment spanning exons 4 to 6 (the last exon). Four HpaII sites in this fragment are paternally methylated in tissues with Igf2 expression, and differential methylation is conserved in the human IGF2 gene (Feil et al., 1994; Reik et al., 1994). Bisulfite analysis refined the definition of DMR2 to a core region of 54 bp, including CpGs 18–25, located at the beginning of exon 6 in the peptide coding region of Igf2 (Figure 1). Consistent paternal methylation occurs in CpGs 18–25 in Igf2-expressing tissues (Forné et al., 1997). Outside this region there are no consistent parental methylation differences. Here we analyse DMR2 by knockout deletions and find that, in contrast to DMR1, it contains an element that augments transcription of Igf2.
Fig. 1. The DMR2 of Igf2. CpG map of DMR2 in Igf2 (GenBank accession No. MMU71085), showing exons 5 and 6 (clear boxes) and the CpGs (lollipops). The core DMR2 in exon 6, between CpGs 18–25, is expanded to show the consistently methylated CpGs (dark circles) on the paternal allele (P) in all tissues where Igf2 is expressed (Forné et al., 1997).
RESULTS AND DISCUSSION
Deletion of DMR2 does not activate the maternal Igf2 allele
The role of DMR2 in imprinting and expression of Igf2 in vivo was examined by gene targeting. A DMR2 knockout (LacZDMR2–) was designed to detect whether the DMR2 contained silencer elements. A 4.65 kb part of the Igf2 gene between exons 4 and 6 containing DMR2 was removed and replaced with a promoterless IRES:lacZneo cassette (Figure 2A and B). With paternal transmission, LacZDMR2– mice were smaller at birth (up to 50%) than their wild-type littermates, as expected from Igf2 deficiency (DeChiara et al., 1990). LacZ expression was observed in tissues that normally express Igf2, e.g. intestine, heart, lung, kidney, tongue, liver and vertebrae, in whole-mount embryos and sagittal sections (Figure 2C). The low level of staining in the liver is an artefact of the particular fixation protocol involving gluteraldehyde. Upon maternal transmission, LacZDMR2– mice were normal in size and did not express significant amounts of LacZ at embryonic days (E) 10, 14 and 17 in any tissue except the choroid plexus in the brain, where Igf2 expression is known to be biallelic (Figure 2C).
Fig. 2. (A) Knockout strategy for deleting DMR2. DMR2 was deleted at the flanking BamHI sites and replaced by IRES:LacZ-neo (grey box). (B) Southern blots with EcoRI digestion and a probe spanning exon 2 distinguished the targeted (8.7 kb) and wild-type (16.5 kb) alleles. (C) Lac Z staining of E 17 embryos. CP, choroid plexus. Scale bar, 1 mm.
Deletion of the whole DMR2, therefore, did not result in activation of the silent maternal allele. Thus, DMR2 is not necessary for silencing the maternal allele of Igf2, in contrast to other elements such as DMR1 (Constância et al., 2000), the H19 DMR (Thorvaldsen et al., 1998; Bell and Felsenfeld, 2000; Hark et al., 2000) and the intergenic region (Ainscough et al., 2000).
The core DMR2 is necessary for high-level transcription of the paternal Igf2 allele
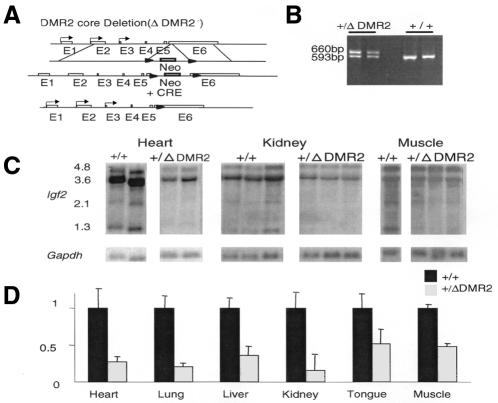
A second knockout (ΔDMR2) was designed to examine the function of the methylated core DMR2. The 54 bp core region of DMR2 (CpG18–CpG25) was deleted in-frame by a Cre loxP strategy (Figure 3A and B). Mice with a maternally inherited ΔDMR2 allele were normal in size and expressed normal levels of Igf2 from their paternal allele, while the maternal allele remained silent (data not shown). Remarkably, however, following paternal transmission, ΔDMR2 offspring were 17% smaller at birth than their wild-type littermates, with the mean of the average weights in 11 litters being 1.51 ± 0.2 and 1.25 ± 0.1 g for +/+ and +/ΔDMR2 genotypes, respectively (P <0.001).
Fig. 3. (A) Knockout strategy for deleting the 54 bp core (ΔDMR2). A targeting vector with neomycin (neo, grey box) and the core DMR2 was flanked by loxP sites (arrowheads). After homologous recombination, Cre-mediated recombination deleted 54 bp from the DMR2 core region and replaced it with the 121 bp loxP sequence. The promoters P1–P3 are indicated as arrows on the corresponding exons. (B) The ΔDMR2 allele was detected by PCR. (C) Northern blots of RNA from tissues of postnatal day 1 mice from +/+ and +/ΔDMR2 pups. Transcript sizes are indicated in kb. The 3.6, 2.1 and 1.3 kb transcripts are from P3; 4.8 kb from P2. (D) Results of densitometric quantification of the 3.6 kb transcript. Expression levels of the wild-type allele in each tissue were normalized to one. At least three samples in each tissue were analysed.
The core deletion is within the translated region of exon 6 that encodes the E peptide or C-terminal propeptide domain of prepro-Igf2. After translation, the E peptide is cleaved and does not form part of the secreted Igf2 protein, and its role is unknown (Stempien et al., 1986). If the E peptide was important in the post-translational cleavage of Igf2, its disruption in the ΔDMR2 mutation could alter the amounts of secreted mature Igf2. We therefore measured Igf2 mRNA levels to determine whether the growth phenotype was likely to be due to reduced Igf2 transcript or peptide levels.
Northern blotting revealed markedly reduced transcripts from all promoters (20–50% of wild-type) of Igf2 in ΔDMR2 mutants in all tissues in which Igf2 is normally transcribed (Figure 3C and D). We also examined northern blots for the 1.8 kb endonucleolytic cleavage fragment with a probe from the 3′ end of exon 6, because of the proximity of the core DMR2 (120 bp upstream) to the 3′ UTR endonucleolytic cleavage sites (Scheper et al., 1996). The amount of 1.8 kb fragment relative to the full-length P3 (3.6 kb) transcript was similar in wild-type and ΔDMR2 tissues (data not shown). Thus, the ΔDMR2 mutation did not affect Igf2 mRNA stability through the endonucleolytic cleavage sites. RT–PCR experiments showed that the expression was from the paternal ΔDMR2 allele in all tissues except brain, in which both alleles were transcribed (data not shown). The loxP site and remaining polylinker sequences did not affect expression in luciferase transfection assays using constructs containing DMR2 deletions with and without loxP (data not shown).
The ΔDMR2 mice were 17% smaller at birth than their wild-type littermates. In contrast, Igf2 null mice are 50% smaller at birth (DeChiara et al., 1990), while enh+/– mice, where the endoderm enhancers have been deleted (Leighton et al., 1995), are 30% smaller at birth. Since the Igf2 null mice make no Igf2 peptide, and the enh+/– mice have reduced transcription only in endodermal tissues, ΔDMR2 mice must make a substantial amount of functional Igf2 peptide despite the mutated E peptide, and most if not all of the growth deficiency can be attributed to reduced transcript levels.
Methylation in the core DMR2 increases expression of a reporter gene
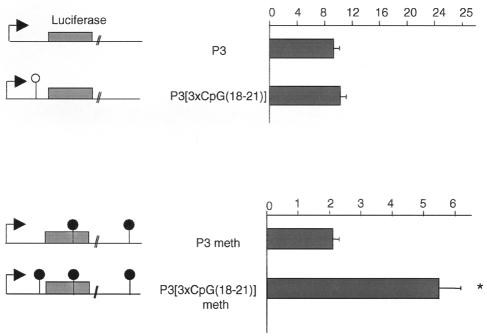
Deletion of the methylated core DMR2 reduced Igf2 transcription on the paternal allele. Hence, the methylated core DMR2 seems to be required for high levels of transcription and could act as a positive regulator on the paternal allele. At present, there is no ideal in vivo system that can address the effect of methylation specifically of the core DMR2. Therefore, we tested the effect of methylation in a transient luciferase transfection system, using HhaI methylase, as this was the only available methylase that did not also methylate the Igf2 promoter. We were particularly interested in defining the minimum sequence requirements that could confer a methylation-dependent increase in transcription. A reporter construct was made with the Igf2 promoter 3 (P3) that expressed high levels of luciferase in Human embryonic kidney (HEK) 293 cells (Figure 4). The core DMR2 contains a single HhaI site corresponding to the methylated CpG21. Because the effect of methylating CpGs 22–25 could not be tested, this part of the sequence was removed, and in order to increase the number of methylatable sites, three copies of the CpG 18–21 sequence (and thus three HhaI sites) were inserted into the P3 construct {P3[3×CpG(18–21)]}. Luciferase activity was not affected by insertion of this sequence (Figure 4). In vitro methylation with HhaI methylase changed the baseline of luciferase activity, such that expression from the P3 construct was reduced 5-fold (Figure 4), presumably because the luciferase gene and the vector also contain a number of HhaI sites. After in vitro methylation, luciferase activity from the P3[3×CpG(18–21)] construct was 2.5-fold higher (P <0.05) than that of the P3 construct (Figure 4). Thus, the sequence including CpG 18–21 seems to be capable of increasing transcript levels when methylated. A substantial series of additional transfection studies was performed in order to pinpoint the activating region of the core DMR2 (A. Murrell et al., unpublished results). These studies suggest that the region between CpG 18–21 is the critical activator, and we are currently dissecting the chromatin structure and potential binding proteins to this region.
Fig. 4. Reporter constructs with luciferase (grey box), the Igf2 P3 promoter (arrow) and CpG18–21 are shown. CpG21 is in a HhaI site (open lollipop). Constructs were methylated in vitro with HhaI methylase (filled lollipops; the luciferase and vector sequences also contain a number of HhaI sites). The histograms show the results of dual luciferase assay experiments. *, P3meth versus P3[3×CpG(18–21)]meth = P <0.05. Results represent the means of six experiments.
These assays suggest that the methylated core DMR2 is needed for high-level expression of Igf2, which is consistent with the observation that DMR2 is, or becomes, methylated in all instances in which Igf2 is expressed substantially. For instance, DMR2 is methylated on both alleles in situations with biallelic expression of Igf2 (Feil et al., 1994; Reik et al., 1995; Forné et al., 1997). Interestingly, two tissues which have previously been shown to have lower levels of methylation at DMR2 (muscle and tongue; Weber et al., 2001) showed the smallest reduction in Igf2 RNA levels in the ΔDMR2 mice (Figure 3D). Thus, other tissue-specific control elements such as enhancers and silencers notwithstanding, methylation at the core DMR2 seems necessary for high-level expression of the Igf2 gene.
The core DMR2 acts at the level of transcription initiation
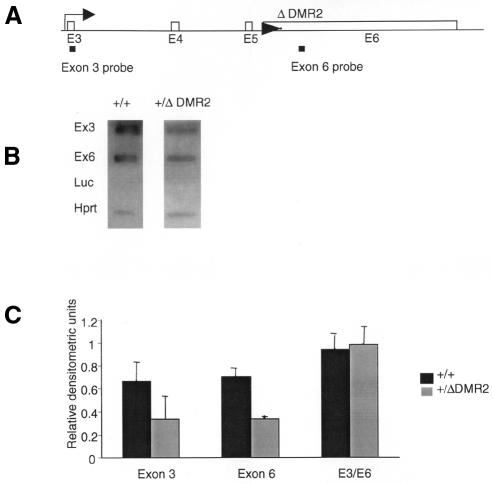
Considering the location of DMR2 at the end of the transcribed region of the gene, it could regulate transcription elongation. In fungi it has been established that methylation downstream of a promoter influences elongation (Rountree and Selker, 1997). In order to address whether the reduced Igf2 mRNA levels were caused by reduced transcription initiation, transcription elongation or RNA stability, nascent RNA was examined by run-on transcription analysis. Nascent RNA from run-on assays in nuclei from the livers of newborn mice with a paternally inherited ΔDMR2 allele and wild-type littermates was hybridized to slot blots containing Igf2 probes located upstream (near the transcription start site of promoter 3, in exon 3) and downstream (exon 6) of the core DMR2 (Figure 5A). Double-stranded probes were used, since we have not found any evidence for antisense transcripts spanning the DMR2 in mouse liver (A. Murrell et al., unpublished results). Normalization of run-on transcripts against the Hprt signal showed that in the +/ΔDMR2 tissue the relative intensities of exons 3 and 6 were 48 and 51.5% of the wild-type intensities, respectively. The ratio of exon 3 to exon 6 (E3/E6) was close to one for both +/ΔDMR2 and wild types (Figure 5B and C).
Fig. 5. (A) The position of probes for initiation (exon 3) and elongation (exon 6) for run-on transcripts. (B) Representative slot blots with probes from exon 3, exon 6, Hprt (positive control) and luciferase (luc, background control), hybridized with nascent RNA from nuclei of livers from newborn day 1 +/ΔDMR2 and +/+ pups. (C) Densitometric analysis of run-on RNA. The densities of the exon 3 and 6 signals were plotted relative to Hprt signals. The densitometic analysis was done on four +/ΔDMR2 and three +/+ samples.
Thus, deletion of the methylated core DMR2 had no impact on elongation or RNA stability, but negatively affected initiation, suggesting that the methylated core DMR2 plays a role in transcription initiation and influences activation of Igf2. The core DMR2 in conjunction with enhancers might be involved in the assembly of basal transcription factors or participate in promoter clearance. Transcription factor initiation complexes can potentially bind to Igf2 promoters on both alleles, since there are identical DNase I hypersensitive sites on both alleles (Sasaki et al., 1992). Promoter clearance involves TFIIH-mediated DNA melting (Holstege et al., 1997), and this process could be influenced by the chromatin structure in the gene.
Igf2 is not unique in having methylation in an intragenic region. Indeed, methylation of CpG islands downstream of the transcription start site occurs in ∼49% of tissue-specific genes (Larsen et al., 1992). These genes are not inactivated by methylation of the CpG islands (Jones, 1999). In some cases, for example Myod1, expression levels actually increase in the presence of methylation downstream of the promoter (Takagi et al., 1995). It has also been shown that some genes that are overexpressed in tumours have acquired methylation in CpG islands located within their transcribed regions (Liang et al., 1998). These results are consistent with the recent observation in Dnmt1– cells that, while 4–10% of all detectable genes were upregulated in the absence of DNA methylation, 1–2% were downregulated (Jackson-Grusby et al., 2001). Therefore, it is possible that a substantial number of genes in the genome require DNA methylation for expression.
Speculation
It has been proposed that imprinted genes can be regulated by ‘epigenetic activation’ as well as by ‘epigenetic inactivation’ (Sleutels et al., 2000; Reik and Walter, 2001). Our results show that the intragenic methylated core DMR2 plays a role in transcription initiation of Igf2. This work reveals a novel type of element in imprinting control whose role is in ‘epigenetic activation’. More generally, the present results highlight the existence of a transcription initiation mechanism that is activated by intragenic methylation, which may be relevant to other imprinted or non-imprinted genes (Jones, 1999). Perhaps the methylated DMR2 influences interactions between the Igf2 promoters and distal enhancers, conceivably by binding factors that mediate physical interaction or transfer of transcription complexes. Alternatively, it could influence the local chromatin structure and facilitate promoter clearance. Although it is generally believed that methylated DNA attracts methyl CpG binding proteins that are repressive for transcription (Bird and Wolffe, 1999), methylation-sensitive binding factors might exist that play a role in activation. We are currently dissecting the chromatin structure and binding proteins of the DMR2 region in order to address these questions.
METHODS
Gene targeting in mice. LacZDMR2– mice were generated by replacing DMR2 with an IRES:lacZneo cassette at the BamHI sites in exons 4 and 6 of Igf2. The targeting vector, a IRES:lacZneo cassette flanked at the 5′ and 3′ ends by 3.5 and 4.5 kb homology arms, respectively, was electroporated into IMT11 embryonic stem (ES) cells (a gift from S. Hunter and M. Evans). ES cell culture and microinjection of targeted ES cells into C57BL/6 blastocysts were as described previously (Plagge et al., 2000). Male chimeras were bred to C57BL/6 for germline transmission; further breeding was with C57BL/6. Genotyping of LacZDMR2– mice was by EcoRI restriction of DNA from tails on Southern blots probed with a 1.5 kb BamHI–BglII probe spanning exon 2.
The ΔDMR2 mice were generated with a cre-lox strategy. A ΔDMR2 targeting construct (Figure 3A) was made using the yeast pRAY1 recombinogenic arms (RA) system (Storck et al., 1996) to delete 54 bp from DMR2 (nucleotides 24 416–24 470) from 7.5 kb of genomic Igf2 subcloned into a yeast shuttle vector (PRS414). PCR primers for the Igf2 gene RA on either side of the 54 bp deletion were: LHRA, 5′-GAA GCT TCG ACC TGG CCC TCC TG; LHRA, 3′-CGC GGA CTG TCT CGA GGT GTC AT; RHRA, 5′-CGC ATG CTT TCG AAA GAG CTC AA; RHRA, 3′-GAG ATC CCC TCG GCC GAT GGT GT. The left and right RAs were inserted into pRAY1 at the XhoI–HindIII and ClaI–NotI sites. The targeting vector was electroporated into CGR ES cells. Male chimeras that gave rise to germline transmission were bred with constitutively expressing Cre926 mice (a gift from M. Bruggemann). Cre excision replaced the 54 bp core DMR2 by a 35 loxP site flanked by 48 bp of pRAY1 polylinker sequence on either side. Mice were genotyped by PCR analysis of DNA from tails with the primers CreXF (5′-TAG GTA GCG AGG TGG GGT TGA-3′) and CreXR (5′-GGA AGG GAA GTG GAG CAG AGA-3′).
RNA and DNA analysis. Southern and northern analyses were as described previously (Feil et al., 1994; Constância et al., 2000; Weber et al., 2001).
β-galactosidase assays. Whole-mount embryos and sagittal sections were fixed and stained for lac Z expression as described previously (Hogan et al., 1994).
Luciferase constructs and transfection assays. The P3 construct contained a 342 bp NheI–StuI fragment (starting 70 bp upstream of the Igf2 P3 promoter) in the NheI–SmaI site in the pGL3 basic vector (Promega). Three tandem 24 bp double-stranded oligonucleotides containing the Igf2 CpG18–21 sequence were inserted into the P3 construct between XhoI and HindIII sites, to make the P3[3×CpG(18–21)] construct. In vitro methylation was with HhaI methylase (New England Biolabs).
HEK 293 cells were seeded into 12-well plates at 1 × 105 cells/ml and transfected with an Effectine kit (Qiagen). Two replicate transfections were done per experiment. Co-transfection was with the pRL-SV40 renilla plasmid to control for transfection efficiency in a Dual-Luciferase Reporter assay system (Promega). pGL3 basic and pGL3 basic promoter (SV40luc) plasmids were standard negative and positive controls. Student’s t-test analysis, one-tailed on paired samples, with unequal variance was performed. A P-value of <0.05 was considered significant.
Run-on transcription. Nuclei extracts were prepared from livers from newborn day 1 mice as described previously (Marzluff and Huang, 1987). Nuclei were suspended in glycerol buffer (40% glycerol, 50 mM Tris–HCl pH 8.3, 5 mM MgCl2, 0.1 mM EDTA) in 50 µl aliquots of 1 × 105 nuclei/µl. Fifty microlitres of transcription buffer (10 mM Tris pH 8.0, 5 mM MgCl2, 0.3 M KCl, 1 mg/ml heparin, 2 mM each ATP, GTP and CTP) and 0.2 mM [α-32P]UTP (800 Ci/mmol) were added to each aliquot of nuclei followed by incubation for 30 min at 30°C. Yeast tRNA (10 mg/ml) and DNase I (RNase free) were added after 25 min. Ten volumes of RLT buffer (RNeasy kit from Qiagen) were added and RNA was extracted using RNeasy Mini protocol and immediately hybridised to slot blot filters as described previously (Marzluff and Huang, 1987).
Igf2 exon and murine Hprt probes for slot blots were PCR amplified. Primers used were: Igf2 exon 3 (Ex3F, 5′-AAC TGG ACA TTA GCT TCT CCT GTG A-3′; Ex3R, 5′-CCG CTG GGG CTG GAA), Igf2 exon 6 (Ex6F, 5′-CAT CGT CCC CTG ATC GTG TT; Ex6R, 5′-TTG CTG GAC ATC TCC GAA GAG-3′) and murine Hprt (F, 5′-TTG CTC GAG ATG TCA TGA AGG A-3′; R, 5′-CCA GCA GGT CAG CAA AGA ACT T-3′).
Acknowledgments
ACKNOWLEDGEMENTS
We thank David Brown and Eurof Walter for help with the statistical analyses, Marianne Bruggemann for providing the Cre926 mice, Toni Plagge for help with yeast recombination, and Andrea Riccio, Robert Feil, Jorn Walter, Anne Ferguson-Smith and members of the laboratory for helpful discussions. This work was supported by the CRC, MRC and BBSRC.
REFERENCES
- Ainscough J.F., John, R.M., Barton, S.C. and Surani, M.A. (2000) A skeletal muscle-specific mouse Igf2 repressor lies 40 kb downstream of the gene. Development, 127, 3923–3930. [DOI] [PubMed] [Google Scholar]
- Bell A.C. and Felsenfeld, G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature, 405, 482–485. [DOI] [PubMed] [Google Scholar]
- Bird A.P. and Wolffe, A.P. (1999) Methylation-induced repression—belts, braces, and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Brandeis M., Kafri, T., Ariel, M., Chaillet, J.R., McCarrey, J., Razin, A. and Cedar, H. (1993) The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J., 12, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan C.I. and Bartolomei, M.S. (1999) Mechanisms of genomic imprinting. Curr. Opin. Genet. Dev., 9, 164–170. [DOI] [PubMed] [Google Scholar]
- Constância M., Dean, W., Lopes, S., Moore, T., Kelsey, G. and Reik, W. (2000) Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nature Genet., 26, 203–206. [DOI] [PubMed] [Google Scholar]
- DeChiara T.M., Efstratiadis, A. and Robertson, E.J. (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature, 345, 78–80. [DOI] [PubMed] [Google Scholar]
- Eden S., Constância, M., Hashimshony, T., Dean, W., Goldstein, B., Johnson, A.C., Keshet, I., Reik, W. and Cedar, H. (2001) An upstream repressor element plays a role in Igf2 imprinting. EMBO J., 20, 3518–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Walter, J., Allen, N.D. and Reik, W. (1994) Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development, 120, 2933–2943. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith A.C. and Surani, M.A. (2001) Imprinting and the epigenetic asymmetry between parental genomes. Science, 293, 1086–1089. [DOI] [PubMed] [Google Scholar]
- Forné T., Oswald, J., Dean, W., Saam, J.R., Bailleul, B., Dandolo, L., Tilghman, S.M., Walter, J. and Reik, W. (1997) Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc. Natl Acad. Sci. USA, 94, 10243–10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr, C.J., Katz, D.J., Ingram, R.S., Levorse, J.M. and Tilghman, S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature, 405, 486–489. [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington, R., Costantini, F. and Lacy, E. (1994) Staining forβ-galactosidase (lacZ) Activity. CSHL Press, New York, NY.
- Holstege F.C., Fiedler, U. and Timmers, H.T. (1997) Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J., 16, 7468–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L. et al. (2001) Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature Genet., 27, 31–39. [DOI] [PubMed] [Google Scholar]
- Jones P.A. (1999) The DNA methylation paradox. Trends Genet., 15, 34–37. [DOI] [PubMed] [Google Scholar]
- Larsen F., Gundersen, G., Lopez, R. and Prydz, H. (1992) CpG islands as gene markers in the human genome. Genomics, 13, 1095–1107. [DOI] [PubMed] [Google Scholar]
- Leighton P.A., Saam, J.R., Ingram, R.S., Stewart, C.L. and Tilghman, S.M. (1995) An enhancer deletion affects both H19 and Igf2 expression. Genes Dev., 9, 2079–2089. [DOI] [PubMed] [Google Scholar]
- Liang G., Salem, C.E., Yu, M.C., Nguyen, H.D., Gonzales, F.A., Nguyen, T.T., Nichols, P.W. and Jones, P.A. (1998) DNA methylation differences associated with tumor tissues identified by genome scanning analysis. Genomics, 53, 260–268. [DOI] [PubMed] [Google Scholar]
- Marzluff W.F. and Huang, R.C. (1987) Transcription of RNA in isolated nuclei. In Hames, B.D. and Higgens, S.J. (eds), Transcription and Translation — A Practical Approach. IRL Press, Oxford, UK, pp. 89–127.
- Moore T., Constância, M., Zubair, M., Bailleul, B., Feil, R., Sasaki, H. and Reik, W. (1997) Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl Acad. Sci. USA, 94, 12509–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagge A., Kelsey, G. and Allen, N.D. (2000) Directed mutagenesis in embryonic stem cells. In Jackson, I.J. and Abbott, C. (ed.), Mouse Genetics and Transgenics: A Practical Approach. IRL Press, Oxford, UK, pp. 247–284.
- Reik W. and Walter, J. (2001) Genomic imprinting: parental influence on the genome. Nature Rev. Genet., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Reik W., Brown, K.W., Slatter, R.E., Sartori, P., Elliott, M. and Maher, E.R. (1994) Allelic methylation of H19 and IGF2 in the Beckwith–Wiedemann syndrome. Hum. Mol. Genet., 3, 1297–1301. [DOI] [PubMed] [Google Scholar]
- Reik W., Brown, K.W., Schneid, H., Le Bouc, Y., Bickmore, W. and Maher, E.R. (1995) Imprinting mutations in the Beckwith–Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum. Mol. Genet., 4, 2379–2385. [DOI] [PubMed] [Google Scholar]
- Rountree M.R. and Selker, E.U. (1997) DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev., 11, 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Jones, P.A., Chaillet, J.R., Ferguson-Smith, A.C., Barton, S.C., Reik, W. and Surani, M.A. (1992) Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev., 6, 1843–1856. [DOI] [PubMed] [Google Scholar]
- Scheper W., Holthuizen, P.E. and Sussenbach, J.S. (1996) Growth-condition-dependent regulation of insulin-like growth factor II mRNA stability. Biochem. J., 318, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F., Barlow, D.P. and Lyle, R. (2000) The uniqueness of the imprinting mechanism. Curr. Opin. Genet. Dev., 10, 229–233. [DOI] [PubMed] [Google Scholar]
- Stempien M.M., Fong, N.M., Rall, L.B. and Bell, G.I. (1986) Sequence of a placental cDNA encoding the mouse insulin-like growth factor II precursor. DNA, 5, 357–361. [DOI] [PubMed] [Google Scholar]
- Storck T., Kruth, U., Kolhekar, R., Sprengel, R. and Seeburg, P.H. (1996) Rapid construction in yeast of complex targeting vectors for gene manipulation in the mouse. Nucleic Acids Res., 24, 4594–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H., Tajima, S. and Asano, A. (1995) Overexpression of DNA methyltransferase in myoblast cells accelerates myotube formation. Eur. J. Biochem., 231, 282–291. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen J.L., Duran, K.L. and Bartolomei, M.S. (1998) Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev., 12, 3693–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Milligan, L., Delalbre, A., Antoine, E., Brunel, C., Cathala, G. and Forné, T. (2001) Extensive tissue-specific variation of allelic methylation in the Igf2 gene during mouse fetal development: relation to expression and imprinting. Mech. Dev., 101, 133–141. [DOI] [PubMed] [Google Scholar]