Abstract
Upregulation of the proto-oncoprotein Myc, a basic, helix–loop–helix, leucin zipper domain transcription factor has profound consequences on cell proliferation, cell growth and apoptosis. Cell cultures of somatic c-myc–/– rat fibroblasts show extremely prolonged doubling times of 52 h. Using time-lapse microscopy, we show here that individual c-myc–/– cells proceeded within ∼24 h through the cell cycle as fast as c-myc+/+ cells. However, c-myc–/– cells were highly sensitive to contact inhibition and readily arrested in the cell cycle already at low density. Activation of conditional MycER overcame cell cycle arrest in c-myc–/– cells and led to continuous proliferation at the expense of increased apoptosis at high cell density. Conditional expression of Mad1, a Myc antagonist, represses proliferation of different cell types including U2OS cells. In analogy to the effect of Myc, this occurs mainly by reducing the probability of cells remaining in the cycle. Our data demonstrate that the Myc/Max/Mad network does not regulate the duration of the cell cycle, but the decision of cells to enter or exit the cell cycle.
INTRODUCTION
The involvement of Myc in tumour development is most convincingly shown by the formation of carcinoma and lymphoma in transgenic mice with targeted overexpression of c-myc in various cell types (Adams and Cory, 1992). The physiological effects of Myc, including promotion of cell proliferation and apoptosis, require association of Myc with its dimerization partner Max and specific binding to DNA (Henriksson and Luscher, 1996). Myc–Max dimers activate a number of target genes implicated in regulation of cell growth and cell cycle progression (Cole and McMahon, 1999; Grandori et al., 2000; Amati et al., 2001; Eisenman, 2001). Importantly, Myc stimulates cyclin expression, activation of cyclin-dependent kinases (Cdks) (Bouchard et al., 1999) and antagonizes the action of Cdk inhibitors, e.g. p15INK4b, p16INK4a and p27Kip1 (Vlach et al., 1996; Alevizopoulos et al., 1997; Perez-Roger et al., 1997, 1999; O’Hagan et al., 2000; Staller et al., 2001), which are upregulated by cell contact inhibition (Polyak et al., 1994) and other mechanisms. Overexpression of Myc has been reported to shorten cell cycle phases and the length of G1 has been correlated directly with Myc levels (Karn et al., 1989).
It has been assumed that Myc is essential for cell proliferation. Unexpectedly, somatic c-myc–/– TGR (rat1) cells were found to be viable. These cells appeared phenotypically flat and spreaded and had doubling times up to 3-fold longer than parental TGR cells. The knockout cells show growth defects with reduced rates of synthesis of ribosomal RNAs and proteins. In addition, c-myc–/– TGR cells show markedly prolonged G1 and G2 phases (Mateyak et al., 1997). Attempts to identify cellular or viral genes capable of re-establishing normal proliferative rates in c-myc–/– cells resulted only in the repeated identification of c-Myc or N-Myc (Nikiforov et al., 2000; Berns et al., 2000), suggesting that Myc is a key-regulator of cell cycle and growth control in rat1 cells. Here we have studied the effect of Myc on cell proliferation at the single-cell level. To our surprise we found that Myc does not regulate the duration of the cell cycle.
RESULTS
Myc knockout cells have normal cell cycle duration
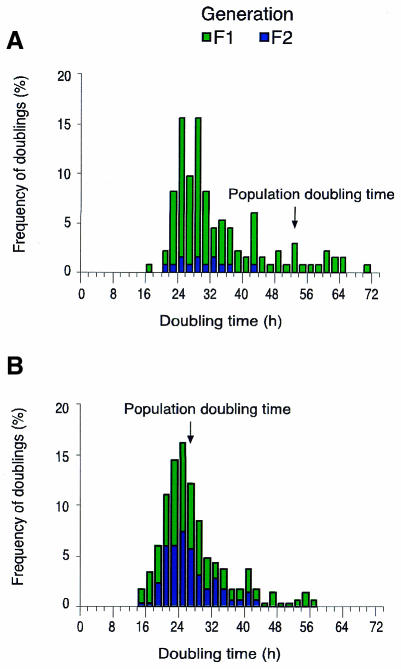
To determine the doubling times of individual cells, c-myc knockout cells were seeded in a living cell chamber and digital photographs were taken every 10 min for a period of 72 h in time-lapse microscopy experiments. Genealogical trees were established for ∼150 cells (F0 generation) and the time intervals between two divisions were determined (see Methods). We confirmed previous studies that the population doubling time of a ∼20% confluent c-myc–/– cell monolayer is in the range of 52 h, if cells were counted every day (data not shown) (Mateyak et al., 1997). However, at the single-cell level the large majority of proliferating cells showed doubling times <52 h with a peak between 24 and 28 h (Figure 1A). Five percent of c-myc–/– cells showed three consecutive doublings with intermitotic intervals each <30 h. Only 10% of cells showed doubling times >52 h. At first view, the short intermitotic intervals observed for doubling c-myc–/– cells are in contradiction to the long doubling time observed for the total c-myc–/– cell population. However, 13% of c-myc–/– cells did not divide and 58% of the first daughter generation (F1) showed no further division, even though >50% of F1 cells were generated in the first 22 h of the 72 h-observation period. In the time-lapse experiment for c-myc–/– cells the population doubling time was measured to be 52 h (Figure 1A). The parental TGR c-myc+/+ cells revealed almost identical doubling times for single cells and for the entire cell population, which were in the range of 26 and 28 h, respectively (Figure 1B). Thus, the major difference between c-myc–/– and c-myc+/+ is that the cells of the former arrest in the cell cycle at much lower density. This suggests a major role for Myc in the control of cell cycle exit and entry rather than in the length of the mitotic cycle.
Fig. 1. Cell cycle duration of rat1 TGR c-myc–/– and c-myc+/+ cells. Cells were cultured to ∼20% density in a living cell chamber and time-lapse microscopy was performed for a period of 72 h. Photographs were taken every 10 min. The time period between two mitoses was determined for c-myc–/– cells (A) and c-myc+/+ cells (B). The arrows mark the doubling time of the total cell population. Approximately 71% of c-myc–/– cells did not double or doubled only once. Cells analysed: 198 randomly selected Ho15.19 c-myc–/– cells revealed 135 intermitotic intervals; 125 TGR-1 c-myc+/+ cells revealed 299 intermitotic intervals. F1, F1 generation; F2, F2 generation.
Activation of MycER in knockout cells does not influence cell cycle duration
To study the function of Myc on cell proliferation more directly, we constructed several knockout cell lines expressing the tamoxifen-regulated MycER fusion protein. Cell lines Smoxi-1, -4, -6 and -11 were selected by the criteria of having normal cell morphology, but undergoing apoptosis after serum starvation in the presence of tamoxifen (for details see Methods). Addition of tamoxifen to confluent Smoxi cells resulted in increased expression of ODC and decreased expression of GADD45, two previously described c-myc target genes (data not shown) (Bello-Fernandez et al., 1993; Marhin et al., 1997).
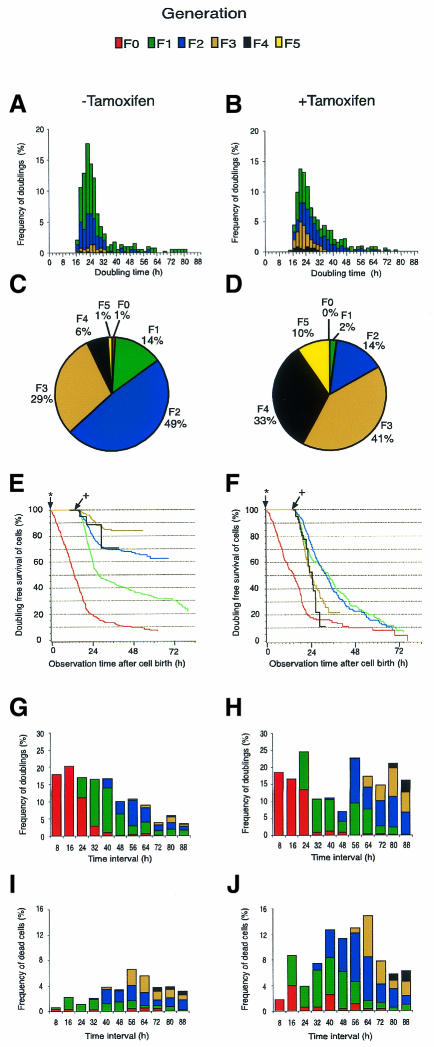
Cells of a representative clone, Smoxi-4, were seeded into a living cell chamber at ∼30% confluency in the absence and ∼45% confluency in the presence of tamoxifen (Figure 2). Time-lapse microscopy was performed and genealogical trees of ∼200 cells were established for each experiment. The time interval between two cell divisions and the time point of cell death was recorded (see Methods). This information was assigned to individual cells in each generation. Algorithms were established to perform statistical analysis of data from the genealogical trees. After an observation period of 88 h, 397 and 502 intermitotic intervals for cells cultured in the absence or presence of tamoxifen, respectively, were analysed. The average intermitotic interval of Smoxi-4 cells was in the range of 24 h, independent of whether the cells were grown in the presence or absence of tamoxifen, or whether cells reached a later generation (Figure 2A and B). The generation to which individual cells belonged was determined at 88 h. In the absence of tamoxifen, many more cells remained in the early generations compared with cells grown in the presence of tamoxifen (Figure 2C and D, compare percentage of cells in each generation). Thus, cells with activated MycER reached significantly higher cell generations.
Fig. 2. Doubling and apoptosis rates of Smoxi-4 cells after activation of MycER. Cells were placed into a living cell chamber in the absence or presence of tamoxifen and photographs were taken every 10 min for a period of 88 h. The fate of 200 cells for each experiment, to undergo division, survival or apoptosis, was determined (see example for a genealogical tree in Methods). (A and B) The time period between two divisions was assigned to single generations for cells grown in the absence or presence of tamoxifen. (C and D) Overview of doublings in individual generations in the absence or presence of tamoxifen. (E and F) Kaplan–Meier presentation of doubling frequencies of individual generation grown in the absence or presence of tamoxifen. A star marks the F0 generation and birth of daughter generations. + represents the first doubling of daughter generations. Frequency of doublings (G and H) and frequency of apoptosis (I and J) in 8 h intervals in the absence or presence of tamoxifen. Cells analysed: 206 randomly selected Smoxi-4 cells (– tamoxifen) revealed 397 intermitotic intervals; 214 Smoxi-4 cells (+ tamoxifen) revealed 502 intermitotic intervals.
This became clearly evident if survival curves of individual cell generations were plotted by the Kaplan–Meier method (see Methods). The method estimates for each daughter cell generation the time-dependent probability to enter the next generation. Cells that underwent apoptosis in the time course of the experiment were censored at the time point of death. The F0 generation of cells (starting cells) was lost almost completely by cell division, regardless of whether tamoxifen was present or not (Figure 2E and F). However, the F1 generation already showed a clear difference; only 77% of cells were lost by cell division in the absence of tamoxifen, whereas 93% of cells were lost in the presence of tamoxifen. This difference became even more pronounced for the F2 and F3 generations. Only 38% of cells of the F2 and 17% of the F3 generations were lost by cell doubling in the absence of tamoxifen, but 90 and 78% of cells, respectively, were lost if tamoxifen was present. These results indicated that, in particular, late generations of cells underwent cell cycle arrest in the absence of tamoxifen, while cells with activated MycER disappeared from late generations with the same kinetics as from the F0 generation. The high frequency of cell doubling with activated MycER was reflected by the high mitosis index for cells between 56 and 88 h (Figure 2G and H).
Importantly, two mechanisms contributed to the tamoxifen-induced shift to higher cell generations: (i) the growth to higher cell density, and (ii) the increased rate of apoptosis in the presence of Myc that made space for further cell proliferation (Figure 2I and J). If cells underwent apoptosis, new cells (see Figure 3E) soon occupied the vacant area. A constantly high rate of mitosis and apoptosis was seen, if a confluent Smoxi-4 cell monolayer was cultured over a period of 12 days (data not shown), indicating that loss of cells by apoptosis in this cell system was balanced by production of new cells. Taken together, the results implied that Myc regulated the decision of cell cycle entry and exit, but not the duration of cell cycling.
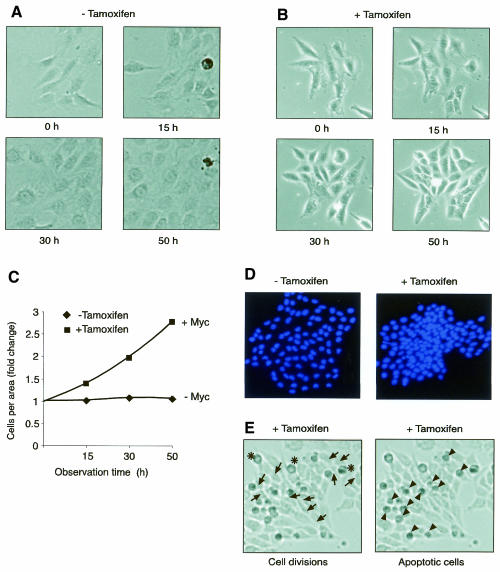
Fig. 3. Growth characteristics of c-myc–/– cells after activation of MycER. Smoxi-4 cells on glass slips were placed into a living cell chamber in the absence or presence of tamoxifen. (A–C) Photographs were taken at indicated time points and the ratio of cells/area was determined. (D) Cell clones with similar area after fixation and staining of nuclei with Hoechst 33258. (E) Cells were grown in the presence of tamoxifen for 88 h in a living cell chamber. A representative cell colony is shown. Black arrows mark places of cell division between 78 and 88 h, asterisks mark mitotic cells and arrowheads mark apoptotic cells. Magnification: (A and B), 100-fold; (D and E), 50-fold.
Myc abrogates contact inhibition and allows proliferation at high cell density
The findings described above suggest that Myc desensitizes cells to contact inhibition and allows growth to higher density. To substantiate this observation, cells were grown to ∼20% confluency on glass plates and then placed into a living cell chamber. Photographs were taken at intervals of 10 min. A representative experiment with Smoxi-4 is shown (Figure 3A and B). In the absence of tamoxifen, cells proliferated at a constant cell/area ratio. After activation of MycER (+ tamoxifen), this ratio changed dramatically so that by 50 h cells reached up to a 3-fold higher density (Figure 3A–C). If cell colonies of similar area were inspected, the strongly increased cell density was particularly evident for the central region of cell clones grown in the presence of tamoxifen (Figure 3D). We analysed the dynamics of cell clones by time-lapse microscopy over a period of 10 h and detected a high rate of apoptosis and mitosis in the region of high cell density, showing that dead cells were immediately replaced by new cells (Figure 3E).
Effect of Mad1 expression on cell cycle duration
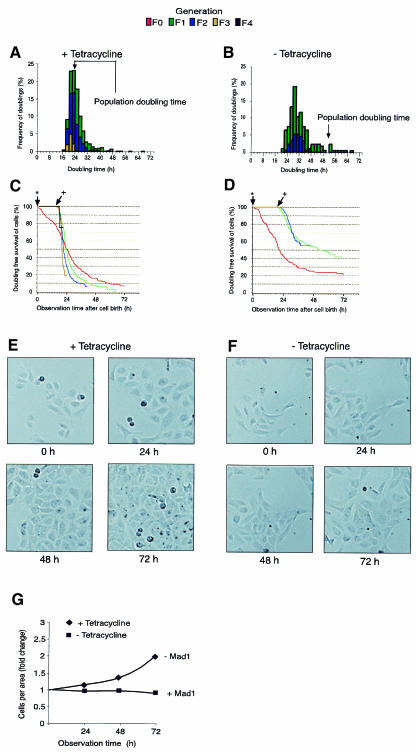
Myc is a member of a larger network of interacting transcription factors with Max at the centre (Henriksson and Luscher, 1996) Grandori et al., 2000). In addition to Myc, Max heterodimerizes with a number of additional basic helix–loop–helix leucin zipper (BHLHLZ) domain proteins, including the transcriptional repressor Mad1 (Roussel et al., 1996; Kiermaier and Eilers, 1997; Sommer et al., 1997; Grandori et al., 2000). Conditional Mad1 expression antagonizes proliferation and Myc-induced apoptosis in the well characterized cell system M47 (a U2OS derivative). Activation of Mad1 in these cells had a profound negative effect on cell doubling (Gehring et al., 2000). We tested the effect of Mad1 expression on proliferation of M47 cells at the single-cell level. Tetracycline was removed from M47 cells 4 days before we started the time-lapse experiment. At this time point all cells are Mad1 positive (Gehring et al., 2000). Without Mad1 expression, most cells doubled within ∼24 h, in the presence of Mad1 within ∼28 h (Figure 4A and B). The shift to a 4 h longer cell cycle, however, could not readily explain the reduced proliferation rate of the entire M47 cell population after Mad1 expression. The major contribution came from a cell cycle arrest. This was already seen for the F0 generation but became more pronounced for the F1, F2 and F3 generations. Forty-two percent of F1 cells and 55% of F2 cells did not double in Mad1 expressing cells, whereas 99% of F1 and 88% of F2 cells doubled in the absence of Mad1 (Figure 4C and D). Expression of Mad1 also inhibited growth to higher cell density (Figure 4E and G). Thus, inversely to activation of MycER in Smoxi-4 cells, expression of Mad1 in M47 cells increased the probability of cells exiting the cell cycle, but had only a minor effect on cell cycle duration. Even though the M47 cell system is not directly comparable with Smoxi-4 cells, the data suggests that Mad1 and Myc have an inverse effect on the cell cycle. Since M47 cells carry a mutated retinoblastoma tumour suppressor gene (Rb) (Iavarone et al., 1994), activation and inactivation of Rb is probably not required for this function of Mad1.
Fig. 4. Effect of Mad expression on cell cycle duration in M47 cells. (A and B) The time period between two duplications was assigned to single generations for cells grown in the absence (+ tetracycline) or presence of Mad1 (– tetracycline) expression for 3 days. (C and D) Kaplan–Meier presentation of doubling frequencies of individual cell generations in the absence or presence of Mad1 expression. (E and F) Cells on glass slips were placed into a living cell chamber in the presence or absence of tetracycline and representative photographs were taken at indicated time points. (G) The ratio of cells/area was determined. Cells analysed (A–D): 192 randomly selected M47 cells (+ tetracycline) revealed 641 intermitotic intervals, 190 M47 cells (– tetracycline) revealed 172 intermitotic intervals.
DISCUSSION
We have shown here that Myc is not required for rapid progression of rat fibroblasts through the cell cycle. As long as inhibitory signals do not act from outside, the cells have the intrinsic capacity to proceed through the cell cycle as rapidly as Myc expressing cells. However, if cells grew densely, negative signals, possibly by direct cell–cell contact, arose and blocked cells in the cycle. The nature of these signals has not yet been identified in detail but may involve the activation of growth arrest (GAS) genes as well as upregulation of inhibitors of cell cycle kinases (Polyak et al., 1994; Vlach et al., 1996; Alevizopoulos et al., 1997; Perez-Roger et al., 1997, 1999; O’Hagan et al., 2000; Staller et al., 2001).
It is conceivable that cell–cell contact is the major driving force to arrest c-myc–/– cells already at low cell density. We were unable to further characterize this phenomenon with regard to the localization of the cells. There are several reasons why our attempts were confounded. First, it is difficult to measure cell–cell contact as an integral over time, as the cells change their positions, turn from inside to outside in small colonies, and are found with or without neighbouring cells varying over time. Furthermore, the cells occupy new areas in the periphery thereby keeping the cells per area ratio relatively constant (Figure 3A). This outgrowth blurs distinguishable properties of localization between inside and outside, as the cell density in the centre is not higher than at the border (Figure 3D, – tamoxifen). It is therefore noteworthy that in the case of activated MycER in Smoxi-4 cells the density increases most prominently in the central part (Figure 3D, + tamoxifen).
Why have earlier studies (Mateyak et al., 1997) suggested that Myc deficient cells have a prolonged cell cycle? First, previous experiments have determined the time period required for quiescent c-myc–/– cells to enter S-phase after mitogenic stimulation. In this special case, expression of Myc probably helps to shorten the transition from G0 to G1. However, here we have measured the time intervals between several consecutive divisions of c-myc–/– cells at low density. In this case Myc had no effect on cell cycle duration.
Secondly, based on the cell cycle distribution of exponentially growing cells the duration of each cell cycle phase was previously calculated by multiplication of the population doubling time with the percentage of cells in each cell cycle phase (Mateyak et al., 1997). This calculation is generally based on the assumption that the percentage of cells found in a cell cycle phase reflects the time that cells need to traverse it. However, this calculation is only valid if the cell population fulfills the following requirements: (i) all cells are cycling, and (ii) all cells divide with similar intermitotic intervals. This is true in the case of TGR-1 c-myc+/+ cells and the calculation is therefore valid (Figure 1B). However, in the case of Ho15.19 c-myc–/– cells we have shown that requirement (i) is not fulfilled (Figure 1A). It is therefore not valid to draw conclusions from the cell cycle distribution to determine the duration of the cell cycle phases G0/G1; S; G2/M in c-myc–/– cells. Thus, the cell cycle distribution of c-myc–/– cells needs to be re-interpreted more carefully. A decreased S-phase fraction reflects less cycling cells. We indirectly confirm this by observing a steadily decreasing mitotic index (Figure 2G and data not shown). The enlarged G0/G1 fraction points to an increased number of arrested cells, as underlined by our time-lapse investigations. Do c-myc–/– cells also arrest in G2/M? Previously, the increase in the number of c-myc–/– cells in G2/M phase has been interpreted in favour of a longer cell cycle time (Mateyak et al., 1997). However, in light of our findings that the mitotic cycle in c-myc–/– cells is about equally long as in the parental cells, it seems more likely that the large fraction of G2/M cells in c-myc–/– cells in the above observation reflects an arrest in G2/M. Thus, our results do not contradict the measured percentages of cells in each cell cycle phase of c-myc–/– cells (Mateyak et al., 1997). However, the results of our investigation required a new interpretation of the latter. In conclusion, the use of flow cytometry to calculate the duration of cell cycle phases needs consideration of the above mentioned requirements. Otherwise the interpretation of the results is misleading.
Myc has been shown to support growth of human B cells (increase in cell mass) (Iritani and Eisenman, 1999; Schuhmacher et al., 1999), probably by upregulation of genes like eIF-4E, whose product is required for translation initiation (Schmidt, 1999). This posed the question of whether the absence of Myc reduces the growth capacity of cells thereby prolonging the cell cycle duration. We found no evidence for marked cell cycle prolongation in c-myc–/– cells. Instead, our data indicate a tight coupling of growth and cell cycle control. Very recent results in transgenic mice carrying a hypomorphic c-myc allele support this notion (Trumpp et al., 2001). Reduction of c-myc expression in the embryo results in reduced body mass. The reduction in body mass is accompanied by hypoplasia (reduced cell number), but not by hypotrophia (reduced cell size) (Trumpp et al., 2001), indicating that the mice have problems producing enough cells, whereas the size of the cells is normal. This finding is in line with a model that Myc in mammalian cells controls the decision to enter or to leave the cell division cycle, and thereby functions as a crucial mediator of signals that determines organ and body size (Trumpp et al., 2001). If cell growth (increase in mass) and cell cycle entry are thightly linked, an effect of Myc on cell growth can be readily demonstrated only if the cell cycle entry is blocked, e.g. by blocking Cdk2 (Schuhmacher et al., 1999). Vice versa, rat1 fibroblasts with an activated MycER do not enter S-phase until cells have reached a defined size (Pusch et al., 1997). A model in which Myc co-ordinates growth and cell cycle entry has been suggested previously (Schmidt, 1999).
Our findings may also have implications for Myc’s role in tumour development. The rat knockout cells carrying MycER do not stop proliferation at high cell density in the presence of tamoxifen. Instead, at high density cells undergo apoptosis and permanently generate small holes in the cell layer, thereby producing space for new cells. In our experiments the rates of proliferation and apoptosis were balanced at high cell density and thus allowed the permanent proliferation of Smoxi cells on dense monolayers. Thus, accidental activation of Myc in vivo must not necessarily result in net apoptosis of cells. Myc activation may release the cell cycle block induced by neighbouring cells and allow local proliferation of cells. The permanent loss of cells by apoptosis at this lesion offers at the same time space for proliferation of new cells. The resulting continuous cell proliferation increases the chance for additional mutagenic events, e.g. at the p53 locus, and thus may promote transformation to a more malignant phenotype. In this scenario, apoptosis is an essential requirement for tumour development.
METHODS
Cell culture. TGR-1 (c-myc wild type), Ho15.19 (c-myc knockout), TGR-1/MycER and Smoxi cells (Smoxi-1, -4, -6, and -11) were cultured in DMEM with 8% calf serum (Life Technologies). M47 cells were maintained in DMEM supplemented with 8% FCS (BioSer) and 1 µg/ml tetracycline. TGR-1/MycER and Smoxi cells were created by lipofectamine (Life Technologies) transfection of TGR-1 and Ho15.19 cells with the LXSH retroviral vector containing a HA-tagged tamoxifen responsive MycER. Smoxi single-cell clones were selected for c-myc knockout appearance in the absence of tamoxifen. MycER expression was monitored several times by western blot analysis using the 9E10 anti-Myc (Santa Cruz) or the 3F10 anti-HA (Roche) antibodies. Immunofluorescence staining was performed with the F10 anti-ER antibody (Santa Cruz). BrdU assays were applied for functional analysis of the MycER construct to detect increased incorporation rates after addition of tamoxifen under serum starvation (data not shown). M47 cells expressing a tetracycline-regulated Mad1 have been described elsewhere (Gehring et al., 2000).
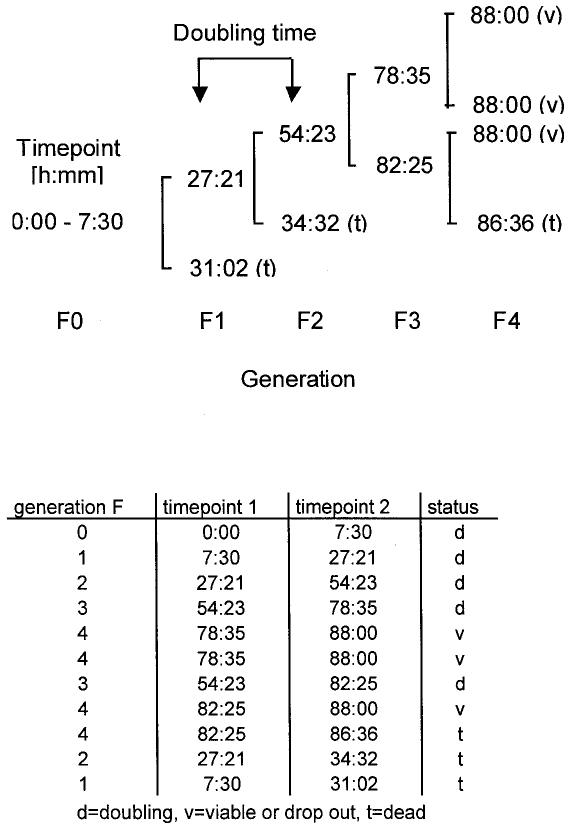
Time-lapse microscopy and movie analysis. A Zeiss Axiovert 200-M microscope equipped with a Hamamatsu 4747-95 digital CCD camera and the Improvision OpenLab 2.1 software was used to take photographs every 10 min for an observation period of 72 or 88 h. Applying the OpenLab presentation module, these series of photographs were displayed as continuous time-lapse movies. Cells were seeded the day before into a POC G-522 life chamber (LaCon, Germany). Temperature was adjusted to 37°C with a tempcontrol-37 (LaCon) and the medium was buffered with 15 mM HEPES. A 50 or 80× magnification was applied to investigate a large area in order to obtain the required number of cases for a representative statistical analysis. Cells were continuously numbered as F0 generation cells on the first photograph (observation time = 0 h) of each movie. F0 cells were then traced to the endpoint of observation recording time point and generation of the following events: (1) d = doubling; (2) t = dead; (3) v = viable if the time point is equal to the endpoint of observation; (4) v = early drop out if the time point is before the endpoint of observation. Cells drop out if they move out of sight before the observation has stopped. In this manner genealogical trees were established for each cell of the F0 generation. An example is shown below. The data from the genealogical tree were then transmitted into an Excel data file. Time points were decimalized and statistical analysis was then performed using the Statistical Analysis Software SAS version 6.12.
The individual doubling time was determined by the measurement of the intermitotic interval of cells in generation F1 or higher. The first division of F0 cells occurred before the onset of observation and therefore no individual doubling time could be attributed to the F0 cells. The population doubling time was calculated by the increase of the total cell number.
Kaplan–Meier method. Probabilities of doubling-free survival were estimated using the Kaplan–Meier method for each generation. Doubling-free survival describes the probability of a cell to remain viable in tissue culture without further division. Only cells leaving the generation by cell division cause a decrease of the curve. Early drop out of viable or apoptotic cells do not affect the curves, as they only reduce the current number of cells under observation but do not fulfil the event ‘doubling’. The curves of the F0 generation decrease immediately, as the time point of the first division is unknown. Therefore, a birth follow up for the F0 generation is not possible. The curves of the F1 and higher generation decrease after a time interval, which corresponds to the shortest individual doubling time in each generation.
Genealogical tree.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Sedivy for c-myc–/– cells, R. Eckel and J. Schoch for help evaluating the data, N. Malek and members of our institute for helpful discussion. We thank LaCon and PeCon (http://www.pe-con.de) for support with the living cell chamber and the incubation system on the inverted microscope. This work was supported by the Deutsche Forschungsgemeinschaft (SFB190 and LU466/6) and Fonds der Chemischen Industrie.
REFERENCES
- Adams J.M. and Cory, S. (1992) Oncogene co-operation in leukaemogenesis. Cancer Surv., 15, 119–141. [PubMed] [Google Scholar]
- Alevizopoulos K., Vlach, J., Hennecke, S. and Amati, B. (1997) Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J., 16, 5322–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B., Frank, S.R., Donjerkovic, D. and Taubert, S. (2001) Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta, 1471, M135–M145. [DOI] [PubMed] [Google Scholar]
- Bello-Fernandez C., Packham, G. and Cleveland, J.L. (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl Acad. Sci. USA, 90, 7804–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K., Hijmans, E.M., Koh, E., Daley, G.Q. and Bernards, R. (2000) A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene, 19, 3330–3334. [DOI] [PubMed] [Google Scholar]
- Bouchard C. et al. (1999) Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J., 18, 5321–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M.D. and McMahon, S.B. (1999) The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene, 18, 2916–2924. [DOI] [PubMed] [Google Scholar]
- Eisenman R.N. (2001) Deconstructing myc. Genes Dev., 15, 2023–2030. [DOI] [PubMed] [Google Scholar]
- Gehring S., Rottmann, S., Menkel, A.R., Mertsching, J., Krippner-Heidenreich, A. and Luscher, B. (2000) Inhibition of proliferation and apoptosis by the transcriptional repressor Mad1. Repression of Fas-induced caspase-8 activation. J. Biol. Chem., 275, 10413–10420. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley, S.M., James, L.P. and Eisenman, R.N. (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol., 16, 653–699. [DOI] [PubMed] [Google Scholar]
- Henriksson M. and Luscher, B. (1996) Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res., 68, 109–182. [DOI] [PubMed] [Google Scholar]
- Iavarone A., Garg, P., Lasorella, A., Hsu, J. and Israel, M.A. (1994) The helix–loop–helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev., 8, 1270–1284. [DOI] [PubMed] [Google Scholar]
- Iritani B.M. and Eisenman, R.N. (1999) c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl Acad. Sci. USA, 96, 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Watson, J.V., Lowe, A.D., Green, S.M. and Vedeckis, W. (1989) Regulation of cell cycle duration by c-myc levels. Oncogene, 4, 773–787. [PubMed] [Google Scholar]
- Kiermaier A. and Eilers, M. (1997) Transcriptional control: calling in histone deacetylase. Curr. Biol., 7, R505–R507. [DOI] [PubMed] [Google Scholar]
- Marhin W.W., Chen, S., Facchini, L.M., Fornace, A.J.,Jr and Penn, L.Z. (1997) Myc represses the growth arrest gene gadd45. Oncogene, 14, 2825–2834. [DOI] [PubMed] [Google Scholar]
- Mateyak M.K., Obaya, A.J., Adachi, S. and Sedivy, J.M. (1997) Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ., 8, 1039–1048. [PubMed] [Google Scholar]
- Nikiforov M.A., Kotenko, I., Petrenko, O., Beavis, A., Valenick, L., Lemischka, I. and Cole, M.D. (2000) Complementation of Myc-dependent cell proliferation by cDNA expression library screening. Oncogene, 19, 4828–4831. [DOI] [PubMed] [Google Scholar]
- O’Hagan R.C., Ohh, M., David, G., de Alboran, I.M., Alt, F.W., Kaelin, W.G.,Jr and DePinho, R.A. (2000) Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev., 14, 2185–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Roger I., Solomon, D.L., Sewing, A. and Land, H. (1997) Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27Kip1 binding to newly formed complexes. Oncogene, 14, 2373–2381. [DOI] [PubMed] [Google Scholar]
- Perez-Roger I., Kim, S.H., Griffiths, B., Sewing, A. and Land, H. (1999) Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27Kip1 and p21Cip1. EMBO J., 18, 5310–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Kato, J.Y., Solomon, M.J., Sherr, C.J., Massague, J., Roberts, J.M. and Koff, A. (1994) p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev., 8, 9–22. [DOI] [PubMed] [Google Scholar]
- Pusch O., Bernaschek, G., Eilers, M. and Hengstschlager, M. (1997) Activation of c-Myc uncouples DNA replication from activation of G1 cyclin-dependent kinases. Oncogene, 15, 649–656. [DOI] [PubMed] [Google Scholar]
- Roussel M.F., Ashmun, R.A., Sherr, C.J., Eisenman, R.N. and Ayer, D.E. (1996) Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol. Cell Biol., 16, 2796–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.V. (1999) The role of c-myc in cellular growth control. Oncogene, 18, 2988–2996. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M., Staege, M.S., Pajic, A., Polack, A., Weidle, U.H., Bornkamm, G.W., Eick, D. and Kohlhuber, F. (1999) Control of cell growth by c-Myc in the absence of cell division. Curr. Biol., 9, 1255–1258. [DOI] [PubMed] [Google Scholar]
- Sommer A., Hilfenhaus, S., Menkel, A., Kremmer, E., Seiser, C., Loidl, P. and Luscher, B. (1997) Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr. Biol., 7, 357–365. [DOI] [PubMed] [Google Scholar]
- Staller P. et al. (2001) Repression of p15INK4b expression by Myc through association with Miz-1. Nature Cell Biol., 3, 392–399. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G.R. and Bishop, M. (2001) c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature, in press. [DOI] [PubMed] [Google Scholar]
- Vlach J., Hennecke, S., Alevizopoulos, K., Conti, D. and Amati, B. (1996) Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J., 15, 6595–6604. [PMC free article] [PubMed] [Google Scholar]