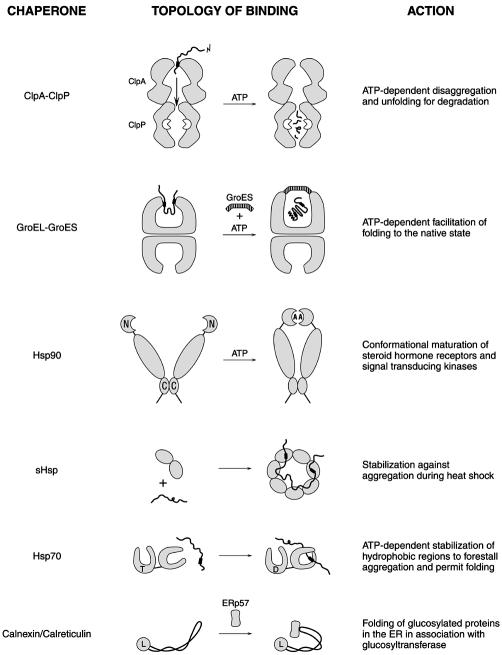
Introduction
A EuroConference/EMBO workshop on molecular chaperones was held May 26–31, 2001, at Sant Feliu de Guixols, Spain, drenched in the sunshine of the Costa Brava with Salvador Dali’s last home and a museum of his work close at hand. Suitably, the cover of the abstract book displayed a detail from one of Dali’s works, featuring a fried egg hanging by its neck. The meeting covered quite a spectrum of subjects, ranging from the involvement of chaperones in cellular physiology on through their structures and mechanisms of action. Figure 1 summarizes some of the chaperones discussed and their actions.
Fig. 1. A summary of some of the chaperones discussed and their actions. The drawings are not to scale. The solid, wavy lines indicate substrate polypeptides, with typically hydrophobic patches involved in binding to chaperones denoted by the thickened segments. N and C refer to the N- and C-terminal domains. In the Hsp90 drawing, A indicates adenine nucleotide; in the Hsp70 drawing, T and D indicate ATP and ADP, respectively. In the calnexin/calreticulin drawing, L indicates the lectin domain and ERp57 the associated PDI.
New cellular actions
From several of the featured talks, it was clear that new roles continue to be recognized for molecular chaperones. For example, a novel role proposed for the molecular chaperone Hsp90 is as a capacitor for evolutionary change (Rutherford and Lindquist, 1998). S. Lindquist (Chicago, IL) reviewed this proposal and some recent experiments in an opening talk. The idea is that Hsp90, implicated in late steps of folding of a host of signal transduction molecules, could normally buffer the phenotypic effects of mutations in such signaling cascades, allowing them to accumulate without apparent effect. During environmental stress, however, the general demand for Hsp90 would increase, as would competition among substrates for binding to Hsp90. As a result of this relative deficiency of Hsp90, multiple previously hidden variations in Hsp90-dependent signaling cascades could now be rapidly expressed. If they afforded an evolutionary advantage, they might become enriched by breeding and fixed, thus providing a mechanism of rapid evolutionary change. Lindquist described some new work on phenotypic expression using Arabidopsis, showing that either the Hsp90 inhibitor, geldanamycin, or heat stress could uncover the same lineage-specific defect. For example, in one plant lineage, either of these treatments led to the production of hairy roots, whereas, in another, both treatments produced pin-shaped leaves.
While the signaling component, Bag1, has been shown to physically associate with the ATPase domain of Hsp70, its low relative abundance (<1% that of Hsp70) has made its role in regulating cell growth difficult to understand. Studies by R. Morimoto (Evanston, IL) and co-workers now explain the mystery: under normal conditions, Bag1 binds to and activates Raf-1/ERK kinase, but, under stress conditions, the large quantity of newly induced Hsp70 competes effectively with Raf-1 for binding Bag1, inactivating Raf-1 and thereby shutting off DNA synthesis and cell growth (Song et al., 2001).
K. Liberek (Gdansk, Poland) and co-workers described a new role for the bacterial hexameric AAA protein, ClpB, in which it cooperates with the DnaK (Hsp70), DnaJ and GrpE chaperones in enabling DNA replication of the broad-host-range plasmid RK2. In vitro, this set of components acted to convert a dimeric form of the RK2 replication initiation protein, TrfA, to its functional monomer. This dissociating action resembles an already described action for ClpB in disrupting protein aggregates.
K. Nagata (Kyoto, Japan) reported a role for an endoplasmic reticulum (ER) stress-induced mannose 8-lectin, called EDEM (ER degradation-enhancing mannosidase-like protein). This 74-kDa type II transmembrane protein has a large lumenal domain with homology to α-1,2 mannosidases, but it does not exhibit mannosidase activity. EDEM could, however, bind man-8 glycans, associate with misfolded α1-antitrypsin and accelerate degradation of the protein in cultured cells. In another talk on ER quality control by G. Lederkremer (Tel Aviv, Israel), ER-retained proteins, such as uncleaved precursors of soluble asialoglycoprotein receptor H2a and unassembled major histocompatibility complex (MHC) Class I heavy chains, were reported to accumulate in the presence of proteasomal inhibitors. They were found in an enclosed compartment adjacent to the centrosome, along with calnexin/calreticulin. The compartment, which did not co-stain with antibodies to β-COP and γ-tubulin, was proposed to be a quality-control compartment located in the vicinity of cytosolic aggresomes.
Chaperone action in membrane insertion and translocation
A contemporary understanding of components mediating targeting and incorporation of integral membrane proteins of Escherichia coli into the lipid bilayer was presented by M. Müller (Freiburg, Germany). The nascent chains of such proteins interact through signal anchor sequences with the signal recognition particle (SRP) system (Ffh,4.5S RNA/FtsY), which directs the chains co-translationally to the SecYE translocon. Subsequent translocation of large hydrophilic segments of integral membrane proteins may require the action of the SecA motor to drive chain insertion into the SecYE translocon. Thus, the SRP and SecA systems may cooperate sequentially in directing membrane targeting and insertion of integral membrane proteins (Neumann-Haefelin et al., 2000). When integration of the multi-spanning mannitol A protein was studied with inside-out membrane vesicles, an additional, recently identified component came into play. Cross-links between transmembrane (TM) domains 1–3 and YidC, an essential 6-TM domain component of the bacterial inner membrane, were observed. YidC may have a role in binding and bundling transmembrane α-helices before their release into the membrane lipids (Beck et al., 2001).
YidC is also critical to the integration of proteins that do not appear to use the SecYE machinery (Samuelson et al., 2000). A. Kuhn (Stuttgart, Germany) reviewed studies of YidC depletion strains, in which M13 procoat could not be inserted across the inner membrane and remained in its precursor form. Despite the absence of YidC, procoat became bound to the membrane, indicating that YidC influences the folded conformation of the procoat to enable it to cross the membrane. Remarkably, purified YidC reconstituted into E. coli phospholipids could support insertion of procoat across the lipid bilayer. Further studies indicated that YidC functions independently of, but cooperatively with, the electrochemical gradient across the inner membrane to enable translocation.
Proteins of the so-called ‘tail-anchored’ class are inserted post-translationally by their C-terminal hydrophobic TM domain. E. Pedrazzini (Milan, Italy), from the group of N. Borgese, reported on studies with cytochrome b5, a protein that exists in two isoforms, one in the ER (Pedrazzini et al., 2000), the other in the outer mitochondrial membrane. The short C-terminal segment that follows the TM domain determines the localization, as shown most convincingly by the fact that the ER form can be redirected to mitochondria by the introduction of positive charges. Using an N-glycosylation site at the C-terminus, it could also be shown that the mitochondrial protein does not travel through the ER to its final destination. The C-terminal signal must therefore be recognized by cytosolic proteins, such as a 70-kDa protein recovered by cross-linking, before the protein is integrated into a membrane.
Mitochondrial translocation mechanisms were discussed by W. Neupert (Munich, Germany), who reviewed the two current hypotheses for the role of mitochondrial Hsp70 and ATP-dependent unfolding: active pulling on the incoming polypeptide by a TIM44-Hsp70 machine resulting in concomitant unfolding, versus Hsp70 trapping of spontaneously unfolding protein and subsequent unraveling by a ‘ratchet’ mechanism involving Brownian oscillation. He described some recent data on the spacing of potential Hsp70 interaction sites on the translocating polypeptide. Mitochondria unfold and translocate chimeric precursors containing 50-residue segments that do not bind to Hsp70 (such as polyGly or polyGlu), although at a somewhat reduced rate, indicating that continuous interaction between TIM44/Hsp70 and the polypeptide is not necessary and supporting a Brownian ratchet model. Neupert also discussed the difference between the force usually available from ATP-driven motors such as kinesin, ∼5 pN, and that required to unfold a polypeptide by pulling on the end, ∼250 pN for domains of titin. Despite the apparent magnitude of the required force, titin domains as passenger proteins can be unfolded and imported into mitochondria, suggesting either that a much greater force than predicted is available or that mitochondria pull slowly with relatively low force but take advantage of conformational ‘breathing’ of the polypeptide to trap partially unfolded states. W. Voos (Freiburg, Germany) reported on import studies using mitochondria with a mutant form of Hsp70, ssc1-2, which has reduced ability to interact with TIM44 but increased binding to translocating preproteins (Voisine et al., 1999). Data were presented that suggest that a trapping function of Hsp70 is sufficient to facilitate import of loosely folded precursors but is not sufficient to drive the unfolding and translocation of tightly folded ones, implying that an active mechanism is required for the latter.
New chaperone structures
E. Vierling (Tucson, AZ) described a new crystal structure of a small heat shock protein (sHsp), Hsp16.9, from wheat (van Montfort et al., 2001). This protein is assembled as a dodecamer composed of two hexamer discs, each itself a trimer of dimers. The α-crystallin domain of this sHsp, a β-sandwich implicated in polypeptide binding, was superposable with that of the earlier-determined archaeal sHsp. But here, the N-terminal domain of one subunit of each dimer was resolvable as an α-helical structure that formed contacts with the same α-helix from an adjacent subunit of the opposite ring. Despite these and many other subunit contacts, facile exchange of subunits of this assembly was demonstrated in vitro. Given the inaccessible positions of the functionally mapped substrate binding sites in the crystal structure, such exchange is apparently required in order to expose these binding sites.
M. Maurizi (Bethesda, MD) reported two crystal structures of the Hsp100 unfoldase, ClpA. One structure was of an intact subunit found in a 65 lattice, where the complex had been distorted into an essentially continuous helical screw instead of the expected hexameric ring. The intact ClpA subunit was observed to be arranged in the order, N-domain–ATPase1–ATPase2–C-domain, with the two ATPases ordered head-to-tail and each composed of the standard bipartite AAA ATPase fold. Nucleotide, in this case ADP, was housed at the interface between the two subdomains. The second structure was of just the N-terminal domain of ClpA, a pseudodimeric α-helical structure. The exact arrangement of the N-domain and ATPase segments in an intact ring awaits electron microscopy (EM) image reconstruction or a crystal structure of an intact ring. Nevertheless, current knowledge allows a rough picture of ClpA action to emerge. Capture/interaction of substrate proteins is initiated at the ‘top’ end of the cylinder by the N-domains. Then, associated but still undescribed movements of this machine ‘rip apart’ the native structure while simultaneously directing the chain down the axial cavity formed by the two AAA ATPases and on into the cognate proteasome-like ClpP protease. The unfolding and translocation steps are thought to be directional, the tagged end of the substrate protein becoming unfolded first, as indicated by studies of chimeric proteins described by A. Matouschek (Evanston, IL; Lee et al., 2001). This part of the protein is the first to proceed into the ClpP protease, as shown in time-dependent fluorescence energy transfer studies described by E. Weber-Ban (Zurich, Switzerland; Reid et al., 2001). Finally, B. Bukau (Freiburg, Germany) reported on the characterization of a new component of the Clp system, called ClpS, whose coding sequence resides upstream in the ClpA operon and is expressed from the same promoter. This 10-kDa protein binds to ClpA, with a stoichiometry of possibly one ClpS per ClpA subunit, and, in the presence of ClpP, prevents both the degradation of ssrA-tagged substrates and the autodegradation of ClpA. However, the presence of ClpS also leads to the ClpA–ClpP degradation of thermally aggregated malate dehydrogenase. These findings suggest a role for ClpS as a specificity factor.
A. Helenius (Zurich, Switzerland) reported on the structure of the P-domain of the ER chaperone, calreticulin, determined by NMR spectroscopy (Ellgaard et al., 2001). This study revealed an unusual 140 Å extended hairpin fold that is flexible in its midportion and covalently connected at one end to a ball-like lectin domain, which recognizes the N-linked glycans of substrate proteins. At its other end, calreticulin binds to the disulfide oxidase/isomerase, ERp57, which acts as a PDI-like folding component in association with calnexin/calreticulin. Thus, glycan-recognizing and folding/disulfide oxidase functions of the two components are brought into proximity via this flexible ‘arm’ structure.
New mechanism studies
A. Fersht (Cambridge, UK) described approaches for studying unassisted protein folding in vitro, particularly for two-state systems. One approach is to study the relative effects of mutations on the stabilities of the native and folding transition states. An equal effect on both states implies that the mutant residue is involved in structure in the transition state. Fast kinetic studies, using temperature jumps to examine unfolding on a nanosecond (real) timescale, were also described. This is a timescale that current molecular dynamics simulation techniques can also reach, making it possible to compare experimental data with computer simulations. Data obtained for an α-helical engrailed homeodomain show remarkable agreement between simulations and solution experiments charting the undocking of the three α-helices by fluorescence (Mayor et al., 2000). A concern has been whether fast folding/unfolding is strictly the province of fast docking/undocking of preformed α-helices. This is apparently not the case, as a similarly fast behavior has now also been observed with a three-stranded antiparallel β-sheet structure, the WW domain of YAP40.
Chaperonins
Chaperonins are the double-ring assemblies of the Hsp60/GroEL and TF55/CCT families that mediate ATP-dependent folding to the native state inside sequestered central cavities. In the case of GroEL–GroES, the well studied bacterial chaperonin system, it has been established that most substrate proteins undergo multiple rounds of binding, folding in the central cavity for a limited time (10–15 s) and release back into solution, whether folded or not. For typical GroEL substrates, only 2–5% of the input protein molecules fold to a native or near-native state in a single cycle. Even though the native state can be reached in the chaperonin chamber, it has remained unknown whether the cavity plays a specific role in the chaperonin reaction. What would happen if a non-native polypeptide, ejected into the bulk solution after a round of attempted folding in the central cavity, could not return to a GroEL molecule? Would productive folding now ensue in the bulk solution? U. Hartl and M. Hayer-Hartl (Martinsreid, Germany) and their co-workers presented a clever experiment to address this question. They biotinylated a cysteine engineered into GroEL near the inlet to the central cavity, showed that this was without effect on the GroEL–GroES reaction and observed by EM that added streptavidin blocked the central cavity and obstructed substrate binding. Then, the effect of streptavidin addition at various times during refolding of a substrate protein by this biotinylated GroEL was determined. Productive folding halted immediately after streptavidin was added, suggesting that the released, non-native substrate proteins that were unable to return to the blocked GroEL could not proceed to the native state in the bulk solution. Thus, the cavity inside GroEL–GroES affords a productive environment for folding as compared to the bulk solution.
In contrast, another study of GroEL-mediated folding seems to have uncovered a mechanism that may rely on productive folding in the bulk solution (A. Horwich, New Haven, CT). This reaction involves the folding of aconitase, an 82-kDa monomer that is too large to fit in the GroEL–GroES cavity. Horwich reported that this protein nevertheless requires both GroEL and GroES for its productive folding, as had already been suggested from earlier studies showing that aconitase needs both Hsp60 and Hsp10 function to achieve a soluble state following import into mitochondria (Dubaquié et al., 1998).
Further analyses of chaperonin action using cryoEM to study various states of both GroEL and the eukaryotic cytosolic chaperonin, CCT, were presented. H. Saibil (London, UK) described new studies examining the conformational changes of an ATP-bound GroEL complex, reporting on novel changes in both the nucleotide-bound and opposite rings. M. Fisher (Kansas City, KS) described the effects of binding a substrate protein, glutamine synthetase, to unoccupied GroEL. This resulted in a difference in the conformations of the two GroEL rings, with the aperture of the distal ring relatively closed, providing an explanation for the 1:1 binding stoichiometry of polypeptide substrate to GroEL complex (Falke et al., 2001).
Hsp90
Structural and mutation studies have recently identified an essential ATP binding and hydrolysis cycle for Hsp90, and this was the focus of several talks. C. Prodromou (London, UK) described effects of deletions at the C-terminus, which blocked normal dimerization of Hsp90 and concordantly abolished ATPase activity. Taken with additional cross-linking and fluorescence studies supporting dimerization of the N-terminus (Prodromou et al., 2000), it would appear that dimerization involving both N- and C-termini is required for ATP binding and hydrolysis activity. Prodromou also presented a model in which the cochaperone Sti1/Hop may stabilize a nucleotide-empty (nucleotide-exchanging) state of the Hsp90 dimer that can accept substrate protein. Subsequent binding of cyclophilin and release of Sti1 would lock both ATP and substrate protein into the complex, and binding of p23 would provide further stabilization. J. Buchner (Munich, Germany) presented studies also supporting a role for C-terminal dimerization in ATP binding/hydrolysis and provided evidence for the roles of the immunophilins in both direct and indirect substrate binding by Hsp90 (Pirkl and Buchner, 2001). S. McLaughlin (Manchester, UK) described experiments on the refolding of the ligand-binding domain of the glucocorticoid receptor (GR), which binds to Hsp90 in the absence of ligand. An α-helical dimer was produced that could bind ligand and that, remarkably, stimulated the ATPase of Hsp90 by 200-fold, supporting the idea that Hsp90 substrates can indeed stimulate activity of its ATPase.
Hsp70
S. Rospert (Halle, Germany) discussed a ribosome-associated complex (RAC) studied in yeast, which contains one subunit of the DnaJ homolog, zuotin, and one subunit of the DnaK homolog, Ssz1 (Gautschi et al., 2001). Studies using cross-linking and immunoprecipitation revealed that RAC is not in direct contact with nascent chains. Interestingly, experiments using ribosome–nascent-chain complexes that were first salt-stripped and then supplemented with RAC showed that RAC is required for cross-linking of the nascent chain to the ribosome-associated Hsp70 homolog, Ssb1/2. This suggests that Ssb1/2p and RAC may functionally interact.
On a related topic, E. Craig (Madison, WI) described studies of the peptide binding domains of yeast cytosolic DnaJ proteins, Ydj1 and Sis1 (Johnson and Craig, 2001). Deletion and domain swapping experiments show that one such peptide binding domain must be in cis to an N-terminal J-domain and a glycine-rich domain to achieve cell viability. Additionally, the most C-terminal domain, implicated in dimerization of J proteins, was not required for viability.
Folding and unfolding in the ER
The size of the pore of the translocation channel in the ER is unclear, but the largest sizes reported (40–60 Å; Hamman et al., 1997) could allow small polypeptides to fold before they have emerged into the ER lumen. To test the effect of the translocon on folding, A. Helenius (Zurich, Switzerland) and co-workers used nascent chains containing the protease domain of the Semliki Forest virus capsid protein. When this domain is folded, it cuts itself off from the growing polypeptide chain. With free ribosomes, folding occurred when the domain was 38–40 residues away from the P-site of the ribosome, but with membrane-bound ribosomes the distance increased to 68 residues. Thus, despite the small size of the protease domain (longest dimension 39 Å), the protein cannot fold inside the translocation channel. Other folded domains that are even smaller (<29 Å) gave the same result. Other studies have shown, however, that an α-helix can form within the translocation channel (Mingarro et al., 2000).
Folding in the ER lumen has been extensively studied over the past years, but a serious obstacle has always been the lack of an in vitro assay in which the folding can be followed in detergent extracts. I. Braakman’s group (Utrecht, The Netherlands) has developed an assay in which pulse-labeled cells are lysed and the completion of folding of a model protein is followed in the extract. Although in vitro folding cannot be accomplished if the protein is completely reduced, the system should at least allow the analysis of late folding stages. B. Tu (San Francisco, CA), from J. Weissman’s group, summarized the recent progress that has been made in understanding how disulfide bridges are formed in the yeast ER (Tu et al., 2000). Protein disulfide isomerase (PDI) is responsible for oxidizing cysteines in substrates, and it is then re-oxidized by the enzyme Ero1p, a flavoprotein containing flavin adenine dinucleotide (FAD). The relay system from FAD via Ero1p to PDI and ultimately to substrate allows rapid disulfide bridge formation even in the presence of reduced glutathione, an environment that permits the reduction and correction of incorrect disulfide bonds. Mammals have two isoforms of Ero1. R. Sitia (Milan, Italy) and co-workers showed that both can oxidize a mammalian immunoglobulin and replace the yeast protein in vivo, but there appear to be differences, with the β-form being more active. Ero1p could form mixed disulfides with PDI, but not with other members of the PDI family, suggesting that they have other oxidizing partners. Previous studies in yeast had shown that PDI was mostly in the oxidized state, but in the mammalian cells that have been tested, it appeared to be mostly reduced. Because there was further reduction upon addition of dithiothreitol (DTT), only one of the two thioredoxin motifs in PDI may have been oxidized (Mezghrani et al., 2001). An unexpected role for PDI was discovered recently in studies on cholera toxin (Tsai et al., 2001). This toxin is taken up by intestinal cells, travels backwards through the secretory pathway and is disassembled in the ER before a fragment of the toxin, the A1-chain, is transported into the cytosol. T. Rapoport (Boston, MA) reported observations that PDI is largely responsible for unfolding of the toxin in the ER. Interestingly, PDI does not act as an oxidoreductase, but rather as a redox-driven chaperone: it binds the substrate in its reduced state, and releases it in its oxidized state.
Concluding remarks
As Dali said in Diary of a Genius (1966), ‘We are all hungry and thirsty for concrete images’. This meeting showed that the abstract concepts of protein folding are becoming more clearly and concretely delineated, both in vivo and in vitro. New insights into the diverse cellular roles of molecular chaperones were presented, several new chaperone structures were described and mechanisms of protein folding and unfolding were further defined.

The workshop was held at Sant Feliu de Guixols, Spain, May 26–31, 2001. It was organized by Bernd Bukau and Ineke Braakman. The poster background is a reproduction of ‘Eggs on a Plate without a Plate’, by Salvador Dali (1930).

Ineke Braakman and Bernd Bukau, the organizers of the meeting
REFERENCES
- Beck K., Eisner, G., Trescher, D., Dalbey, R.E., Brunner, J. and Muller, M. (2001) YidC, an assembly site for polytopic Escherichia coli membrane proteins located in the immediate proximity to the SecYE translocon and lipids. EMBO rep., 2, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubaquié Y., Looser, R., Fünfschilling, U., Jenö, P. and Rospert, S. (1998) Identification of in vivo substrates of the yeast mitochondrial chaperonins reveals overlapping by non-identical requirement for hsp60 and hsp10. EMBO J., 17, 5868–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Riek, R., Braun, D., Herrmann, T., Helenius, A. and Wüthrich, K. (2001) Three-dimensional structure topology of the careticulin P-domain based on NMR assignment. FEBS Lett., 488, 69–73. [DOI] [PubMed] [Google Scholar]
- Falke S., Fisher, M.T. and Gogol, E.P. (2001) Structural changes in GroEL effected by binding a denatured protein substrate. J. Mol. Biol., 308, 569–577. [DOI] [PubMed] [Google Scholar]
- Gautschi M., Lilie, H., Funfschilling, U., Mun, A., Ross, S., Lithgow, T., Rucknagel, P. and Rospert, S. (2001) RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl Acad. Sci. USA, 98, 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman B.D., Chen, J.C., Johnson, E.E. and Johnson, A.E. (1997) The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational protein translocation at the ER membrane. Cell, 89, 535–544. [DOI] [PubMed] [Google Scholar]
- Johnson J.L. and Craig, E.A. (2001) An essential role for the substrate-binding region of Hsp40s in Saccaromyces cerevisiae. J. Cell Biol., 152, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Schwartz, M.P., Prakash, S., Iwakura, M. and Matouschek, A. (2001) ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell, 7, 627–637. [DOI] [PubMed] [Google Scholar]
- Mayor U., Johnson, C.M., Daggett, V. and Fersht, A.R. (2000) Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation. Proc. Natl Acad. Sci. USA, 97, 13518–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezghrani A., Fassio, A., Benham, A., Simmen, T., Braakman, I. and Sitia, R. (2001) Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J., 20, 6288–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingarro I.I., Nilsson, I.I., Whitley, P. and von Heijne, G. (2000) Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol., 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C., Schafer, U., Muller, M. and Koch, H-G. (2000) SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J., 19, 6419–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E., Villa, A., Longhi, R., Bulbarelli, A. and Borgese, N. (2000) Mechanism of residence of cytochrome b(5), a tail-anchored protein, in the endoplasmic reticulum. J. Cell Biol., 148, 899–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkl F. and Buchner, J. (2001) Functional analysis of the Hsp90-associated human peptidyl prolyl cis/trans isomerases, FKBP51, FKBP52, and Cyp40. J. Mol. Biol., 308, 795–806. [DOI] [PubMed] [Google Scholar]
- Prodromou C., Panaretou, B., Chohan, S., Siligardi, G., O’Brien, R., Ladbury, J.E., Roe, S.M., Piper, P.W. and Pearl, L.H. (2000) The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J., 19, 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B.G., Fenton, W.A., Horwich, A.L. and Weber-Ban, E.U. (2001) ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc. Natl Acad. Sci. USA, 98, 3768–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S.L. and Lindquist, S. (1998) Hsp90 as a capacitor for morphological evolution. Nature, 396, 336–342. [DOI] [PubMed] [Google Scholar]
- Samuelson J.C., Chen, M., Jiang, F., Moller, I., Wiedmann, M., Kuhn, A., Phillips, G.J. and Dalbey, R.E. (2000) YidC mediates membrane protein insertion in bacteria. Nature, 406, 637–641. [DOI] [PubMed] [Google Scholar]
- Song J., Takeda, M. and Morimoto, R.I. (2001) Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nature Cell Biol., 3, 276–282. [DOI] [PubMed] [Google Scholar]
- Tsai B., Rodighiero, C., Lencer, W.I. and Rapoport, T.A. (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell, 104, 937–948. [DOI] [PubMed] [Google Scholar]
- Tu B.P., Ho-Schleyer, S.C., Travers, K.J. and Weissman, J.S. (2000) Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science, 290, 1571–1574. [DOI] [PubMed] [Google Scholar]
- van Montfort R.L.M., Basha, E., Friedrich, K.L., Slingsby, C. and Vierling, E. (2001) Crystal structure and assembly of an eukaryotic small heat shock protein. Nature Struct. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Voisine C., Craig, E.A., Zufall, N., von Ahsen, O., Pfanner, N. and Voos, W. (1999) The protein import motor of mitochondria: unfolding and trapping of preproteins are distinguished and separable functions of matrix Hsp70. Cell, 97, 565–574. [DOI] [PubMed] [Google Scholar]