SUMMARY
The gut epithelium has a remarkable ability to recover from damage. We employed a combination of high throughput sequencing approaches, mouse genetics, and murine and human organoids, and identified a role for TGFB signaling during intestinal regeneration following injury. At 2 days following irradiation (IR)-induced damage of intestinal crypts, a surge in TGFB1 expression is mediated by monocyte/macrophage cells at the location of damage. Depletion of macrophages or genetic disruption of TGFB-signaling significantly impaired the regenerative response. Intestinal regeneration is characterized by induction of a fetal-like transcriptional signature during repair. In organoid culture, TGFB1-treatment was necessary and sufficient to induce the fetal-like/regenerative state. Mesenchymal cells were also responsive to TGFB1 and enhanced the regenerative response. Mechanistically, pro-regenerative factors, YAP/TEAD and SOX9, are activated in epithelium exposed to TGFB1. Finally, pre-treatment with TGFB1 enhanced the ability of primary epithelial cultures to engraft into damaged murine colon, suggesting promise for cellular therapy.
Keywords: intestine, regeneration, fetal reversion
Graphical Abstract
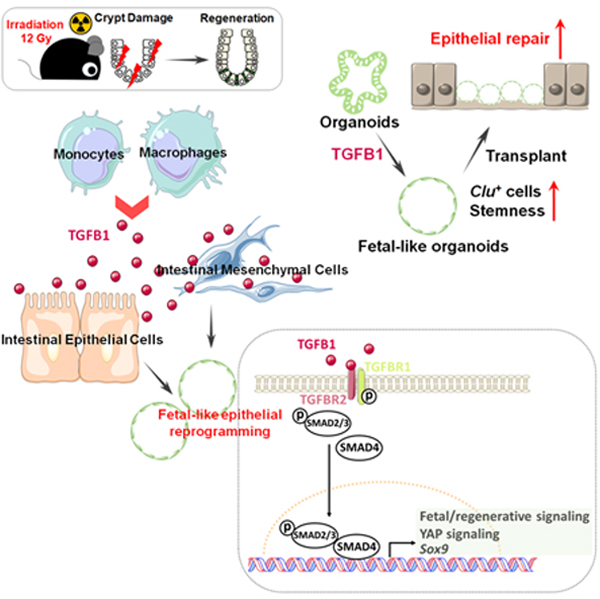
Table of Contents Statement (eTOC)
Chen et al. identify TGFB1 as a key facilitator of intestinal regeneration, revealing that TGFB1 is sufficient to induce epithelial cells to acquire a fetal-like state typical of regenerating intestine. This fate shift enhances organoid transplants into damaged mouse colons. The TGFB-induced regenerative circuit may have applications in cellular therapy.
INTRODUCTION
The intestinal epithelium is a single layer of cells organized into crypts and villi. Under homeostatic conditions, the intestinal epithelium displays a remarkably high turnover rate, where the majority of epithelial cells are replaced every 3 to 5 days, fueled by proliferation of stem and progenitor cells localized in the crypts. The high-rate of crypt cell proliferation makes crypts sensitive to damage by acute inflammation, chemotherapy, or radiotherapy. Upon damage, stem cells are often lost, and differentiated epithelial cell lineages exhibiting plasticity can respond to tissue damage and restore resident intestinal stem cells (ISCs), including crypt base columnar stem cells (CBCs), which are important for regeneration following injury.1 For example lineage tracing studies have shown that, Dll1+2 and Atoh1+ secretory3 progenitor cells, Lyz1+ Paneth cells4, and Alpi+ enterocyte progenitor cells5 can all de-differentiate to Lgr5+ ISCs in response to injury, as can cells believed to be slower cycling reserve stem cell populations.6–10
A remarkable property of regenerating intestinal epithelium is its acquisition of a fetal-like transcriptome. The fetal-like, regenerative state is observed in response to helminth infection, irradiation, ablation of CBCs, or DSS-induced colonic damage.11,12 Regenerating epithelium in response to virtually all types of tissue damage rely upon activation of the hippo-YAP/TAZ signaling pathway.8,12,13 Transcription factors SOX9 and ASCL2 have also been shown to be important in driving the regenerative response,9,14 and the fetal-like/regenerative signature appears to be coopted by colon cancers,6,15 further highlighting the importance of this phenomenon. Despite our understanding of cellular plasticity in the intestine, the signaling mechanisms leading to the fetal-like regenerative state and subsequent regenerative process are less well understood.
In this study, we aimed to uncover cells driving the regenerative response, as well as the signaling mechanisms underlying the regenerative ability of intestinal tissues in hopes of finding new therapeutic avenues for intestinal regeneration. Using genetic mouse models, epigenetics, bulk and single cell RNA-sequencing and tissue/organoid culture approaches, we explored the interplay among intestinal epithelial cells, mesenchymal cells and immune cells. We found that TGFB signaling is activated in the intestine post-irradiation, and the main source of TGFB1 is monocytes/macrophages. We further explored the relationship between TGFB signaling and the regenerating intestinal epithelium, as well as the ability of intestinal mesenchyme to promote regeneration after TGFB1 exposure. We discovered a critical role for TGFB1 in promoting intestinal regeneration by inducing a fetal-like, regenerative state in the epithelium via activation of pro-regeneration transcription factors YAP and SOX9. Therapeutically, TGFB1-treated organoids support a more robust tissue engraftment into a mouse model of colitis, suggesting that activation of the regenerative response enhances cellular therapy. These results suggest that modulation of TGFB-signaling could enhance regenerative strategies or cellular therapies in the human intestine.
RESULTS
Monocyte/Macrophages deliver TGFB1 ligands to intestinal crypts damaged by irradiation
To better appreciate a time-resolved intestinal response to ionizing radiation, we performed a detailed study across the 5 days post-12 Gy whole body irradiation of mice. Loss of intestinal crypt architecture was widespread by 2 days post-irradiation, with notable loss of proliferative (Ki67 and BrdU incorporation) and stem cell (OLFM4) markers (Figure 1A–B, Figure S1A). Between days 2 and 3 post-IR, cellular proliferation in the crypt resumed, most frequently near the crypt-villus junction (Figure 1B and Figure S1A–D), with highly proliferative, regenerating crypts observed by day 4–5 (Figure 1A). Consistent with published reports, transcriptome analysis of isolated crypt epithelium at day 2–3 post-IR (GSE16515716) identified elevated levels of genes associated with a fetal/regenerative epithelium;11,12,17 a corresponding decrease was observed in transcripts associated with the crypt-base-columnar stem cell18 (Lgr5, Olfm4, Figure 1C–D, Figure S1E). Similar patterns are observed at the single-cell level (GSE1177838) when comparing crypt epithelial cells from control mice or mice at 3 days post-IR (Figure 1E). To specifically focus on cells driving regeneration at an earlier time point in regeneration, we isolated proliferative cells at 56h post-IR, a moment when proliferative cells begin to reappear towards the upper regions of the former crypts. To isolate proliferative cells, we employed Ki67-RFP transgenic mice19 and used Fluorescence-Activated Cell Sorting (FACS) followed by scRNA-seq. Compared to Ki67+cells in homeostatic crypt epithelium, Ki67+ cells within the regenerating epithelium formed a separate cluster when visualized using Uniform Manifold Approximation and Projection (UMAP) and expressed elevated transcript levels of markers associated with fetal/regenerative intestinal epithelium, including Ly6a and Clu (Figure 1F–G, Figure S1F). Within the regenerative cell cluster, Olfm4+ and Clu+ cells appeared at opposite ends, with Clu hi cells expressing lower levels of cell cycle genes than Olfm4 hi cells (Figure S1G–H), consistent with the previous report of Clu+ cells marking a more quiescent revival population.8 Taken together, these data indicate that a regenerative response following 12 Gy irradiation initiates around day 2 post-IR with the re-emergence of Ki67+ and BrdU+ cell populations that express hallmark transcripts of the fetal/regenerative epithelium.
Figure 1. Crypt regeneration mainly starts 3 days after irradiation.
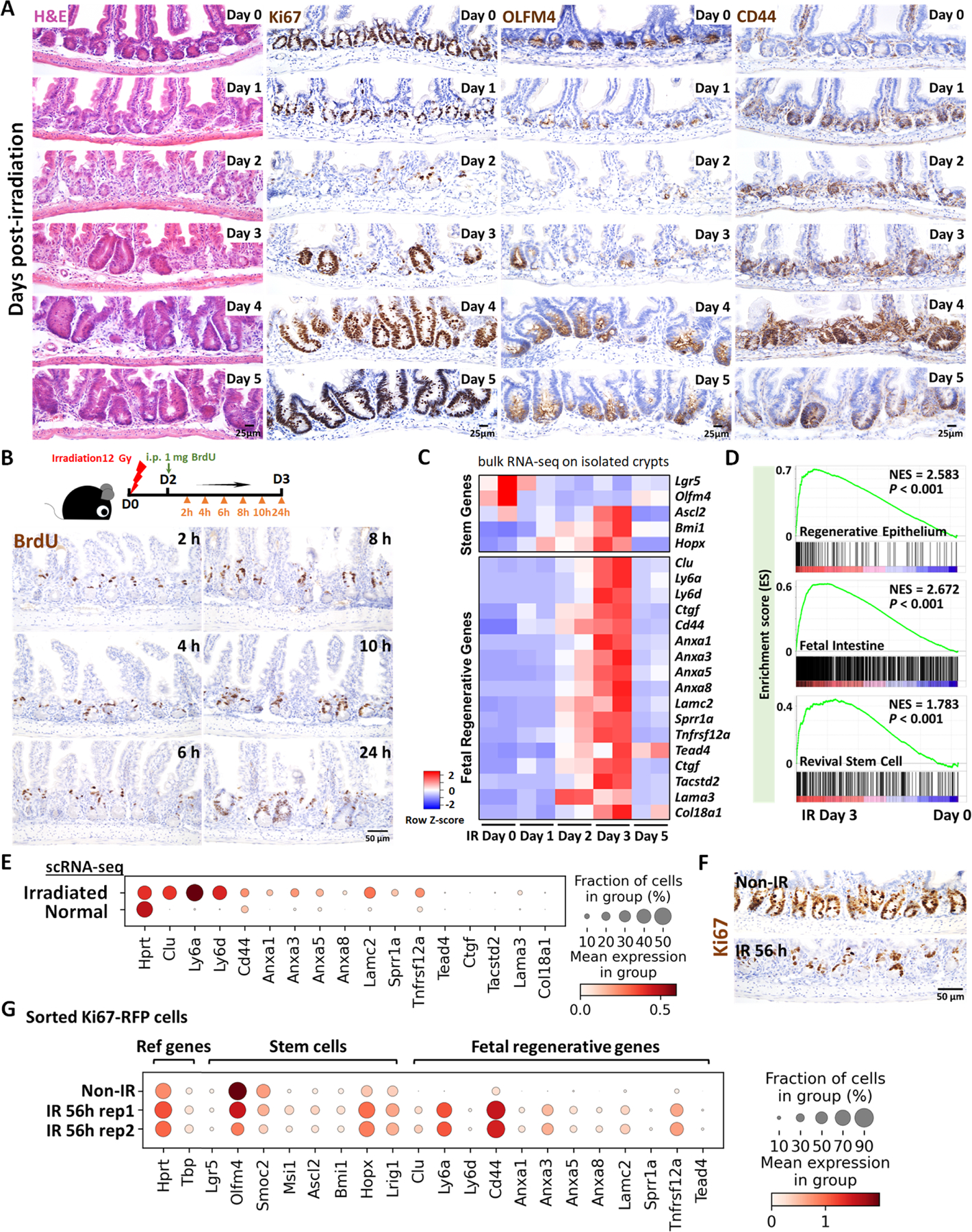
(A) Demonstration of intestinal regeneration following 12 Gy of irradiation of mice. Crypt cells are lost by 2 days post-irradiation (12 Gy) but restoration begins at Day 3 after irradiation, when highly proliferative regenerative clusters of cells expand, as evidenced by H&E staining and immunohistochemistry staining of stem/proliferative markers (brown color) including Ki67, OLFM4 and CD44 (representative of 3 biological replicates). (B) Immunostaining of BrdU (proliferative marker, brown color; representative of 3 biological replicates). Mice were injected with 1 mg of BrdU at Day 2 post-irradiation. Intestinal tissues were harvested after 2, 4, 6, 8, 10 and 24 hours of BrdU injection. (C) Heatmap depicts transcript levels of fetal/regenerative marker genes and regenerative stem cell-associated genes that are highly expressed at Day 3 post-irradiation (GSE165157,16 RNA-seq, n=2 biological replicates per time-point). (D) GSEA confirms that gene signatures of regenerative epithelium, fetal spheroid and revival stem cells8,12,17 are elevated at Day 3 post-irradiation (GSE165157,16 crypt cells, n=2 biological replicates per time-course, Kolmogorov-Smirnov test, P < 0.001). See expanded panel in Figure S1E. (E) scRNA-seq reveals that fetal/regenerative transcripts are elevated in irradiated crypt cells after 3 days of irradiation. Cell numbers per condition (GSE1177838) for irradiated crypt cells: n=4252 and normal crypt cells: n=4328. (F) Immunostaining of Ki67 after 56 hours of irradiation vs. non-irradiation (proliferative marker, brown color; representative of 3 biological replicates). (G) scRNA-seq reveals that fetal/regenerative and reserve stem cell transcripts are elevated in sorted Ki67-RFP positive cells after 56 hours of irradiation. Number of cells in each condition was Non-IR Ki67-RFP positive cells: n=1739; IR 56h Ki67-RFP positive cells replicate 1: n=677; IR 56h Ki67-RFP positive cells replicate 2: n= 669. Ki67-RFP positive cells were isolated and sorted from crypt cells of Mki67tm1.1Cle/J mice after 56 hours of IR vs. non-IR control. IR: irradiation; Non-IR: non-irradiation (normal control). Also see Figure S1F–H.
We next sought to define key cells and signaling pathways that support the regenerative epithelium. We first conducted scRNA-seq analysis from whole intestinal tissues post-IR (GSE165318), and interrogated ligands known to regulate prominent pathways important for epithelial homeostasis, including the TGFB/BMP and WNT pathways. Of the ligands we analyzed for transcript expression associated with regenerative time points, Tgfb1 stood out with robust expression, particularly at day 3 post-IR (Figure 2A). We next conducted a time-resolved series of experiments to explore potential TGFB1 contribution to intestinal regeneration (Figure S2A). Validation qRT-PCR analysis of Tgfb1 expression in whole intestine confirmed elevated levels at days 2–3 post-IR, and ELISA assays showed corresponding increase in TGFB1 protein levels at day 3 post-IR in whole intestinal lysates (Figure 2B–D, Figure S2B–E). Increase in TGFB1 corresponded to elevated levels of p-SMAD2/3, the downstream transcriptional effectors of TGFB signaling (Figure S2F–G). Further analysis of scRNA-seq data identified a cluster of cells expressing monocyte/macrophage markers as the most prominent source of Tgfb1 transcripts in the gut post-IR (Figure 2E, Figure S2H). Macrophages have previously been implicated in intestinal repair/regeneration,20 and these results suggest production of TGFB may play a key role in this process. Increases in monocyte/macrophage-associated transcripts (Figure 2F), cells expressing macrophage protein markers (F4/80, Figure 2G, Figure S2I), and co-expression of macrophage markers and Tgfb1 transcripts (Figure 2H, Figure S2J) collectively pointed to monocyte/macrophages being recruited to the damaged crypts and producing TGFB1 ligands at days 2–3 post-IR.
Figure 2. TGFB1 is highly enriched in Day 3 irradiated mouse intestine, and monocytes/macrophages are likely the main cell source of TGFB1.
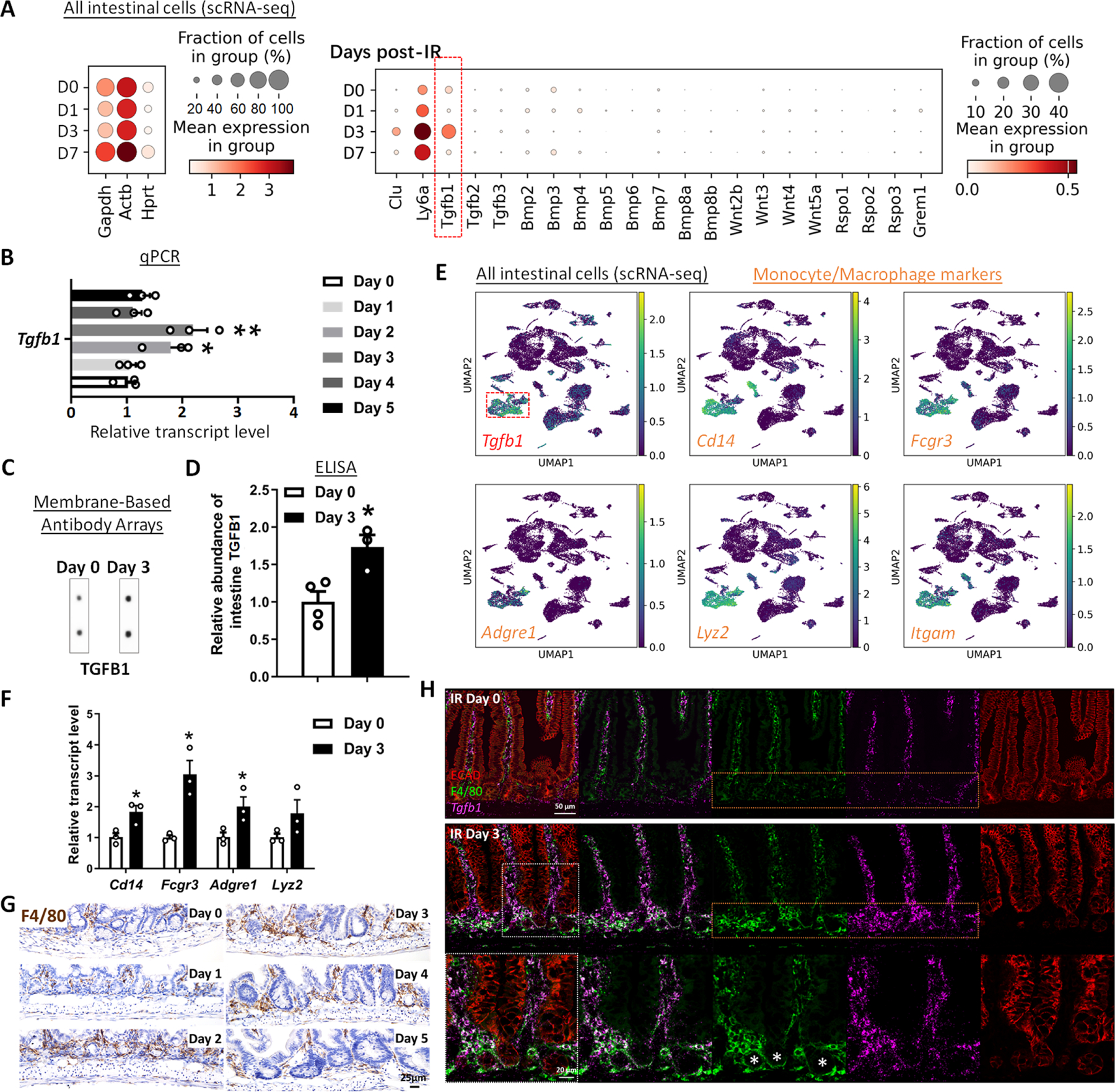
(A-C) Tgfb1 transcript is notably enriched in the intestine at 3 days post-irradiation. (A) Dot plots of scRNA-seq data following mouse irradiation at days 0, 1, 3, and 7: GSE165318, duodenum/jejunum boundary, n=3–4 biological replicates per time point) indicate that among secreted regulators of the TGF/BMP/WNT signaling pathways, transcripts corresponding to Tgfb1 are the most upregulated during regeneration of the gut (red box) and overlap expression of epithelial regenerative markers Clu and Ly6a. (B) Elevated Tgfb1 transcript levels are observed at days 2 to 3 post-irradiation. The qRT-PCR data are presented as mean ± SEM (n=3 biological replicates, whole duodenal fragments). Transcript levels are relative to Day 0 (before IR), statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post at P < 0.01** or P < 0.05*. See schematic of experimental design in Figure S2A. (C-F) TGFB1 protein levels are elevated in the intestine at Day 3 post-irradiation. (C) Membrane-based antibody arrays: n=2 independent experiments, 2 technical replicates per membrane (See full blots in Figure S2B). (D) ELISA to measure TGFB1: Data are presented as mean ± SEM (n=3–4 biological replicates, duodenal fragments, Student’s t-test at P < 0.05*). (E) UMAP projection of all cells identifies a cell cluster expressing highest levels of Tgfb1, and these Tgfb1-expressing cells co-express markers of monocytes/macrophages. Cells per time point: D0: n=4415; D1: n=2995; D3: n=7368; D7: n=3783; D14: n=2220 (GSE165318). (F) qRT-PCR corroborates elevation of transcript levels of monocyte/macrophage marker genes at 3 days post-IR. All qRT-PCR data are presented as mean ± SEM (n=3 biological replicates, duodenal fragments, Student’s t-test at P < 0.05*). (G) Tissues from mice at different days post-12 Gy irradiation were probed for the monocyte/macrophage marker F4/80 using immunohistochemistry. An increase in F4/80 cells (brown color) occurs when the tissue begins to heal at 2 days post-IR (representative of 3 biological replicates, see quantification in Figure S2I). (H) RNAscope localized Tgfb1 transcripts relative to immunostaining signal from ECAD (epithelial marker) and F4/80 (representative of 3 biological replicates). Co-stains reveal that F4/80-marked macrophages are associated with damaged crypt epithelium at day 3 post-IR and overlap with regions of elevated levels of Tgfb1, suggesting that monocyte/macrophages are recruited to the damaged tissue and produce TGFB1.
TGFB1 signaling and monocyte/macrophages are required for epithelial regeneration following irradiation
To further explore a role for TGFB1 in epithelial regeneration, we sought to define TGFB-receptor expression. TGFB receptors are expressed in the epithelium, and Tgfbr2 is specifically elevated at 3 days post-IR, specifically at a time when homeostatic stem cell markers are reduced and fetal/regenerative marker transcripts are elevated in the epithelium (Figure 3A). Tgfbr2 transcripts are particularly enriched in epithelial cells co-expressing regenerative markers such as Clu and Ly6a (Figure 3B–C, Figure S3A), and enriched near the crypt zone when assayed in situ using RNAscope (Figure 3D). Other scRNA-seq datasets of regenerating intestinal epithelium (GSE14586621) independently confirm the elevation of Tgfbr2 in the epithelium following IR and correspond to the pattern of transcripts associated with the regenerative cell state (Figure 3E, Figure S3B–C).
Figure 3. TGFB pathway is required for epithelial regeneration in the intestine after irradiation.
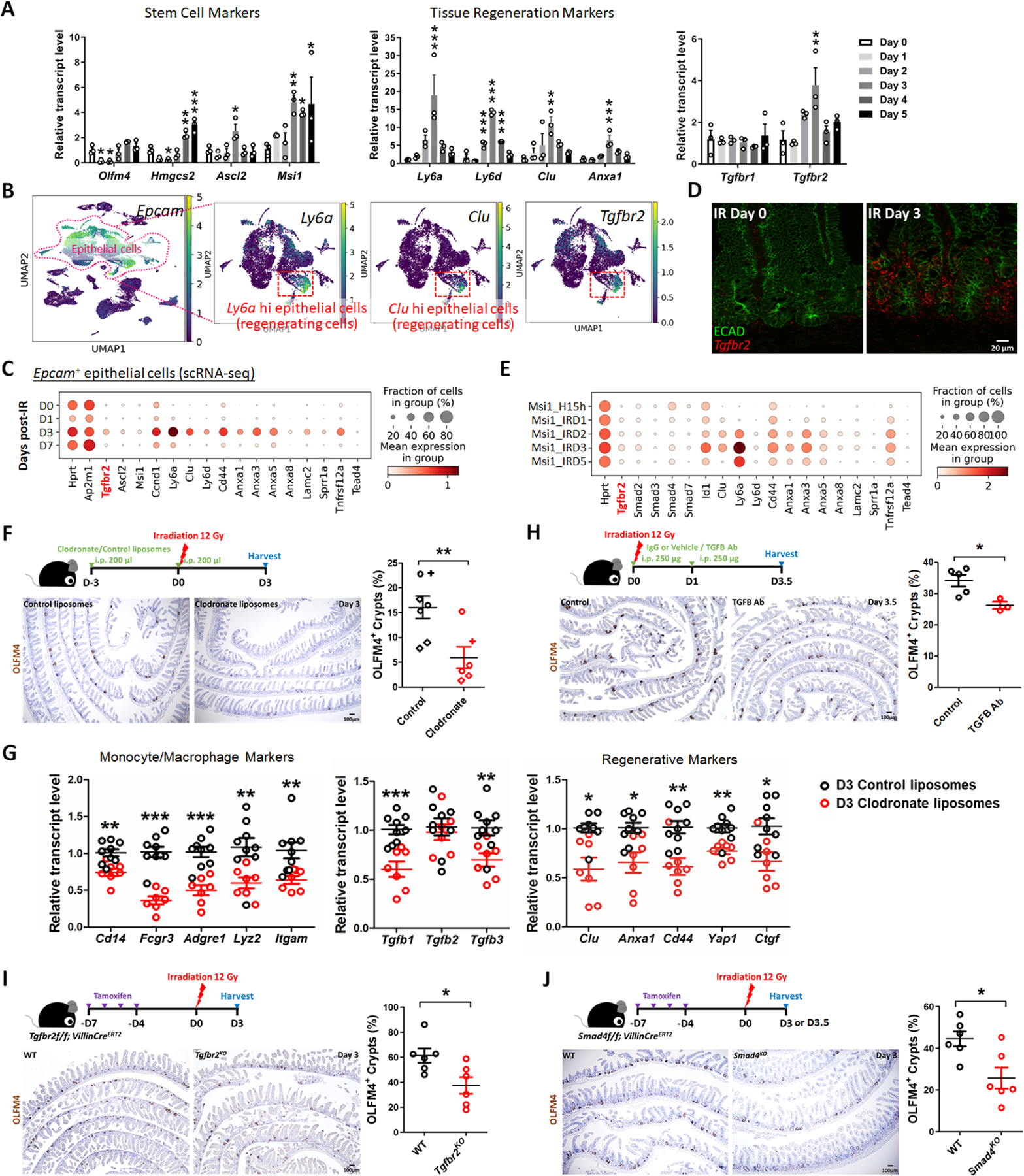
(A) qRT-PCR analysis indicates that transcript levels of stem cell marker genes, tissue regeneration marker genes and Tgfbr2 are dynamic in during intestinal regeneration post-irradiation. The qRT-PCR data are presented as mean ± SEM (n=3 biological replicates, duodenal fragments). Transcript levels relative to Day 0 before irradiation. Statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post at P < 0.001***, P < 0.01** or P < 0.05*. (B) scRNA-seq of mouse intestines across a time-course post-irradiation (GSE165318). Of all the epithelial cells in the dataset (marked by Epcam expression), there is a strong correlation between Tgfbr2-expressing cells and the subset of epithelial cells expressing regenerative markers (Ly6a and Clu, see expanded panel in Figure S3A). (C) Dot plots of epithelial cells from the same dataset reveal that Tgfbr2 is highly enriched at Day 3 post-irradiation and correlated with fetal/regenerative gene profiling. Hprt and Ap2m1 were used as reference genes. (D) RNAscope reveals elevated Tgfbr2 transcripts in the Day 3 irradiated intestine (representative of 3 biological replicates). ECAD: epithelial marker. (E) An independent scRNA-seq (GSE145866 21) dataset also reveals that transcripts related to TGFB pathway and fetal/regenerative genes are elevated at Day 3 post-irradiation in sorted Msi1-GFP positive cells (irradiation-resistant) and their progeny cells. Msi1-CreERT2; R26-mTmG mice were treated with tamoxifen for 15 hours and then used as controls or further irradiation for 1, 2, 3, and 5 days. Number of cells in each condition was Msi1_H15h: n= 2281; Msi1_IRD1: n= 1257; Msi1_IRD2: n= 1312; Msi1_IRD3: n= 2989; Msi1_IRD5: n= 1792. (F) Monocytes/Macrophages were depleted using clodronate-containing liposomes (2 treatments of 200 µl i.p. injections 72 hours pre- and day of irradiation). Tissues were assessed for regenerative cell clusters using OLFM4 immunostaining. Clodronate-treated samples shows a significant reduction in the number of OLFM4 positive regenerating cell clusters. Different symbols (circle, diamond and cruciform) represent biological replicates from three different batches of experiments (n=6–7 biological replicates, distal duodenum to proximal jejunum, Student’s t-test at P < 0.01**). (G) qRT-PCR confirms downregulated transcript levels of monocyte/macrophage marker genes, Tgfb genes, and regenerative marker genes in the intestine upon clodronate treatment (n=7–9 biological replicates, duodenal fragments, Student’s t-test at P < 0.001***, P < 0.01** or P < 0.05*). Tissues were collected 3 days post-irradiation. (H) Mice treated with 2 doses of neutralizing antibodies directed against TGFB were less efficient at regenerating post irradiation compared to control-treated mice, as measured by counting the number of proliferative foci as marked by OLFM4 immunostaining (n=3–5 biological replicates, distal duodenum to proximal jejunum, Student’s t-test at P < 0.05*). IgG or vehicle treated mice were used as control mice. Tissues were collected 3.5 days post-irradiation. (I) Tgfbr2 intestine-specific knockout restricts regeneration after irradiation (n=6 biological replicates, duodenum, Student’s t-test at P < 0.05*). (J) Smad4 intestine-specific knockout restricts regeneration after irradiation (n=6 biological replicates, Jejunum, Student’s t-test at P < 0.05*). Mice were treated with tamoxifen to inactivate Tgfbr2 or Smad4 in the intestinal epithelium 7 days before 12 Gy of irradiation. Intestine was collected 3 or 3.5 days post-IR and scored for regenerative foci using OLFM4 immunostaining.
Increased expression levels of fetal/regenerative transcripts were observed in organoids co-cultured with IR macrophages compared to organoids co-cultured with non-IR macrophages or organoids only (Figure S3D). To better appreciate the role of monocyte/macrophages and TGFB1 signaling in intestinal regeneration in vivo, we perturbed recruitment of these cells to damaged intestines or blocked TGFB-signaling following 12 Gy irradiation. We depleted monocyte/macrophage populations by treating mice with clodronate-containing liposomes. Clodronate liposome treatment led to a visible reduction in F4/80+ cells in intestines at 3 days post-IR compared to animals treated with control liposomes (Figure S3E), and led to a corresponding decrease in regenerative epithelial foci in the intestine (OLFM4 immunostain, Figure 3F). This is consistent with the report that a block in monocyte recruitment to the damaged intestine results in less regeneration after IR in a CCR2KO model.22 Additionally, transcript levels for markers of monocyte/macrophages were decreased by clodronate treatment, as well as Tgfb1 transcripts and protein (Figure 3G, Figure S3F). A reduction in regenerative cell markers was also observed upon clodronate treatment (Figure 3G) consistent with a role for monocyte/macrophage cells in both promoting regeneration and with supplying TGFB1 to the damaged crypts. To specifically target TGFB signaling in the regenerative process, we either treated mice with TGFB-neutralizing antibodies, or genetically inactivated Tgfbr2 or Smad4. In each case, perturbations of the TGFB-signaling pathway led to reduction of regenerative crypts following IR (Figure 3H–J). These experiments suggest a mechanism in which monocyte/macrophages promote epithelial regeneration by promoting TGFB1 signaling activity in the intestinal epithelium.
TGFB1 is necessary and sufficient to induce fetal reversion in intestinal organoid cultures
The previous experiments perform loss-of-function assays to support a role for TGFB1 in driving epithelial regeneration. We next investigated whether exogenous treatment of TGFB1 would be sufficient to induce a regenerative epithelial state in the context of intestinal organoid culture.23 We exposed intestinal organoids to 4 Gy IR four days after initiation of crypt culture, and 48 hours later we treated cultures with a single, 24-hour dose of TGFB1 (Figure 4A) to approximate the timeline of TGFB1 enrichment in the regenerating gut (Figures 2–3). Compared to controls, organoids treated with TGFB1 exhibited a spheroid morphology (Figure 4B–C) and a notable elevation in fetal and regenerative cell transcripts (Figure 4D–E). TGFB1-induced expression of the fetal/regenerative/revival cell/YAP signaling signatures was i) rapid, occurring within 24 hours, ii) sustained, lasting at least 5 days following the single TGFB1 treatment (Figure 4F–G, Figure S4A), iii) dependent upon epithelial Tgfbr2 expression, as the response did not occur in TGFB1-treated Tgfbr2KO organoids (Figure 4G, Figure S4A–C), and iv) sensitive to TGFBR-inhibitors (Figure S4D–F). To better appreciate the timeline of the organoid response to TGFB1 treatment, we performed scRNA-seq analysis of organoids treated with vehicle or TGFB1 for 6, 15, or 24 hours (Figure 4H). TGFB1-treatment induced increasingly elevated levels of canonical TGFB1-pathway targets such as Id1, Smad7, and Tgif1 over time, and these increases were accompanied by a marked induction of regenerative markers such as Clu and Anxa-family genes (Figure 4I). Approximately half of the cells expressed transcripts consistent with differentiated enterocytes, and the remainder expressed transcripts associated with progenitor cell populations (Figure 4J–K). These analyses point to an induction of the regenerative marker Clu as early as 6 hours post-TGFB1 treatment, and maximal expression at the latest time point collected, 24h. RNA velocity analysis and UMAP visualization indicated that Clu-expressing cells at the later time points appear most closely related to Lgr5-expressing progenitor cells, suggesting a potential origin of these cells. Collectively, these data indicate that TGFB1 potently induces a shift in the morphology and transcriptome of intestinal organoids to acquire properties of regenerating epithelium.
Figure 4. TGFB1 is sufficient to induce fetal/regenerative gene signatures and increase Clu positive cells in organoid culture.
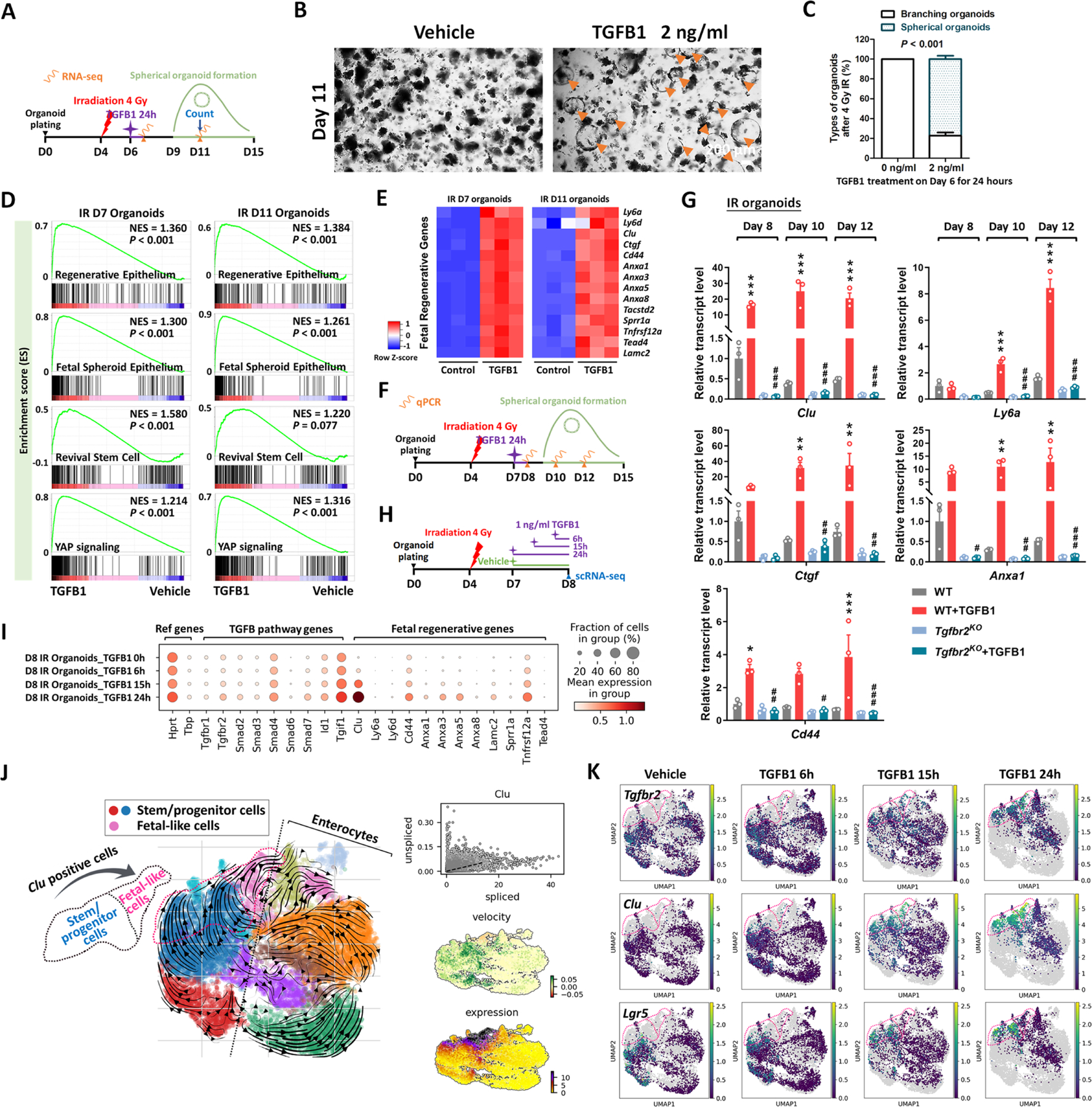
(A-E) Organoids treated with TGFB1 for 24 hours acquire a spheroid morphology (see orange arrows in panel B) and maintain expression of regeneration marker genes for at least 5 days post-TGFB1 treatment. (A) Schematic for the experiments to score morphology, counts, and bulk RNA-seq. Primary intestinal organoids were exposed to 4 Gy of irradiation on Day 4, followed by TGFB1 treatment on Day 6 for 24 hours. Organoids were collected for bulk RNA-seq on Day 7 and Day 11. (B) Representative images of irradiated organoids treated with vehicle or TGFB1. (C) Percentage of branching and spherical organoids upon TGFB1 treatment. Organoids were counted at Day 11, which is 5 days post-TGFB1 treatment (n=3 independent organoid cultures, Student’s t-test at P < 0.001). (D) Bulk RNA-seq of intestinal organoids cultured with TGFB1 for 24 hours show strong correlation with published gene signatures associated with intestinal regeneration post-DSS injury, fetal spheroid, revival stem cells and YAP signaling,8,12,13,17,89 as measured by GSEA (n=3 independent organoid cultures, Kolmogorov-Smirnov test). (E) Heatmaps display that RNA-seq expression levels of fetal/regenerative genes are highly expressed upon TGFB1 treatment compared to the vehicle controls (n=3 independent organoid cultures). (F) Schematic for the qRT-PCR experimental design. Primary intestinal organoids cultured from Tgfbr2KO mice (4 days post-tamoxifen) and their littermate WT controls, were exposed to 4 Gy of irradiation on Day 4, followed by TGFB1 treatment (1 ng/ml) on Day 7 for 24 hours. Organoids were collected on Day 8, Day 10 and Day 12 for qRT-PCR. (G) qRT-PCR indicates that expression of regeneration marker genes increases within 24 hours and is sustained for at least 5 days post-TGFB1 treatment in the WT organoids, but not in the Tgfbr2KO organoids. See more examples in Figure S4A. All the data are presented as mean ± SEM (n=3 independent organoid cultures). Statistical comparisons were performed using one-way ANOVA followed by Tukey’s multiple comparisons test at P < 0.001***, P < 0.01** or P < 0.05* (WT+TGFB1 vs WT); P < 0.001###, P < 0.01## or P < 0.05# (Tgfbr2KO+TGFB1 vs WT+TGFB1). (H-K) scRNA-seq of organoids post-irradiation and upon TGFB1 treatment across time points. (H) Schematic of scRNA-seq experimental design. Primary intestinal organoids were exposed to 4 Gy of irradiation on Day 4, followed by TGFB1 treatment (1 ng/ml) on Day 7. Organoids were collected for scRNA-seq after 6, 15, and 24 hours of TGFB1 treatment. Organoids treated with vehicle were used as control. (I) Dot plots show that TGFB1 activates TGFB pathway and fetal/regenerative genes in a time-dependent manner. (J) RNA velocity analysis identifies that cells in Lgr5-expressing clusters are synthesizing new Clu transcripts. (K) UMAP plots indicate that, across time points, there is a close correlation between Clu and Tgfbr2 expression. Lgr5-expressing clusters begin to overlap with Clu-expressing cells. Number of cells in each condition was Vehicle: n= 2815; TGFB1 6h: n= 4071; TGFB1 15h: n= 2788; TGFB1 24h: n=2177.
The stromal mesenchyme responds to TGFB1 to promote fetal reversion of the epithelium
Pdgfra-expressing cells have demonstrated roles regulating epithelial growth.24–26 We therefore analyzed Pdgfra positive cells from the scRNA-seq dataset from whole intestinal tissues post-IR (Figure S2H), and found that Tgfbr2 transcripts were enriched in Pdgfra+ mesenchymal cell populations (Figure 5A, Figure S5A). To define the role of intestinal mesenchyme in promoting TGFB1-dependent regeneration, we cultured Pdgfra-Lo mesenchymal fibroblasts from Pdgfra-H2B-EGFP transgenic mice27 according to published isolation strategies.28 Cultured mesenchyme changed morphology in response to TGFB1-treatment, aggregating into clusters of cells in a dose-dependent response to TGFB1 (Figure 5B–C), confirming their ability to respond to the ligand. It seems mesenchymal cells are more contractile after TGFB1 treatment, which may facilitate recruiting mesenchymal cells to damaged tissue. To identify the role of TGFB1 signaling in the interactions between the epithelium and mesenchyme, we co-cultured these cell types and manipulated TGFB-signaling (Figure 5D). Organoids were cultured for 3 days in 3D matrix bubbles and then floated above mesenchyme monolayers for 2 days before collecting the epithelium for qRT-PCR. Pre-treating the mesenchyme cultures with TGFB1 influenced the subsequent co-cultures, with a dose-dependent induction of transcripts associated with regeneration in the epithelium (Figure 5E). Conversely, pre-treatment with inhibitors of TGFB receptors suppressed the same markers of regeneration in the epithelium (Figure 5E). In response to TGFB1 pre-treatment, mesenchymal cells exhibited higher levels of transcripts expected to promote regeneration/wound healing such as Ptgs2, Wnt5a, and Lif;29–31 lower levels of transcripts that promote homeostatic epithelial growth, such as Grem1 and Rspo324,32 were observed in response to TGFB1 treatment (Figure 5F), suggesting that TGFB1 reshapes the signaling environment to favor regenerative growth.
Figure 5. TGFB1-treated mesenchyme promotes fetal-like conversion of intestinal organoids.
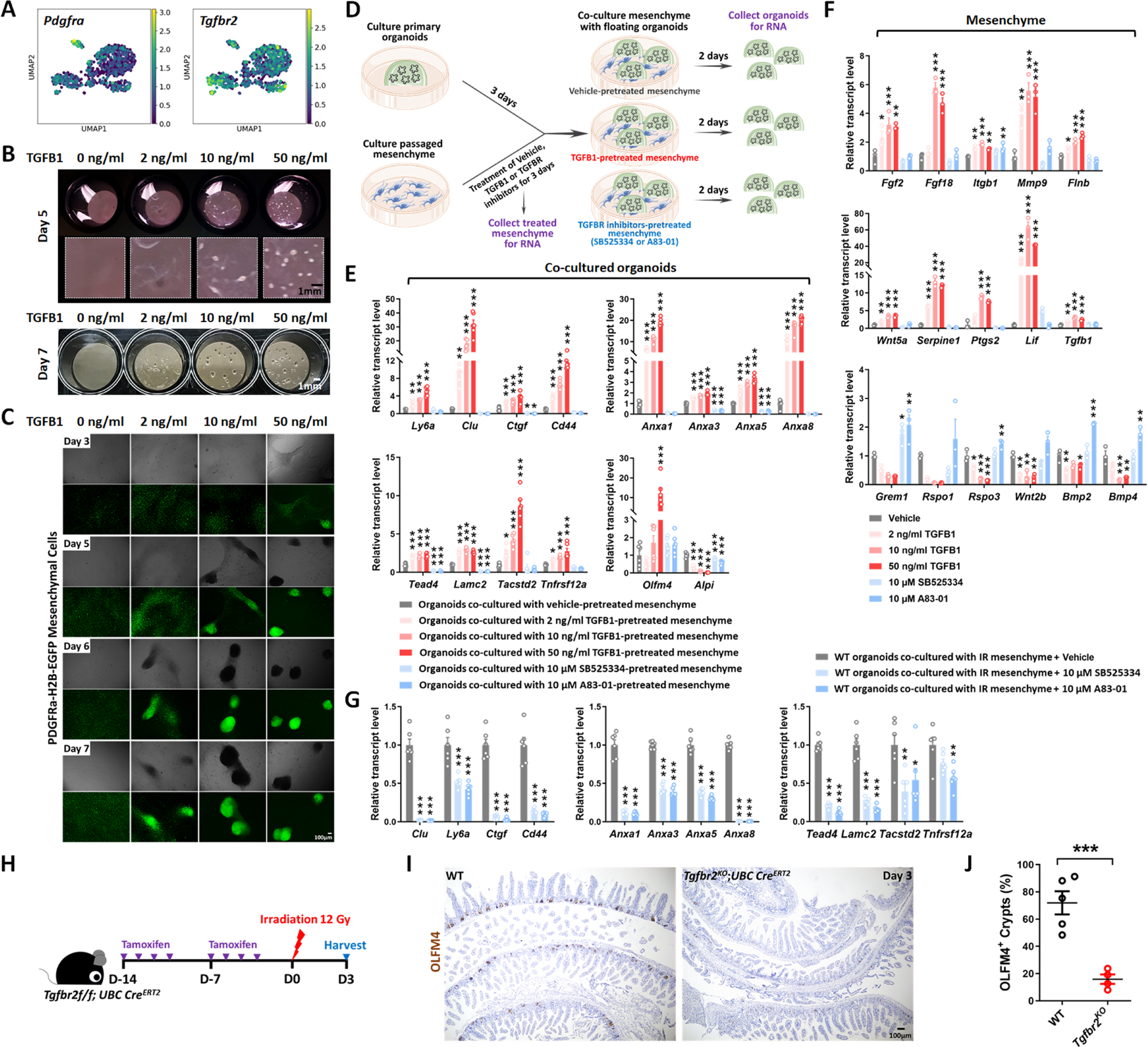
(A) UMAP indicates Pdgfra-positive mesenchymal cells express Tgfbr2. Pdgfra positive mesenchymal cells were subset from the scRNA-seq data set featured in Figure S2H (GSE165318). (B-C) TGFB1-induces aggregation of Pdgfra-positive mesenchymal cells in a dose- and time-dependent manner (n=3 independent experiments, passaged mesenchyme). (D-F) TGFB1-treated mesenchyme induces fetal-like gene signatures in intestinal organoids upon co-culture. (D) Schematic of experimental design of co-culture. Passaged intestinal mesenchyme cells were pre-treated with vehicle, TGFB1 or TGFBR inhibitors for 3 days, and then co-cultured as overlaid matrigel bubbles containing primary organoids at day 3 post isolation. After 2 days of co-culture, organoids were collected in their matrix bubbles for qRT-PCR (n=6 independent organoid cultures with 2 different cell densities of mesenchyme). TGFB1 was removed during co-culture and only used for pre-treatment. TGFBR inhibitors were either kept (E) or removed (Figure S5E) in co-culture. (F) Mesenchyme cells pre-treated with vehicle, TGFB1 or TGFBR inhibitors for 3 days were also collected for qRT-PCR (n=3 independent mesenchyme cultures). (G) Presence of TGFBR inhibitors suppresses fetal-like conversion of intestinal organoids co-cultured with mesenchyme isolated from mice 3 days post-irradiation (n=6 independent organoid cultures with 2 different cell lines of mesenchyme). All the qRT-PCR data are presented as mean ± SEM. Transcript levels relative to vehicle control, and statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post at P < 0.001***, P < 0.01** or P < 0.05*. (H-J) Tgfbr2 knockout via UBC-CreERT2 restricts regeneration after irradiation. 5-week old mice were treated with tamoxifen to inactivate Tgfbr2 in the whole body. Intestine was collected 3 days post-IR and scored for regenerative foci using OLFM4 immunostaining (n=4–5 biological replicates, duodenum, Student’s t-test at P < 0.001***). (H) Schematic of experimental design. (I) Representative images. (J) Quantification.
Similar changes in transcriptional profiles were observed in vivo within the Pdgfra-expressing populations in response to IR (Figure S5A), and mesenchyme isolated from irradiated intestines expressed higher levels of transcripts for pro-regenerative growth ligands and reduced levels of ligands associated with homeostatic growth (Figure S5B). A pro-regenerative signature was also robustly reduced when mesenchymal cultures derived from irradiated mice were pre-treated with TGFB-inhibitors before epithelial overlay (Figure 5G, Figure S5C–E). Finally, to define the role of TGFB in both mesenchyme and epithelial cells in response to IR in vivo, we utilized Tgfbr2f/f;UBC-CreERT2 mice to inactivate the receptor across all cell populations. We found loss of Tgfbr2 in mesenchyme and epithelial cells led to a dramatic reduction of regenerative crypts following IR (Figure 5H–J). The impaired regeneration is more pronounced in the UBC-CreERT2 model of Tgfbr2 loss in both mesenchyme and epithelial cells than in epithelial-specific Tgfbr2 knockout (Figure 3I). These experiments implicate stromal cells in TGFB-mediated intestinal regeneration.
TGFB1 induces a YAP-SOX9 regenerative circuit
To better appreciate the epithelial response to TGFB1, we conducted ATAC-seq and defined 1,647 genomic regions that gained chromatin accessibility in organoids treated with TGFB1 (Figure 6A, Figure S6A–C, Table S1, Diffbind FDR < 0.01). Examples of regeneration marker genes with increased chromatin accessibility in response to TGFB1 treatment include Anxa1, Cd44 and Wnt5a (Figure 6B). Regions of the genome with increased accessibility were enriched in transcription factor motifs known to bind SOX, TEAD, and SMAD families of transcription factors (Figure 6C, HOMER). By contrast, the 3,900 genomics regions more accessible in the vehicle-treated condition are enriched in transcription factor binding motifs that are associated with function of the homeostatic intestinal epithelium (Figure 6C). This chromatin accessibility analysis suggests that Hippo-TEAD, SOX9, and TGFB-SMAD signaling activity or expression are elevated in response to TGFB1 treatment. Notably, SOX9 protein levels are induced after IR of the intestine and SOX9-expressing cells coincide with cells expressing the regenerative factor YAP (Figure 6D, Figure S6D–E). In scRNA-seq data reanalyzed from Figure 3B to focus on epithelial cells, Sox9 is elevated in response to IR, and corresponds to elevation of genes involved in YAP signaling and organization of the extracellular matrix (Figure 6E–G). Importantly, Sox9 is co-expressed in cells producing Tgfbr2, suggesting a potentially direct connection between TGFB1 and Sox9 regulation (Figure 6H). In mice treated with clodronate liposomes to deplete monocyte/macrophages, SOX9 protein and transcript levels were notably reduced in the crypt domain at 3 days post-IR compared to controls (Figure 6I, Figure S6F).
Figure 6. YAP-SOX9 circuit responds to TGFB1-induced open chromatin and epithelial regeneration in the intestine.
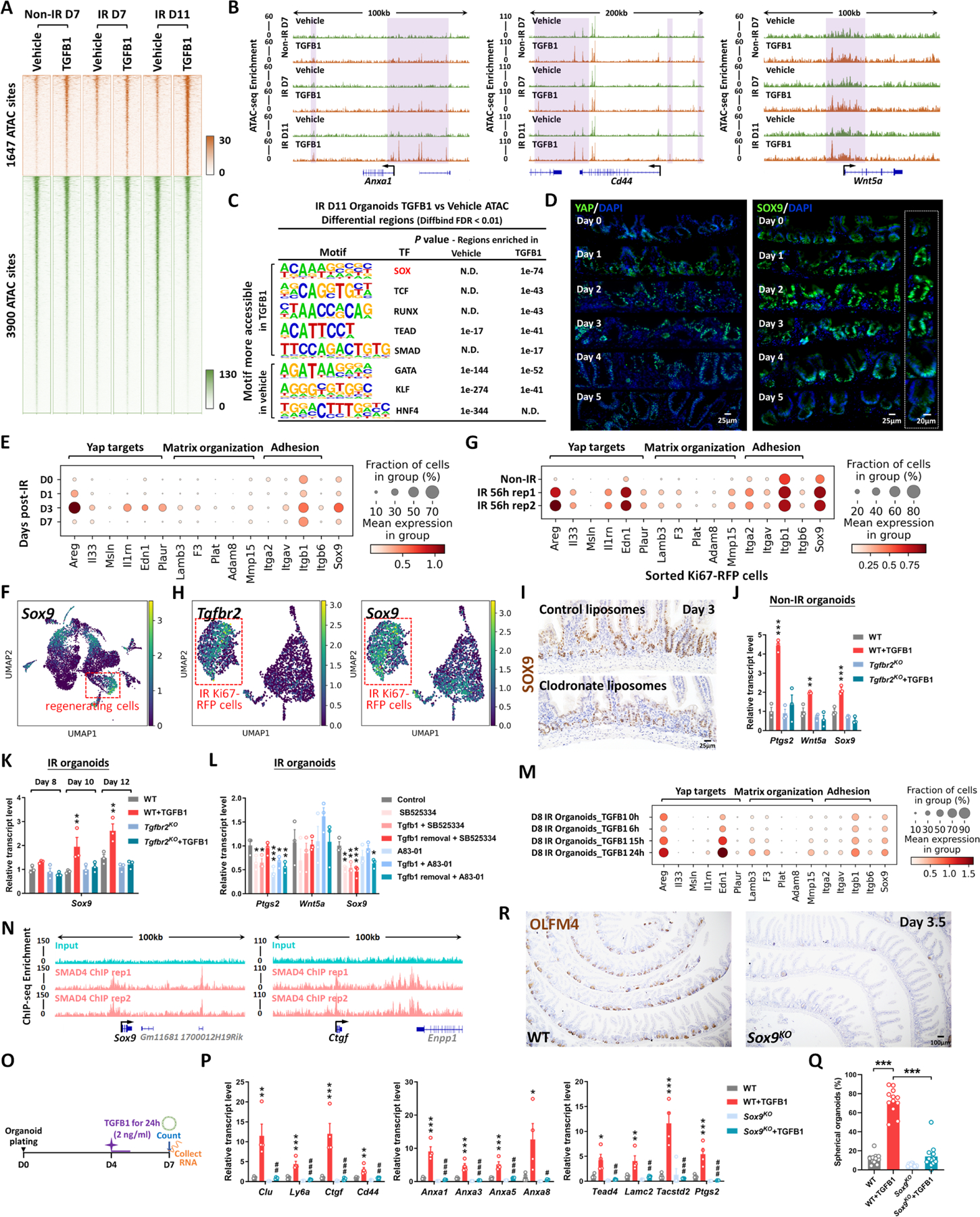
(A) TGFB1-induced accessible chromatin identified using ATAC-seq Day 11 TGFB1-treated vs. vehicle-treated organoids (Diffbind FDR < 0.01, n=3 independent cultures). The experimental design is the same as for bulk RNA-seq and shown in Figure 4A. (B) Examples of genes harboring TGFB1-induced open chromatin visualized using IGV. (C) HOMER de novo DNA-motif enrichment analysis of ATAC-seq regions (Diffbind FDR < 0.01) shows that SOX, TCF, RUNX, TEAD and SMAD binding sequences are more prevalent in accessible regions of TGFB1-treated organoids, whereas GATA, KLF and HNF4 binding sequences are more prevalent in accessible regions of vehicle-treated organoids. N.D.: Not Detectable. (D) Immunofluorescence staining of YAP and SOX9 across a time course post-irradiation (representative of 3 biological replicates). Dot plots (E) and UMAP (F) reveal that YAP-related gene signatures and Sox9 are elevated during regeneration post-irradiation, as evidenced by scRNA-seq (subset of intestinal epithelial cells, GSE165318). (G) scRNA-seq dot plots reveal that YAP related gene signatures and Sox9 are also elevated in sorted Ki67-RFP positive cells after 56 hours of irradiation. (H) scRNA-seq UMAP reveals that Tgfbr2-positive cells express Sox9 in sorted Ki67-RFP positive cells after 56 hours of irradiation. (I) Depletion of monocytes/macrophages (main cell sources of TGFB1 secretion) results in a downregulation of SOX9, as evidenced by immunostaining (representative of 3 biological replicates). (J-L) qRT-PCR indicates that TGFB1 induces expression of YAP related genes (Ptgs2 and Wnt5a) and Sox9 in WT organoids, and these effects are blocked in Tgfbr2KO or TGFBR inhibitor-treated organoids. All the qRT-PCR data are presented as mean ± SEM (n=3 independent organoid cultures). Transcript levels are relative to WT, and statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post at P < 0.001***, P < 0.01** or P < 0.05*. (M) Dot plots show that TGFB1 activates YAP related gene signatures and Sox9 in a time-dependent manner (see experimental design in Figure 4H). (N) ChIP-seq shows that SMAD4 binds to gene loci of Sox9 and Ctgf in mouse intestinal epithelium (GSE112946 33). (O-Q) Loss of Sox9 blocked the induction of fetal/regenerative phenotypes induced by TGFB1 in organoids. (O) Schematic of experimental design. (P) qRT-PCR of fetal/regenerative markers. (Q) Percentage of spherical organoids upon TGFB1 treatment. Organoids were counted and collected for RNA extraction at Day 7, which is 3 days post-TGFB1 treatment. All the data are presented as mean ± SEM (n=4 independent organoid cultures). Statistical comparisons were performed using one-way ANOVA followed by Tukey’s multiple comparisons test at P < 0.001***, P < 0.01** or P < 0.05* (WT+TGFB1 vs WT); P < 0.001###, P < 0.01## or P < 0.05# (Sox9KO+TGFB1 vs WT+TGFB1). (R) Loss of Sox9 restricts regeneration after irradiation, as evidenced by OLFM4 immunostaining. Mice were treated with tamoxifen to inactivate Sox9 in the intestinal epithelium 7 days before 12 Gy of irradiation. Intestine was collected 3.5 days post-IR (representative of 3 biological replicates).
Treatment of organoids with TGFB1 induced Sox9 expression, and this induction was dependent upon the expression of Tgfbr2, could be blocked by TGFBR2-inhibitors, and increased with time after exposure to TGFB1 (Figure 6J–M). Organoid cells expressing Sox9 in response to TGFB1 treatment (Figure S6G) were co-localized to cells expressing Clu (Figure 4K). ChIP-seq data (GSE11294633) indicate that SMAD4 can directly bind to the Sox9 and Ctgf loci, suggesting a direct pathway between TGFB1 and the regenerative response (Figure 6N). Furthermore, organoid cultures were refractory to TGFB1-induced spheroid formation in the absence of SOX9, and reduced levels of regenerative gene expression were also observed in TGFB-treated Sox9KO organoids compared to TGFB-treated control organoids (Figure 6O–Q). SOX9 is critical for the regenerative response, as epithelial-specific inactivation of Sox9 led to a severely reduced number of OLFM4+ regenerative crypts at 3 days post-IR (Figure 6R), similar to what has been reported by others.9 These data are consistent with a model in which TGFB1 exposure leads to direct transcriptional activation of Sox9 and the YAP pathway, with subsequent activation of the regenerative response.
Pre-treatment of organoid cultures with TGFB1 enhances engraftment efficiency into damaged colon
Epithelial transplants hold tremendous promise for cellular therapy to correct genetic disorders affecting the intestine or to accelerate healing of damaged mucosa. Given the importance of TGFB1 in promoting regenerative characteristics in the intestinal epithelium, we suspected that TGFB1 pre-treatment of epithelial organoid cultures could improve transplant efficiency. Since pre-irradiating organoids would not be conducive to use in the clinic, we revisited the strategy of treating non-irradiated organoids with TGFB1 and monitored the response using RNA-seq and qRT-PCR. We found TGFB1-induced fetal/regenerative gene expression is independent of IR-induced damage (Figure 7A–D, Figure S7A), and conserved in human organoids (Figure S7B–E). Loss of Tgfbr2 suppresses fetal/regenerative gene expression in these organoids as well (Figure S7F). Given a robust induction of regenerative marker gene expression in response to TGFB1 treatment in non-IR organoids (Figure 7A–D), we next assayed the effects of TGFB1 pre-treatment of organoids in a well-developed engraftment assay in which the host epithelium is damaged through exposing animals to Dextran Sulfate Sodium (DSS34,). We noted that there were variations of intestinal damage among DSS-treated mice, but the cohort receiving TGFB1 pre-treated organoids showed improved histological and pathological features compared to control groups (Figure S7G–K). To minimize the variations inherent in the DSS model, we next conducted a competitive transplantation assay in which organoids treated with vehicle or TGFB1 would be co-transplanted via enema. Organoids were collected from transgenic mice expressing fluorescent reporters that could be traced following implantation (Figure 7E–F). Excitingly, TGFB1-treated organoids were significantly more likely to colonize the host colon than vehicle treated controls, as visualized by fluorescence (Figure 7G, Figure S7L). Engraftment, quantified either by the size of the individual graft or the average size of the organoid graft per mouse was significantly enhanced in the TGFB1-treated condition (Figure 7H–I). These data reflect the promise of TGFB1 pre-treatment in supporting the use of epithelial cultures for cellular therapy in the GI tract.
Figure 7. Transplantation of TGFB1-treated organoids enhances engraftment into DSS-treated mice.
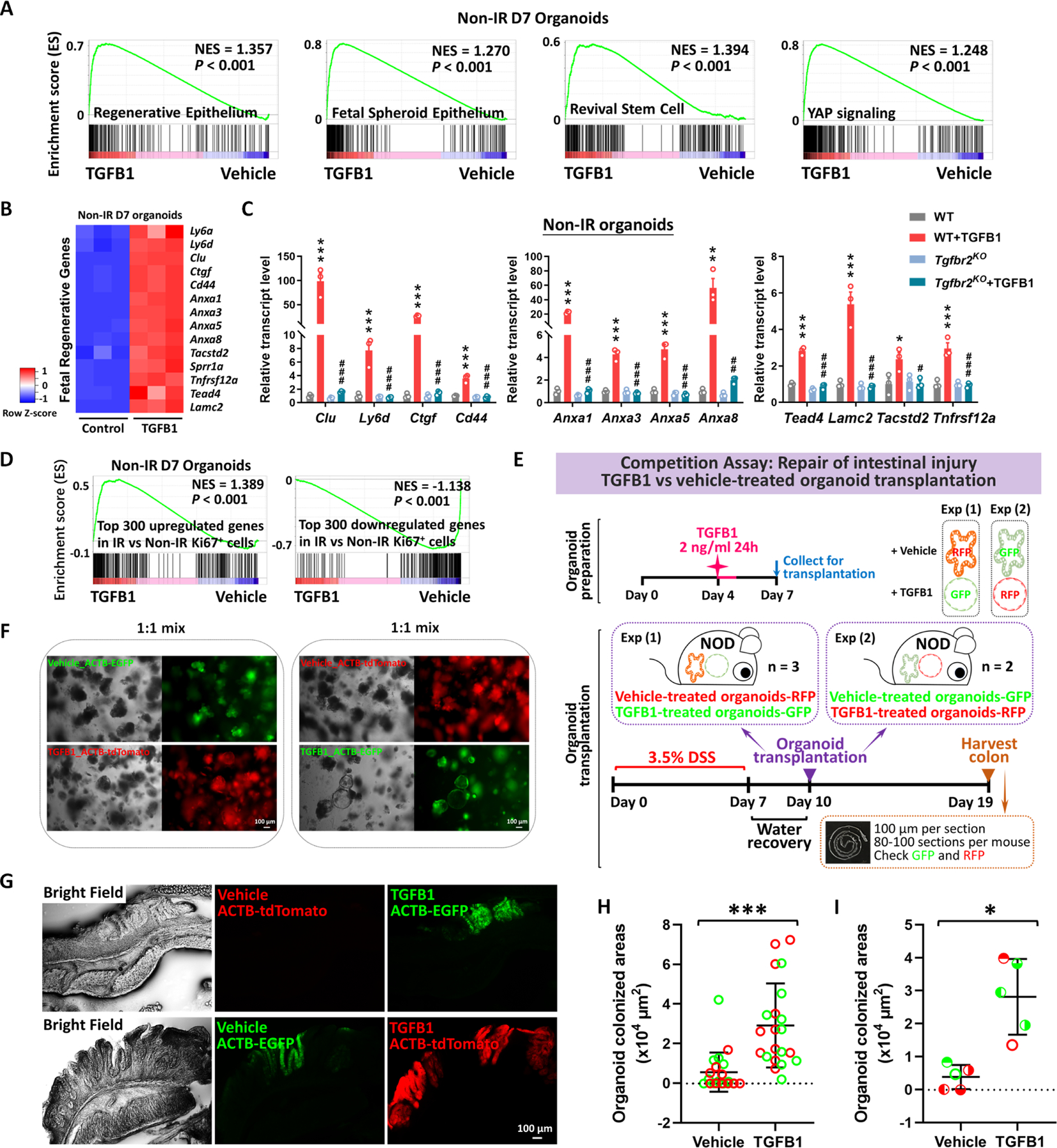
(A) Gene signatures of regenerative epithelium, fetal spheroids, revival stem cells and YAP signaling8,12,13,17,89 are each elevated post-TGFB1 treatment, as assayed by GSEA in non-irradiated conditions (n=3 independent organoid cultures, Kolmogorov-Smirnov test, P < 0.001). (B) Heatmaps display that RNA-seq expression levels of fetal/regenerative genes are highly expressed upon TGFB1 treatment compared to the vehicle controls (n=3 independent organoid cultures). Schematic of experimental design for bulk RNA-seq for panels A-B is depicted in Figure S7A. (C) For non-IR organoids, to deplete Tgfbr2, the primary organoids were treated with 1 μM tamoxifen for 12 hours on Day 3, followed with TGFB1 (2 ng/ml) treatment on Day 6. Organoids were collected 24 hours after TGFB1 treatment. All the data are presented as mean ± SEM (n=3 independent organoid cultures). Statistical comparisons were performed using one-way ANOVA followed by Tukey’s multiple comparisons test at P < 0.001***, P < 0.01** or P < 0.05* (WT+TGFB1 vs WT); P < 0.001###, P < 0.01## or P < 0.05# (Tgfbr2KO+TGFB1 vs WT+TGFB1). (D) GSEA reveals that genes upregulated or downregulated upon TGFB1 treatment strongly correlate with transcriptional changes in Ki67-RFP cells from the intestine of mice with irradiation vs. non-irradiation, respectively, as described in Figure 1 (Kolmogorov-Smirnov test, P < 0.001, n=3 independent organoid cultures). (E) Experimental design for organoid transplantation assay, to determine the ability of TGFB1 to prime organoids in culture prior to transplantation for engrafting into damaged colonic tissue. Transgenic organoid lines were used to later help visualize transplants. Organoids were treated with either vehicle or with TGFB1 to induce regenerative properties. To induce epithelial damage in the mouse intestine, 3.5% DSS was prepared in drinking water and fed to NOD mice for 7 days. After a period of water recovery, the treated and control organoids were mixed 1:1 and used for enema-based transplant. Either vehicle-treated organoids with RFP were mixed with TGFB1-treated organoids with GFP; or vehicle-treated organoids with GFP were mixed with TGFB1-treated organoids with RFP. Organoid mixtures were transferred into DSS-treated mice on Day 10. Colon tissues were collected on Day 19, and cryosections were prepared for checking GFP or RFP under fluorescence microscope. (F) Representative images of organoids used for transplantation. (G-I) Representative images and quantification of transplant efficiency upon TGFB1 pre-treatment. (G) Fluorescent micrographs demonstrating transgenic organoid grafts into mice. (H) The size of grafts observed. (I) The average area of organoid grafts per mouse. Color indicates whether transplanted organoids derived from red or green fluorescent lines. Symbol type represents a single mouse used in the competition assay (n=21 grafts from 5 mice, Student’s t-test at P < 0.001*** or P < 0.05*).
DISCUSSION
Radiotherapy, chemotherapy, inflammatory bowel disease, and other maladies of the gut induce extensive damage of the intestinal epithelial lining. These conditions could be alleviated by transplant of healthy epithelial cells. Genetic disorders affecting intestinal epithelial cell functions, such as microvillus inclusion disorders or congenital diarrhea or nutrient transporter deficiencies could also be corrected by replacement of defective epithelium with genetically corrected epithelial transplants. In this study, we discover a critical role for TGFB1 in promoting intestinal regeneration. Dysregulation of tissue repair can trigger excessive extracellular matrix deposition and lead to fibrosis, which replaces normal parenchymal tissue. Myofibroblasts secrete large amounts of extracellular matrix and are regarded as the culprits of fibrosis after injury.35 TGFB signaling drives fibrosis in many organs including the intestine;36–42 however, our strategy of ex-vivo treatment could avoid tissue fibrosis. We propose an application of these findings to enhance intestinal engraftment by using TGFB1 ligands to induce a fetal-like regenerative-state in intestinal organoids. We demonstrate that TGFB1-treated organoids support more robust tissue engraftment in a mouse model of ulcerative colitis.
YAP, a downstream transcriptional effector of Hippo, is important for regeneration of intestinal epithelium after IR.13 We highlighted a number of YAP target genes that are induced by TGFB1, including Areg.43 As a transcriptional target of YAP, amphiregulin (AREG) contributes to YAP-mediated cell proliferation and migration.44 Loss of Areg impaired intestinal regeneration after radiation injury,45 while administration of AREG ameliorated colitis upon DSS insult.46 Edn1 is another well-established YAP/TAZ target gene.47 scRNA-seq identified a set of stem-cell specific markers for the least differentiated intestinal stem cells, including Lgr5 and Edn1,48 indicating the potential role of Edn1 in intestinal stemness during YAP-dependent recovery. Furthermore, YAP is known to promote wound healing through its functions in cell adhesion, cell mechanics, and regulation of the actin cytoskeleton.49–51 As a major adhesion receptor, ITGB1 plays an important role in cell migration and adhesion,52 which may contribute to the wound healing process of intestinal regeneration. Areg, Edn1, and Itgb1 are upregulated in the intestinal epithelial cells during regeneration after IR. These transcripts are also upregulated upon TGFB treatment in the intestinal organoids. Altogether, these data suggest TGFB induces a gene program overlapping with YAP targets involved in the wound healing process.
Mechanistically, we demonstrate pleiotropic functions for TGFB1 during regeneration. These TGFB1 functions include the induction of Clu+ cells, the promotion of fetal/regenerative gene signatures, the stimulation of mesenchymal cells to secrete pro-regenerative ligands, and the subsequent induction of a YAP-SOX9 circuit in the intestinal epithelium. Clu+ cells are very rare in homoeostatic intestine, but are activated in damaged intestine and can rapidly expand to reconstitute Lgr5+ ISCs and promote regeneration of the intestinal epithelium.8 These Clu+ cells undergo a YAP-dependent transient expansion upon intestinal injury,8 and will go on to restore homeostatic stem cells. While our study was focused on the earliest stages of the regenerative process, we presume that homeostasis will be restored following TGFB1-induction, as we see TGFB1-treated organoids will transition back to branched structures associated with the homeostatic state. A fetal-like reversion of the regenerative epithelium and re-initiation of a fetal-like developmental transcriptional program were observed during intestinal regeneration after injury caused by DSS-induced colitis or helminth infection.11,12 Upon injury, YAP transiently reprograms Lgr5+ ISCs by inducing a regenerative program while suppressing the Wnt-dependent homeostatic program.13 We bring insight into the function of TGFB1 in the regeneration process, and suggest a clear advantage for TGFB1-treated organoids to engraft in DSS-damaged intestines compared to canonically-treated intestines.
Recent studies suggest mesenchymal cells as important sources of niche signals.24–26,53–55 We reveal that TGFB1 pushes mesenchymal cells from homeostatic RSPO/Grem1 signals to regeneration/wound healing signals. Elevated levels of Ptgs2 and Wnt5a were observed in TGFB1-treated mesenchymal cells. Ptgs2-expressing fibroblasts process arachidonic acid into prostaglandin E2 (PGE2), which enhances YAP activity through the prostaglandin EP4 receptor and drives expansion of Sca-1+ reserve-like stem cells.29 PGE2 triggers cell fate plasticity by promoting a switch from differentiated enterocytes to wound-associated epithelial (WAE) cells.56 Additionally, Wnt5a, a noncanonical Wnt ligand, potentiates TGFB signaling and is required for crypt regeneration through the WAE cells.30 Interestingly, Trem2-expressing macrophages associate with the WAE layer,57 and WNT-producing macrophages enhance epithelial regeneration following tissue damage.58 While previous studies have demonstrated TGFB1 production by circulating macrophages following phagocytosis of dying cells,59 here we demonstrate that macrophages are the primary TGFB1 source in intestinal regeneration, and the TGFB1 is necessary for full regeneration and sufficient to induce the epithelial regenerative state. Future studies will be required to genetically dissect the role of TGFB signaling pathway in stromal cell subpopulations of the intestine by using different Cre drivers. TGFB1 is not the only signal that can trigger the regeneration-promoting molecules from the stroma,32,60 and future studies will be required to study the crosstalk of TGFB and other signals.
Limitations of the Study
Further studies using lineage tracing methods should be done to corroborate the finding inferred using RNA velocity, specifically to define the source of the Clu+ cells induced by TGFB1. The DSS model of ulcerative colitis is used to assay for organoid engraftment, however the variable degree of tissue ulceration in response to DSS treatment hinders future evaluation and optimization of the organoid engraftment procedure. Development of more consistent methodologies to de-epithelialize the host colon will be important for future development of cellular therapies. SOX9 and YAP/TAZ function in numerous cellular contexts, but it remains unclear how their transcriptional targets may differ in regenerating cells compared to homeostatic intestinal epithelial cells and could be important to further dissect the mechanisms through which regenerating cells acquire a distinct transcriptome.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael Verzi (verzi@biology.rutgers.edu).
Materials availability
This study did not generate any unique reagents and the mouse lines generated in this study are available upon execution of a suitable Materials Transfer Agreement.
Data and code availability
Single-cell RNA-seq, RNA-seq and ATAC-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Ki67 antibody | Abcam | Cat#: ab16667, RRID:AB_302459 |
| Anti-OLFM4 antibody | Cell Signaling | Cat#: 39141, RRID:AB_2650511 |
| Anti-CD44 antibody | BD Biosciences | Cat#: 558739, RRID:AB_397098 |
| Anti-BrdU antibody | Bio-Rad | Cat#: MCA2060, RRID:AB_323427 |
| Anti-F4/80 antibody | Cell Signaling | Cat#: 70076, RRID:AB_2799771 |
| Anti-SOX9 antibody | Cell Signaling | Cat#: 82630, RRID:AB_2665492 |
| Anti-YAP antibody | Cell Signaling | Cat#: 4912, RRID:AB_2218911 |
| Anti-p-SMAD3 antibody | Cell Signaling | Cat#: 9520, RRID:AB_2193207 |
| Anti-p-SMAD2/3 antibody | Cell Signaling | Cat#: 8828, RRID:AB_2631089 |
| Anti-β-actin antibody | Abcam | Cat#: ab8227, RRID:AB_2305186 |
| Anti-Ecadherin antibody | BD Transduction Labs | Cat#: 610181 |
| Anti-Rabbit-750 | Sigma Aldrich | Cat#: SAB4600373 |
| Anti-Mouse-594 | Jackson Immuno Labs | Cat#: 715-585-150 |
| TGF beta-1,2,3 Monoclonal Antibody (1D11) | Invitrogen | Cat#: MA5-23795, AB_2609812 |
| Mouse IgG1 Isotype Control | R&D | Cat#: MAB002, RRID:AB_357344 |
| Biological samples | ||
| Human duodenal organoid lines | Michigan Translational Tissue Modeling Laboratory | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Biotium | 40043 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| Human TGF-B1 | Peprotech | 100-21 |
| OCT compound | Tissue-Tek | 4583 |
| Hematoxylin | VWR | 95057-858 |
| Eosin | Sigma Aldrich | HT110180 |
| SB525334 | Selleckchem | S1476 |
| A83-01 | Tocris | 2939 |
| Trizol | Invitrogen | 15596018 |
| SYBR Green PCR Master Mix | Applied Biosystems | 4309155 |
| Dextran Sulfate sodium salt (DSS) | Affymetrix | J14489 |
| Cultrex reduced growth factor basement membrane matrix, Type R1 | R&D | 3433-010-R1 |
| Corning Matrigel | Corning | 356231 |
| Y-27632 2HCl | Selleckchem | S1049 5mg |
| CHIR99021 | Axon Medchem | 1386 |
| Murine Noggin | Peprotech | 250-38 |
| Advanced DMEM/F-12 | Gibco | 12634-010 |
| GlutaMax | Gibco | 35050-061 |
| HEPES | Gibco | 15630-080 |
| Penicillin-Streptomycin | Invitrogen | 15140-122 |
| N-2 supplement | Gibco | 17502048 |
| B-27 supplement | Gibco | 12587-010 |
| Recombinant Human EGF Protein | R&D | 236-EG |
| Recombinant Murine EGF | Peprotech | 315-09 |
| Primocin | InvivoGen | ant-pm-1 |
| TrypLE | Thermo Fisher | 12604-013 |
| Dispase | Stem Cell Technologies | 07913 |
| DNAse I | Sigma Aldrich | D4513 |
| HBSS, no Calcium, no Magnesium | Thermo Fisher | 14170120 |
| Collagenase IV | Worthington Biochemical Corporation | LS004188 |
| Collagenase D | Roche | 11088858001 |
| Dispase II | Gibco | 17105041 |
| Fetal Bovine Serum (FBS) | Gibco | 26140-095 |
| TSA Buffer | ACD | 322809 |
| Clondronate Liposomes | FormuMax Scientific | F70101C-NC-10 |
| DAB | Amresco | 0430 |
| Critical commercial assays | ||
| KAPA Mouse Genotyping Kits | KAPA Biosystems | KK7352 |
| Illumina Tagment DNA Enzyme and Buffer Large Kit | Illumina | 20034198 |
| QIAGEN RNeasy Micro Kit | Qiagen | 74004 |
| SuperScript III First-Strand Synthesis SuperMix | Invitrogen | 18080-400 |
| Pierce BCA Protein Assay kit | Thermo Fisher | 23225 |
| Mouse TGF beta 1 ELISA kit | Abcam | ab119557 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 Assay | ACD | 323110 |
| Anti-F4/80 MicroBeads and MS column-based cell separation kit | Miltenyi Biotec | 130-110-443 |
| Mouse growth factor array C3 kit | RayBiotech | AAM-GF-3-4 |
| PIPseq T2 3’ Single Cell Capture and Lysis Kit v2.1 | Fluent Biosciences | N/A |
| Vectastain ABC HRP Kit | Vector Labs | PK-6101 |
| MinElute PCR purification kit | Qiagen | 28004 |
| Deposited data | ||
| RNA-seq, scRNA-seq and ATAC-seq data | This study | GEO: GSE222505 |
| RNA-seq of crypts upon IR | Qu et al., 2021 | GEO: GSE165157 |
| scRNA-seq of normal crypts and irradiated crypts | Ayyaz et al., 2019 | GEO: GSE117783 |
| scRNA-seq of duodenum/jejunum boundary samples upon IR | N/A | GEO: GSE165318 |
| scRNA-seq of sorted Msi1-GFP positive cells (irradiation-resistant) and their progeny cells upon IR | Sheng et al., 2020 | GEO: GSE145866 |
| SMAD4 ChIP in mouse intestinal epithelium | Chen et al., 2019 | GEO: GSE112946 |
| Experimental models: Organisms/strains | ||
| Mouse: Villin-Cre-ERT2 | el Marjou et al., 2004 | JAX: 020282 |
| Mouse: UBC-Cre-ERT2 | Ruzankina et al., 2007 | JAX: 007001 |
| Mouse: Villin-Cre | Madison et al., 2002 | JAX: 004586 |
| Mouse: Tgfbr2f/f | Leveen et al., 2002 | JAX: 012603 |
| Mouse: Smad4f/f | Yang et al., 2002 | JAX: 017462 |
| Mouse: Sox9f/f | Akiyama et al., 2002 | JAX: 013106 |
| Mouse: Mki67tm1.1Cle/J (Ki-67 RFP) | Basak et al., 2014 | JAX: 029802 |
| Mouse: PDGFRa-H2B-EGFP | Hamilton et al., 2003 | JAX: 007669 |
| Mouse: ACTB-EGFP | Okabe et al., 1997 | JAX: 006567 |
| Mouse: ROSA26mT/mG (ACTB-tdTomato-EGFP) | Muzumdar et al., 2007 | JAX: 007676 |
| Mouse: NOD SCID | Blunt et al., 1995 | JAX: 001303 |
| Mouse: C57BL/6 | The Jackson Laboratory | JAX: 000664 |
| Software and algorithms | ||
| Scanpy | Wolf et al., 2018 | https://scanpy.readthedocs.io |
| Velocyto | La Manno et al., 2018 | http://velocyto.org/ |
| GSEA | Subramanian et al., 2005 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Heatmapper | Babicki et al., 2016 | http://www.heatmapper.ca |
| BEDTools | Quinlan, 2014 | https://bedtools.readthedocs.io/en/latest/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Deeptools | Ramirez et al., 2016 | https://github.com/deeptools/deepTools |
| MACS | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| MACS2 | Zhang et al., 2008 | https://github.com/topics/macs2 |
| CutAdapt | Martin, 2011 | https://cutadapt.readthedocs.org |
| Picard | Broad Institute, 2019 | http://broadinstitute.github.io/picard |
| Haystack | Pinello et al., 2018 | https://github.com/pinellolab/haystack_bio |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/motif |
| IGV | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv |
| DiffBind | Stark and Brown, 2011 | http://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Cufflinks | Trapnell et al., 2012 | http://cufflinks.cbcb.umd.edu |
| Kaluza | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| ImageJ | NIH | https://imagej.nih.gov/ |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
The Villin-CreERT2 transgene,61 UBC-CreERT2 transgene,62 embryonic-onset Villin-Cre,63 Tgfbr2f/f,64 Smad4f/f,65 and Sox9f/f66 alleles were integrated to generate the conditional compound-mutants and controls. For Villin-CreERT2;Smad4f/f, mice (8–12 weeks old) were treated with tamoxifen (Sigma T5648) at 50 mg/kg/day for 4 consecutive days by intraperitoneal injection. For Villin-CreERT2;Tgfbr2f/f and Villin-CreERT2;Sox9f/f, mice (8–12 weeks old) were treated with tamoxifen at 100 mg/kg/day for 4 consecutive days. The knockout efficiency varied in Sox9KO, and only the mice with more than 75% of Sox9 depletion were used for downstream analysis. For UBC-CreERT2;Tgfbr2f/f, 5-week-old mice were treated with tamoxifen at 100 mg/kg/day for 4 consecutive days at week 5. The same treatment was repeated at week 6. Embryonic-onset Villin-Cre;Sox9f/f mice were used for the organoid culture experiment when evaluating the requirement of Sox9 upon TGFB1 treatment. C57BL/6, Mki67tm1.1Cle/J (also known as Ki67-RFP),19 PDGFRa-H2B-EGFP,27 ACTB-EGFP,67 ROSA26mT/mG (ACTB-tdTomato,-EGFP),68 and NOD SCID69 mice were also used in this study. KAPA Mouse Genotyping Kits (Kapa Biosystems, KK7352) were used to identify genotypes of mice.
For IR treatment, mice were subjected to 12 Gy whole-body IR with a 137 Cs γ-source irradiator at a dose rate of 60 or 85 cGy/minute. Experiments were analyzed within groups exposed to the same irradiator treatment. For the BrdU pulse-chase experiment, mice were injected with 1 mg BrdU by intraperitoneal injection. To deplete monocytes/macrophages, C57BL/6 mice were treated with clodronate-containing liposomes (FormuMax Scientific, SKU: F70101C-NC-10) by intraperitoneal injection (2 treatments of 200 µl, 72 hours pre- and day of IR). Treatment with control liposomes (same treatment course and dose) was performed as a control. To neutralize TGFB, C57BL/6 mice were treated with TGFB antibody (1D11, MA5–23795, Invitrogen) by intraperitoneal injection (250 µg per dose, two doses, on the day right after IR and 1 day after IR). Mouse IgG1 Isotype (MAB002, R&D) or vehicle were used as control. All mouse protocols and experiments were approved by the Rutgers Institutional Animal Care and Use Committee.
METHOD DETAILS
Histology and immunostaining
Freshly harvested intestinal tissues were fixed overnight with 4% paraformaldehyde at 4°C, and then washed with PBS. For paraffin embedding, tissues were then dehydrated through ascending alcohols and processed with xylene prior to embedding. For cryo-embedding, tissues were then processed with 15% sucrose and 30% sucrose until tissues sunk prior to freezing in OCT compound (Tissue-Tek 4583). 5 μm-thick paraffin sections and 10 μm-thick cryosections were used for immunohistochemistry and immunofluorescence using standard procedures, respectively. Hematoxylin (VWR, 95057–858) and eosin (Sigma, HT110180) staining was performed using standard procedures. Immunohistochemistry was performed using primary antibodies against Ki67 (Abcam ab16667, 1:300), OLFM4 (Cell Signaling 39141, 1:2500), CD44 (BD 558739, 1:300), BrdU (Bio-Rad MCA2060, 1:500), F4/80 (Cell Signaling 70076, 1:500), and SOX9 (Cell Signaling 82630, 1:600). After incubating with secondary antibody and the Vectastain ABC HRP Kit (Vector Labs), slides were developed using 0.05% DAB (Amresco 0430) and 0.015% hydrogen peroxide in 0.1 M Tris, and then counterstained with hematoxylin. The slides were mounted and viewed on a Nikon Eclipse E800 microscope. Images were photographed with a Lumenera INFINITY3 camera and infinity capture imaging software (v6.5.6). A Zeiss Observer Z1 microscope was used to image the immunofluorescence staining of YAP (Cell Signaling 4912, 1:100), SOX9 (Cell Signaling 82630, 1:100) and DAPI (Biotium 40043, 1:5000). ImageJ and Adobe Photoshop were used to adjust contrast and brightness. When adjustments of sharpness, contrast, or brightness were made, they were applied uniformly across comparative images.
Intestinal crypt isolation
Freshly harvested intestine was flushed with cold PBS, opened longitudinally, cut into 1 cm pieces, and then rotated in 3 mM EDTA in PBS at 4 °C for 5 minutes, 10 minutes and 40 minutes (refreshing EDTA/PBS every time). The tissue was then vigorously shaken to release the epithelium, and crypts passed through a 70-μm cell strainer. Cells were pelleted by centrifugation at 200 g for 3 minutes at 4 °C and then washed with cold PBS. Cell pellets were used for organoid culture and single cell dissociation for ATAC-seq as described in later sections.
Organoid culture and treatment
Primary crypt-derived organoids were isolated from proximal half of mouse small intestine and cultured in Cultrex® reduced growth factor basement membrane matrix, Type R1 (R&D, 3433-010-R1) or Corning Matrigel (356231) according to established methods.23 Organoid medium was changed every 2 days. The organoids were treated with 1 μM tamoxifen dissolved in ethanol for 12 h. Vehicle-treated organoids served as a control. Tamoxifen was added into culture medium of organoids on Day 3 after seeding. Recombinant TGFB1 (Peprotech 100–21) or 10 μM TGFB receptor inhibitors including SB525334 (Selleckchem S1476) and A83–01 (Tocris 2939) were prepared according to the supplier’s instructions, and vehicle controls were used in organoid culture. For IR treatment, organoids on Day 4 after seeding were subjected to 4 Gy IR with a 137 Cs γ-source irradiator at a dose rate of 60 or 85 cGy/minute. 1–2 ng/ml TGFB1 was added on Day 6 or Day 7 after seeding, and was removed 24 hours after treatment. For non-IR treatment, 2 ng/ml TGFB1 was added on Day 4 or Day 6 after seeding, and was removed 24 hours after treatment. Details of each experimental design can be found in figure schematics and figure legends. TGFB1 activity was tested before each experiment and the dose of TGFB1 was adjusted by its ability to induce spherical organoids, targeting at ~60–80% of spherical organoids after irradiation in this study, according to the decline in TGFB1 activity over time in storage as determined (Table S2).
Human duodenal organoid lines were cultured as previously reported22,43 with slight modifications. Briefly, human organoids were plated in Corning Matrigel with L-WRN complete medium, which contains 50% L-WRN conditioned medium. Every 220 μL Matrigel stock was supplemented with 59 μL cold L-WRN complete medium, plus an additional 0.6 μL 2.5 mM Y27632 (Selleckchem S1049), and 5.5 μL 100 μM CHIR99021 (Axon 1386). Conditioned media from L-WRN cells containing Wnt3a, Rspondin3, and Noggin was mixed 1:1 with 2x Basal media comprised of 43.8 mL advanced DMEM/F-12, 1 mL 200 mM GlutaMAX, 1 mL 1 M HEPES, 1 mL N-2 supplement, 2 mL B-27 supplement, 200 μL 0.5 M N-Acetyl-L-cysteine, and 1 mL penicillin/streptomycin. To make L-WRN complete medium, the mixed media mentioned above was further supplemented with 50 ng/mL human EGF, 100 μg/mL primocin, 2.5 μM CHIR99021 and 10 μM Y27632. To investigate the conserved function of TGFB1 in human, 2 ng/ml recombinant TGFB1 and vehicle controls were used in human duodenal organoid culture for 24 hours. The human organoids were collected 24 hours or 72 hours after TGFB1 initiation.
Single cell dissociation of organoids for ATAC-seq and scRNA-seq
Mouse organoids were cultured and treated as mentioned above. Primary cultured organoids were collected by removing Matrigel using cold PBS. Matrigel droplets containing organoids were disrupted. Organoids were pelleted by centrifugation at 300 g for 3 minutes at 4 °C and washed with cold PBS. After removing the PBS and Matrigel, organoids were resuspended in 1 mL pre-warmed (37 °C) TrypLE, and rotated at 37 °C for 15 to 30 minutes until organoids were dissociated into single cells (confirmed via microscope). Cells were pelleted at 300 g for 3 minutes at 4 °C, and washed with cold PBS. Cells were passed through SP Bel-Art Flowmi 40 μm cell strainer and collected into a new protein LoBind tube.
30,000 cells were prepared as mentioned above and used for ATAC-seq as described previously70,71 with slight modifications. Briefly, cells were centrifuged at 500 g for 5 minutes at 4°C and resuspended in ice-cold lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% NP-40). Cells were then centrifuged at 500 g for 10 minutes at 4°C. The isolated nuclear pellets were incubated with a 50 μl reaction of Tn5 Transposase (Illumina Tagment DNA Enzyme and Buffer Large Kit 20034198) for 30 minutes at 37°C. The transposed chromatin was purified with MinElute PCR purification kit (QIAGEN REF 28004), and PCR was amplified with high-fidelity 2x PCR Master Mix (New England Biolabs M0541). One-third of the maximum fluorescent intensity during a qPCR trial run was used to determine the additional cycles for library prep. The PCR amplified libraries were purified and sent to Novogene America for sequencing.
Cells used for scRNA-seq were prepared according to PIPseq Milli 3’ Single Cell Capture and Lysis User Guide. Briefly, a cell suspension at a concentration of 1000 live cells/μL was prepared in Cell Suspension Buffer provided from the PIPseq™ T2 3’ Single Cell Capture and Lysis Kit (v2.1, Fluent BioSciences). 5000 cells (total 5 μL) were added into Pre-templated Instant Partitions (PIPs). Stable emulsions carrying captured mRNA were sent to Fluent Biosciences for downstream processing. A visual inspection was performed at sample receipt to assess the emulsion quality before initiating downstream sample processing. Satisfactory samples were carried through mRNA isolation, cDNA generation, cDNA amplifications, sequencing ready library preparation, library pooling, and sequencing according to the PIPseq™ T2 Single Cell Sequencing Kit (v2.1) specifications. cDNA and library quality were assessed by using ThermoFisher Qubit 4 Fluorometer and Agilent 4200 TapeStation System. cDNA libraries were sequenced on an Illumina NextSeq 2000 instrument to a minimum sequencing depth of 50,000 reads per cell.
Fluorescence-Activated Cell Sorting (FACS) for scRNA-seq
Crypts were isolated from proximal half of small intestine of Mki67tm1.1Cle/J mice after 56 hours post-irradiation and their non-IR littermate control, as described in the previous section. 100-μm cell strainer was used in IR samples, as crypts were expanded during regeneration after IR. Crypts were pelleted by centrifugation at 300 g for 3 minutes at 4 °C and then washed with cold PBS. To dissociate single cells for FACS, isolated crypts were further rotated with 5U/ml dispase (Stem Cell 07913) and 200 U/ml of DNase I (Sigma D4513) at 37 °C for 30 minutes, and were then washed twice with 1% BSA/PBS, and filtered with a 40-μm cell strainer. Cells were prepared in 1% BSA/PBS with 200 U/ml of Dnase I for sorting. Ki67-RFP+ DAPI− cells from mice 56 hours post-IR and non-IR condition were detected and sorted with Beckman Coulter Astrios EQ High Speed Cell Sorter, respectively. Dead cells were eliminated using 0.5 μg/ml DAPI. Kaluza analysis 2.1.3 software was used for FACS data analysis (Table S3). Total 5500 cells were sorted and concentrated into 5 μL for initiating the PIPseq pipeline (PIPseq™ T2 3’ Single Cell Capture and Lysis Kit v2.1, Fluent BioSciences). Cells were captured and lysed according to the manufacturer’s instructions. Samples were sent to Fluent BioSciences for downstream processing as described above.
Isolation of intestinal macrophages
Intestinal macrophages were isolated from C57BL/6J control mice or mice at 3 days post-IR. Intestine was flushed with cold PBS, opened longitudinally and cut into 2 cm pieces. Pieces of tissue were rotated in pre-warmed 10% FBS, 10 mM HEPES, 5 mM EDTA in HBSS for 10 minutes, three times at 37 °C to remove epithelium. Tissues were vortexed lightly after each time point. Tissues were washed with 2% FBS, 10 mM HEPES in HBSS twice, followed by an HBSS wash, and then gently minced with scissors and placed into pre-warmed 1% penicillin/streptomycin, 10% FBS, 15 mM HEPES, 1 mg/ml Collagenase D (Roche 11088858001) and 0.35 mg/ml Dnase I (Sigma D4513) mixed in RPMI-1640. Tissues were rotated for 30 minutes at 37 °C, and vortexed lightly every 10 minutes. Following digestion, cells were passed through a 70-μm cell strainer and spun down at 300 g for 5 minutes at 4 °C.
Cells were passed through a 40-μm cell strainer, counted, and then spun down at 300g for 5 minutes. Cells were resuspended in 180 ul of magnetic-activated cell sorting (MACS) buffer (0.5% BSA and 2 mM EDTA in PBS), and 20 ul of Anti-F4/80+ Microbeads UltraPure (Miltenyi Biotec 130-110-443) was added to the mixture. Cells were incubated for 15 minutes at 4 °C, washed with 2 ml of MACS buffer post incubation, centrifuged at 300 g for minutes, and then resuspended in 500 ul of MACS buffer before placing over columns. MS columns (Miltenyi Biotec 130-042-201) were used for cell separation, and were washed with 500 ul of MACS buffer prior to applying cell suspension onto the column. MACS columns were then washed three times with 500 ul of MACS buffer, then removed from the OctoMACS (Miltenyi Biotec 130-042-109) separator. 1 ml of MACS buffer was applied onto the column, and cells complexed with F4/80+ beads were plunged into a collection tube. Cells were diluted in macrophage media, consisting of 2mM GlutaMAX, 20 mM HEPES, 100 U/ml penicillin/streptomycin, 1 mM Sodium Pyruvate and 10% FBS in RPMI-1640, and counted. Primary macrophages were used for co-culture experiments.
Co-culture of intestinal macrophages and organoids
Primary organoids were taken at 4 days post initial seeding, and gently resuspended into cold PBS. Organoids were then spun down at 300 g for 5 minutes at 4 °C. Organoid pellets were resuspended into Cultrex matrix, and primary intestinal macrophages eluted from MACS columns were added to the mixture and plated onto plates for co-culture in 25 ul bubbles (around 5x104 macrophages per co-culture organoid bubble). For reference, organoids were grown in organoid medium as described in the previous section. Organoids were also grown in macrophage media as a second reference point, yielding similar results. Media for the co-cultures was changed every day. After 2 days in co-culture, matrix bubbles were resuspended in cold PBS and allowed to sit on ice for 7–10 minutes. This allowed separation of organoids from macrophages, with organoids collecting at the bottom of the tube and macrophages remaining in the supernatant. Organoid pellets were resuspended in 1 ml Trizol and further processed for RNA extraction.
Isolation and culture of intestinal mesenchymal cells
Mesenchymal cells were isolated from proximal half of small intestine from C57BL/6 or PDGFRa-H2B-EGFP mice as previously described28 with slight modifications. Freshly harvested intestine was flushed with cold PBS, opened longitudinally, cut into 2 cm pieces, and then rotated in 30 mL pre-digestion buffer (pre-warmed, HBSS containing 10% FBS, 10 mM HEPES and 5 mM EDTA) at 37°C for 20 minutes twice (refresh pre-digestion buffer every time). After washing tissues in wash buffer (HBSS containing 2% FBS, 10 mM HEPES), the tissues were further transferred into 20 mL of pre-warmed digestion buffer (RPMI medium containing 10% FBS, 1% P/S, 15 mM HEPES, 25 U/mL of collagenase IV (Worthington LS004186), 100 U/ml of Dnase I (Sigma D4513), 0.3 g/100 mL of Dispase II (Gibco 17105041)) and rotated at 37°C for 30 minutes. After vortexing the cell solution intensely for 20 sec every 10 minutes, the cell solution was passed through a 40 μm cell strainer. Cells were pelleted by centrifugation at 400 g for 5 minutes at 4 °C and then resuspended in RPMI medium containing 10% FBS. Remaining tissues were incubated with 20 mL of fresh digestion buffer, and the above steps were repeated. Cells were combined, seeded at a desired density (see below), and cultured in Advanced DMEM/F12 (Gibco 12634–010) medium, containing 10% FBS (Gibco 26140–095), 1% penicillin and streptomycin (Invitrogen 15140–122), 1% HEPES (Gibco 15630–080) and 1% Glutamax (Gibco 35050–061).
Co-culture of intestinal mesenchymal cells and organoids
To set up a co-culture system, 3 intact 25ul Matrigel droplets were gently taken out from a cultured primary organoid plate 3 days after seeding. These Matrigel droplets containing organoids were transferred and floated above the cultured mesenchyme for 2 days. Co-cultured organoids were then collected for RNA extraction. Primary or passaged mesenchymal cells were cultured in 12-well plates. Primary mesenchymal cells isolated from IR and non-IR C57BL/6 mice were seeded at three densities, including 4x105 cells, 106 cells, 2x106 cells. The cultures were refreshed at Day 3 to remove any debris. Co-culture was initiated on Day 4. For passaged mesenchymal cells, cells were seeded at two densities, including 105 cells and 2.5x105 cells. These cells were pre-treated with vehicle, TGFB1 or TGFBR inhibitors for 3 days, starting at Day 1 after seeding. After TGFB1 was removed and cells were washed extensively, these pre-treated mesenchymal cells were then used for co-culture. The detailed schematics of co-culture experiments are shown in Figure 5D and Figure S5C.
RNA extraction, bulk RNA-seq, and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
For tissues, 2 cm of mouse duodenum was homogenized in Trizol (Invitrogen 15596018), and processed for RNA extraction according to the manufacturer’s instructions. For cultured organoids or mesenchymal cells, QIAGEN RNeasy Micro Kit was used to extract RNA according to the manufacturer’s instructions. For bulk RNA-seq, RNA samples were prepared and sent to BGI Americas. For qRT-PCR, cDNA was synthesized from total RNA with Oligo(dT)20 primers using SuperScript III First-Strand Synthesis SuperMix (Invitrogen 18080–400). qRT-PCR analysis was performed using gene-specific primers and SYBR Green PCR Master Mix (Applied Biosystems, 4309155). The sequences of the primers used are available upon request. The 2-ΔΔCt method was applied to calculate the fold change of relative transcript levels, and Hprt was used for normalization.
Protein extraction and western blot
2 cm of mouse duodenum was homogenized in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, protease inhibitor cocktails, and phosphatase inhibitors) and rotated at 4°C for 30 min for protein extraction. Protein concentration was determined by Pierce BCA Protein Assay Kit (Thermo). Immunodetection was performed using specific antibodies against p-SMAD3 (Cell signaling 9520, 1:1000), p-SMAD2/3 (Cell signaling 8828, 1:1000), SMAD2/3 (Cell signaling 8685, 1:1000), and β-actin (Abcam ab8227, 1:5000). The intensity of signal was quantified by ImageJ.
Membrane-based antibody arrays
Protein lysates were extracted from 2 cm of duodenal fragment of mice after 3 days of irradiation, and non-IR mice were used as controls. The protein lysates (200 μg) were applied to Mouse Growth Factor Array C3 (RayBiotech, CODE: AAM-GF-3-4) according to the manufacturer’s instructions. Each growth factor was represented in duplicate on the membrane. Two independent experiments were performed to evaluate the expression level of various growth factors. The intensity of signal was quantified by ImageJ. The positive control was used to normalize the results from different membranes being compared.
ELISA of TGFB1
Protein lysates were extracted from 1) 2 cm of duodenal tissues of mice after 3 days of irradiation, and non-IR mice were used as controls; 2) 2 cm of duodenal tissues of mice with clodronate liposomes, and mice treated with control liposomes were used as controls. Blood was collected from abdominal aorta of experimental mice mentioned above with heparinized syringes. Blood samples were centrifuged at 1500g for 15 minutes at 4 °C and the supernatant was collected as the plasma. To detect the relative abundance of TGFB1 in the intestine tissues and plasma, the protein lysates or plasma were applied to Mouse TGF beta 1 ELISA Kit (ab119557, abcam) according to the manufacturer’s instructions.
RNAscope in situ hybridization and immunofluorescence
RNA localization was performed according to the RNAscope Multiplex Fluorescent Reagent Kit v2 Assay (ACD 323110) as previously described.72 Briefly, mouse intestines were linearized and twisted into a swiss roll and prepared by 24-hour fixation in 10% normal buffered formalin at room temperature followed by dehydration through a methanol series and rehydration to 70% ethanol before paraffin infusion. Paraffin sections were cut at 5 μm, baked for one hour at 60°C, deparaffinized and then pretreated with hydrogen peroxide (ACD kit) for 10 minutes, antigen retrieval (ACD 322000) for 15 minutes and protease plus treatment (ACD kit) for 30 minutes. Probes were Mm-Tgfb1 (407751) and Mm-Tgfbr2-C2 (406241-C2) diluted 1:50 into the channel 1 probe. TSA fluorophores were diluted in multiplex TSA buffer (ACD 322809) Cy5 (Akoya TB-000203) at 1:2000, Cy3 (Akoya TB-000202) diluted 1:2500 and OPAL-488 (Perkin Elmer FP1168) at 1:2000. Following RNAscope, sections were co-stained with rabbit anti-F4/80 (Cell Signaling 70076, 1:100) and mouse-anti-Ecadherin (BD Transduction Labs 610181 1:500). Secondary antibodies were anti-Rabbit-750 (Sigma SAB4600373) and anti-Mouse-594 (Jackson ImmunoLabs 715-585-150) both diluted at 1:500. Slides were mounted with Prolong Gold and imaged on an Olympus FV3000 confocal microscope.
Organoid transplantation, imaging and quantification
Colitis was induced in NOD SCID female mice by administration of 3.5% Dextran sulfate sodium salt (DSS, Affymetrix J14489) in drinking water for 7 days followed by normal water for 3 days. Transplantation was performed 10 days after DSS initiation. The procedure was performed as previously described12,34 with slight modifications. Donor crypts from C57BL/6, ACTB-EGFP or ROSA26mT/mG (ACTB-tdTomato) mice were isolated from proximal half of small intestine and cultured in Cultrex® reduced growth factor basement membrane matrix, Type R1 (Trevigen) according to established methods.23 On Day 4 of primary culture, 2 ng/ml TGFB1 or vehicle was added into culture medium and removed 24 hours later. On Day 7 of primary culture, organoids were collected by removing Matrigel using cold PBS. For comparison of mice with transplantation of vehicle-treated organoids or TGFB1-treated C57 organoids, approximately 600 organoids were suspended in 50 μl of 5% Matrigel in PBS for transplantation of each mouse. Sham surgery controls were also included in this comparison, which have the same procedures but without organoids in the 50 μl of 5% Matrigel in PBS for transplantation. For the competition assay, to minimize variations, vehicle- and TGFB1-treated organoids were 1:1 mixed as follows and transferred to the same mouse: vehicle-treated organoids expressing RFP (ACTB-tdTomato) were mixed with TGFB1-treated organoids expressing GFP (ACTB-EGFP); or vehicle-treated organoids expressing GFP (ACTB-EGFP) was mixed with TGFB1-treated organoids expressing RFP (ACTB-tdTomato). In this competitive assay of repair of intestinal injury, approximately 600 organoids were suspended in 50 μl of 5% Matrigel in PBS for transplantation of each mouse.
Immune compromised NOD SCID female mice were anesthetized with isoflurane, and their colons were flushed using a gentle sterile PBS enema, followed by gentle massage of the abdomen to expel enema fluid and colon contents. The cell suspension mix of organoids was subsequently infused into the colonic lumen. The anus was sealed with surgical skin glue, which was removed after 6 hours. For the competitive colonization assay, colon tissues were collected after 9 days of organoid colonization. Colons were washed with PBS, opened longitudinally and pinned on a wax plate. After soaking in 4% paraformaldehyde for 10 minutes, swiss rolls were made for these colon tissues, and fixed for 2 hours with 10 ml of 4% paraformaldehyde at 4°C, and then washed with PBS. For cryo-embedding, tissues were then processed with 15% sucrose for 1 hour and 30% sucrose overnight at 4°C prior to freezing in OCT compound. The whole cryo block from each mouse was processed with sequential 100 μm-thick cryosections, 80–100 sections per mouse. These sequential cryosections were imaged for GFP or RFP colonized regions of organoids under a Zeiss Observer Z1 microscope. ImageJ was used to quantify the organoid colonized areas from each grafts observed. For comparison of mouse histopathology following DSS treatment, transplantation of vehicle-treated organoids or TGFB1-treated organoids or sham controls were conducted as above. Colon tissues were collected 4 days after organoid transplantation. The colon lengths were measured and H&E staining was used to evaluate the regeneration/recovery of each mouse.
Bioinformatics
For RNA-seq, raw sequencing reads (fastq) were first quality checked using fastQC (v0.11.3) and were further aligned to mouse (mm9) genomes using Tophat2 (v2.1.0) to generate bam files. Cuffquant (v2.2.1) was used to generate cxb files from bam files. Cuffnorm (v2.2.1) was performed to calculate FPKM values using quartile normalization. Cuffdiff (v2.2.1) was applied to identify differentially expressed genes between groups using quartile normalization and per-condition dispersion. Gene set enrichment analysis (GSEA v4.0.3)73 was performed as described, and the gene signature lists used in this study were shown in Table S4. Heatmapper 74 was used to display relative transcript levels of genes of interest by using normalized FPKM values from Cuffnorm.
For scRNA-seq of organoid samples, prior to processing, a mouse STAR reference was generated using a mouse genome FASTA build from the Ensembl 102 release and GTF annotations from GENCODE (GRCm39 vM29 2022.04) using STAR’s genomeGenerate function. BCLs derived from Illumina sequencing of PIPseq samples were demultiplexed and output in FASTQ format using BCL-convert v4.0.3. Barcode whitelisting and error correction were performed using PIPseeker v0.55. Reads passing this stage were then aligned to the mouse reference genome using STARsolo (STAR v2.7.10a). An aligned, sorted BAM file was generated by STAR, which included required information for RNA velocity analysis. Cell calling was performed using PIPseeker’s transcript-count thresholding approach and sensitivity 3 was chosen for all samples, which most closely targeted the first inflection point after the knee point on the barcode-rank plot. For sorted Ki67-RFP+ DAPI− cells, samples were processed in the same way, but using salmon v1.46 instead of STAR. A reference genome was first built from the same sources as above using the salmon index command with a 19-bp k-mer size. The cell barcodes files from the filtered matrix, corresponding to the cell fraction, were then processed with Scanpy v1.9.1.75 RNA velocity was analyzed by Velocyto 0.17.76
For ATAC-seq, paired-end ATAC-seq fastq file was quality checked using fastQC. ATAC-seq adapter sequences were removed from each read file using CutAdapt (v1.9.1).77 Each read file was then aligned to the mouse mm9 genome using bowtie2. Picard (v2.18.27) was used to determine the median alignment size of each alignment bam file. Peak region bed files were called from each alignment bam file using MACS2 (v2.1.0). MACS2 was run with a “shift” distance of −0.5 times the median alignment size and an “extsize” distance equal to the median alignment size for each alignment bed file. The peak files were filtered against ENCODE blacklist, which is known to yield false ChIP-seq signals due to the inaccuracies of a particular genome assembly.78 BEDtools79 was used to subtract the sites on the ENCODE blacklist. DiffBind (v2.4.7)80 was used to identify differential signals of ATAC-seq between vehicle- or TGFB1-treated organoids, and FDR < 0.01 was used as the cutoff for significance. Haystack (v0.4.0)81 quantile normalized bigwigs were used to plot heatmaps using computeMatrix and plotHeatmap from deeptools (v2.4.2)82. SitePro (v1.0.2)83 was used to visualize the average signals of ATAC-seq in the desired genomic regions. Homer findMotifsGenome.pl (v4.8.3, homer de novo Results)84 was used to identify transcription factor motifs enriched at peaks. The Integrative Genomics Viewer (IGV 2.4.13)85 was used to visualize ATAC-seq bigwig tracks.
For ChIP-seq, FastQC (v0.11.3) was used to check the quality of raw sequencing reads (fastq), and bowtie2 (v2.2.6) was used to align the sequences to mouse (mm9) genomes and generate bam files. Model-based Analysis of ChIP-Seq86 (MACS 1.4.1) was used for peak calling and to generate bed files from aligned reads. The shiftsize parameter used in MACS was based on the fragment size of Pippin Prep library size selection. SMAD4 ChIP-seq of mouse duodenal epithelium are at a MACS p-value of 10-5. IGV 2.4.1385 was used to visualize ChIP-seq bigwig tracks.
QUANTIFICATION AND STATISTICAL ANALYSIS
The data is presented as mean ± SEM, and statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post test or Tukey’s multiple comparisons test with the GraphPad Prism version 8.0.2 or Student’s t-test at P < 0.001***, P < 0.01** or P < 0.05*. Other bioinformatics related statistical analysis was completed with the embedded statistics in each package, including MACS or MACS2,86 HOMER,84 DiffBind,80 Cuffdiff,87 GSEA73,88 and Scanpy.75 P < 0.05 (95% confidence interval) was considered statistically significant.
Supplementary Material
Table S1. Diffbind analysis and de novo motifs of TGFB1 vs. vehicle-enriched chromatin-accessible regions as defined by HOMER, related to Figure 6A–C.
Table S2. The measurement of TGFB1 activity in this study, related to Methods.
Table S3. Sorted Ki67+DAPI- cells from crypts of Mki67tm1.1Cle/J mice 56h post IR and non-IR controls, related to Methods.
Table S4. Gene signature lists used in this study, related to Methods.
Highlights.
TGFB1 signaling is required for epithelial regeneration following irradiation.
TGFB1 promotes fetal reversion in intestinal organoid cultures.
TGFB1 induces a YAP-SOX9 regenerative circuit.
TGFB1-pretreated organoids enhance engraftment efficiency into damaged colon.
ACKNOWLEDGMENTS
M.P.V is supported by grants from the National Institutes of Health (NIH R01 DK126446 and R01 DK121915). M.P.V. and J.R.S are supported by the Intestinal Stem Cell Consortium from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and National Institute of Allergy and Infectious Diseases (NIAID) of the NIH under grant number U01 DK103141. L.C. is supported by grants from Start-up Research Fund of Southeast University RF1028623015, National Natural Science Foundation of China 32270830 and Jiangsu Provincial Science Fund for Distinguished Young Scholars BK20230026. K.D.W. is supported by grant from NIH R01 DK121166. J.B. and S.T.M. were supported, in part, by a grant from the National Institutes of Health, P30 DK034987. S.T.M is supported by R01DK109559 and the Katherine E. Bullard Charitable Trust for Gastrointestinal Stem Cell and Regenerative Research. N.G. is supported by grant from NIH R01 DK132885 and R01 DK119198. The authors acknowledge the Office of Advanced Research Computing (OARC) at Rutgers University for providing access to the Amarel cluster and associated research computing resources that have contributed to the results reported here. We thank the Michigan Medicine Translational Tissue Modeling Laboratory for providing the human duodenal organoid lines. The research was also supported by flow cytometry/cell sorting core facility at Environmental and Occupational Health Sciences Institute (EOHSI) at Rutgers University. This work benefited from the Cancer Center Support Grant (CCSG, P30CA072720) from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests. L.C. and M.P.V are listed inventors of a Provisional Patent Application 63392365.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Metcalfe C, Kljavin NM, Ybarra R, and de Sauvage FJ (2014). Lgr5(+) Stem Cells Are Indispensable for Radiation-Induced Intestinal Regeneration. Cell stem cell 14, 149–159. 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 2.van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee ANY, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. (2012). Dll1(+) secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14, 1099-+. 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomic G, Morrissey E, Kozar S, Ben-Moshe S, Hoyle A, Azzarelli R, Kemp R, Chilamakuri CSR, Itzkovitz S, Philpott A, and Winton DJ (2018). Phospho-regulation of ATOH1 Is Required for Plasticity of Secretory Progenitors and Tissue Regeneration. Cell stem cell 23, 436-+. 10.1016/j.stem.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu S, Tong K, Zhao Y, Balasubramanian I, Yap GS, Ferraris RP, Bonder EM, Verzi MP, and Gao N (2018). Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell stem cell 23, 46–59 e45. 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. (2016). Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell stem cell 18, 203–213. 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Bala P, Rennhack JP, Morris C, Aitymbayev D, Moyer SM, Duronio GN, Doan P, Liang XY, Hornick JL, Yurgelun MB, et al. (2022). Developmental reprogramming mediates aberrant cell state plasticity in colorectal cancer initiation. bioRxiv, 10.1101/2022.1109.1122.509032. [DOI] [PMC free article] [PubMed]
- 7.Okamoto R, Shimizu H, Suzuki K, Kawamoto A, Takahashi J, Kawai M, Nagata S, Hiraguri Y, Takeoka S, Sugihara HY, et al. (2020). Organoid-based regenerative medicine for inflammatory bowel disease. Regenerative therapy 13, 1–6. 10.1016/j.reth.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. 10.1038/s41586-019-1154-y. [DOI] [PubMed] [Google Scholar]
- 9.Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H, and Magness ST (2015). SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology 149, 1553–1563 e1510. 10.1053/j.gastro.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yousefi M, Li N, Nakauka-Ddamba A, Wang S, Davidow K, Schoenberger J, Yu Z, Jensen ST, Kharas MG, and Lengner CJ (2016). Msi RNA-binding proteins control reserve intestinal stem cell quiescence. The Journal of cell biology 215, 401–413. 10.1083/jcb.201604119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113. 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, et al. (2018). YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell stem cell 22, 35–49 e37. 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, and Wrana JL (2015). Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718. 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 14.Murata K, Jadhav U, Madha S, van Es J, Dean J, Cavazza A, Wucherpfennig K, Michor F, Clevers H, and Shivdasani RA (2020). Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells. Cell stem cell 26, 377–390 e376. 10.1016/j.stem.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han T, Goswami S, Hu Y, Tang F, Zafra MP, Murphy C, Cao Z, Poirier JT, Khurana E, Elemento O, et al. (2020). Lineage Reversion Drives WNT Independence in Intestinal Cancer. Cancer discovery 10, 1590–1609. 10.1158/2159-8290.CD-19-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu M, Xiong L, Lyu Y, Zhang X, Shen J, Guan J, Chai P, Lin Z, Nie B, Li C, et al. (2021). Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res 31, 259–271. 10.1038/s41422-020-00453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Chiang IL, Ohara TE, Fujii S, Cheng J, Muegge BD, Ver Heul A, Han ND, Lu Q, Xiong S, et al. (2019). Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells. Cell 179, 1144–1159 e1115. 10.1016/j.cell.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. (2012). The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. The EMBO journal 31, 3079–3091. 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basak O, van de Born M, Korving J, Beumer J, van der Elst S, van Es JH, and Clevers H (2014). Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. The EMBO journal 33, 2057–2068. 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruder B, and Becker C (2020). At the Forefront of the Mucosal Barrier: The Role of Macrophages in the Intestine. Cells 9. 10.3390/cells9102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng X, Lin Z, Lv C, Shao C, Bi X, Deng M, Xu J, Guerrero-Juarez CF, Li M, Wu X, et al. (2020). Cycling Stem Cells Are Radioresistant and Regenerate the Intestine. Cell Rep 32, 107952. 10.1016/j.celrep.2020.107952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibowitz BJ, Zhao G, Wei L, Ruan H, Epperly M, Chen L, Lu X, Greenberger JS, Zhang L, and Yu J (2021). Interferon b drives intestinal regeneration after radiation. Science advances 7, eabi5253. 10.1126/sciadv.abi5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–U147. 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S, et al. (2020). Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell stem cell 26, 391–402 e395. 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Toth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Subepithelial telocytes are an important source of Wnts that supports intestinal crypts (vol 557, pg 242, 2018). Nature 560, E29–E29. 10.1038/s41586-018-0286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, and Virshup DM (2018). PDGFR alpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. P Natl Acad Sci USA 115, E3173–E3181. 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton TG, Klinghoffer RA, Corrin PD, and Soriano P (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Molecular and cellular biology 23, 4013–4025. 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JE, Fei L, Yin WC, Coquenlorge S, Rao-Bhatia A, Zhang X, Shi SSW, Lee JH, Hahn NA, Rizvi W, et al. (2020). Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nature communications 11, 334. 10.1038/s41467-019-14058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roulis M, Kaklamanos A, Schernthanner M, Bielecki P, Zhao J, Kaffe E, Frommelt LS, Qu R, Knapp MS, Henriques A, et al. (2020). Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature 580, 524–529. 10.1038/s41586-020-2166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, and Stappenbeck TS (2012). Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113. 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Wang J, Zhao Y, Zhang X, Liu J, Zhang C, Haffty B, Verzi M, Zhang L, Gao N, et al. (2020). LIF is essential for ISC function and protects against radiation-induced gastrointestinal syndrome. Cell death & disease 11, 588. 10.1038/s41419-020-02790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto N, Goto S, Imada S, Hosseini S, Deshpande V, and Yilmaz OH (2022). Lymphatics and fibroblasts support intestinal stem cells in homeostasis and injury. Cell stem cell 29, 1246–1261 e1246. 10.1016/j.stem.2022.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Toke NH, Luo S, Vasoya RP, Fullem RL, Parthasarathy A, Perekatt AO, and Verzi MP (2019). A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nature genetics 51, 777–785. 10.1038/s41588-019-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe S, Kobayashi S, Ogasawara N, Okamoto R, Nakamura T, Watanabe M, Jensen KB, and Yui S (2022). Transplantation of intestinal organoids into a mouse model of colitis. Nat Protoc 17, 649–671. 10.1038/s41596-021-00658-3. [DOI] [PubMed] [Google Scholar]
- 35.Gurtner GC, Werner S, Barrandon Y, and Longaker MT (2008). Wound repair and regeneration. Nature 453, 314–321. 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 36.Vallance BA, Gunawan MI, Hewlett B, Bercik P, Van Kampen C, Galeazzi F, Sime PJ, Gauldie J, and Collins SM (2005). TGF-beta1 gene transfer to the mouse colon leads to intestinal fibrosis. American journal of physiology. Gastrointestinal and liver physiology 289, G116–128. 10.1152/ajpgi.00051.2005. [DOI] [PubMed] [Google Scholar]
- 37.Sato M, Muragaki Y, Saika S, Roberts AB, and Ooshima A (2003). Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. The Journal of clinical investigation 112, 1486–1494. 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clouthier DE, Comerford SA, and Hammer RE (1997). Hepatic fibrosis, glomerulosclerosis, and a lipodystrophy-like syndrome in PEPCK-TGF-beta1 transgenic mice. The Journal of clinical investigation 100, 2697–2713. 10.1172/JCI119815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sime PJ, Xing Z, Graham FL, Csaky KG, and Gauldie J (1997). Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. The Journal of clinical investigation 100, 768–776. 10.1172/JCI119590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts AB, Russo A, Felici A, and Flanders KC (2003). Smad3: a key player in pathogenetic mechanisms dependent on TGF-beta. Annals of the New York Academy of Sciences 995, 1–10. 10.1111/j.1749-6632.2003.tb03205.x. [DOI] [PubMed] [Google Scholar]
- 41.Zanninelli G, Vetuschi A, Sferra R, D’Angelo A, Fratticci A, Continenza MA, Chiaramonte M, Gaudio E, Caprilli R, and Latella G (2006). Smad3 knock-out mice as a useful model to study intestinal fibrogenesis. World journal of gastroenterology 12, 1211–1218. 10.3748/wjg.v12.i8.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Shi W, Wang YL, Chen H, Bringas P Jr., Datto MB, Frederick JP, Wang XF, and Warburton D (2002). Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. American journal of physiology. Lung cellular and molecular physiology 282, L585–593. 10.1152/ajplung.00151.2001. [DOI] [PubMed] [Google Scholar]
- 43.Meyer AR, Brown ME, McGrath PS, and Dempsey PJ (2022). Injury-Induced Cellular Plasticity Drives Intestinal Regeneration. Cellular and molecular gastroenterology and hepatology 13, 843–856. 10.1016/j.jcmgh.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, and Haber DA (2009). YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol 11, 1444–1450. 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao J, and Sheng H (2010). Amphiregulin promotes intestinal epithelial regeneration: roles of intestinal subepithelial myofibroblasts. Endocrinology 151, 3728–3737. 10.1210/en.2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen F, Yang W, Huang X, Cao AT, Bilotta AJ, Xiao Y, Sun M, Chen L, Ma C, Liu X, et al. (2018). Neutrophils Promote Amphiregulin Production in Intestinal Epithelial Cells through TGF-beta and Contribute to Intestinal Homeostasis. Journal of immunology 201, 2492–2501. 10.4049/jimmunol.1800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes & development 22, 1962–1971. 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320 e1322. 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, and Sahai E (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 15, 637–646. 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nardone G, Oliver-De La Cruz J, Vrbsky J, Martini C, Pribyl J, Skladal P, Pesl M, Caluori G, Pagliari S, Martino F, et al. (2017). YAP regulates cell mechanics by controlling focal adhesion assembly. Nature communications 8, 15321. 10.1038/ncomms15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neto F, Klaus-Bergmann A, Ong YT, Alt S, Vion AC, Szymborska A, Carvalho JR, Hollfinger I, Bartels-Klein E, Franco CA, et al. (2018). YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. eLife 7. 10.7554/eLife.31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shafaq-Zadah M, Gomes-Santos CS, Bardin S, Maiuri P, Maurin M, Iranzo J, Gautreau A, Lamaze C, Caswell P, Goud B, and Johannes L (2016). Persistent cell migration and adhesion rely on retrograde transport of beta(1) integrin. Nat Cell Biol 18, 54–64. 10.1038/ncb3287. [DOI] [PubMed] [Google Scholar]
- 53.Aoki R, Shoshkes-Carmel M, Gao N, Shin S, May CL, Golson ML, Zahm AM, Ray M, Wiser CL, Wright CV, and Kaestner KH (2016). Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cellular and molecular gastroenterology and hepatology 2, 175–188. 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stzepourginski I, Nigro G, Jacob JM, Dulauroy S, Sansonetti PJ, Eberl G, and Peduto L (2017). CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A 114, E506–E513. 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, and Basler K (2018). GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453. 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 56.Miyoshi H, VanDussen KL, Malvin NP, Ryu SH, Wang Y, Sonnek NM, Lai CW, and Stappenbeck TS (2017). Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. The EMBO journal 36, 5–24. 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, and Stappenbeck TS (2009). Efficient colonic mucosal wound repair requires Trem2 signaling. Proc Natl Acad Sci U S A 106, 256–261. 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saha S, Aranda E, Hayakawa Y, Bhanja P, Atay S, Brodin NP, Li J, Asfaha S, Liu L, Tailor Y, et al. (2016). Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nature communications 7, 13096. 10.1038/ncomms13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, and Henson PM (1998). Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. The Journal of clinical investigation 101, 890–898. 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harnack C, Berger H, Antanaviciute A, Vidal R, Sauer S, Simmons A, Meyer TF, and Sigal M (2019). R-spondin 3 promotes stem cell recovery and epithelial regeneration in the colon. Nature communications 10, 4368. 10.1038/s41467-019-12349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193. 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 62.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, and Brown EJ (2007). Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell stem cell 1, 113–126. 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL (2002). Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. The Journal of biological chemistry 277, 33275–33283. 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 64.Leveen P, Larsson J, Ehinger M, Cilio CM, Sundler M, Sjostrand LJ, Holmdahl R, and Karlsson S (2002). Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood 100, 560–568. 10.1182/blood.v100.2.560. [DOI] [PubMed] [Google Scholar]
- 65.Yang X, Li C, Herrera PL, and Deng CX (2002). Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–81. 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 66.Akiyama H, Chaboissier MC, Martin JF, Schedl A, and de Crombrugghe B (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & development 16, 2813–2828. 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okabe M, Ikawa M, Kominami K, Nakanishi T, and Nishimune Y (1997). ‘Green mice’ as a source of ubiquitous green cells. Febs Lett 407, 313–319. Doi 10.1016/S0014-5793(97)00313-X. [DOI] [PubMed] [Google Scholar]
- 68.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 69.Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, and Jackson SP (1995). Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 80, 813–823. 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 70.Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current protocols in molecular biology 109, 21 29 21–29. 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218. 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holloway EM, Czerwinski M, Tsai YH, Wu JH, Wu A, Childs CJ, Walton KD, Sweet CW, Yu Q, Glass I, et al. (2021). Mapping Development of the Human Intestinal Niche at Single-Cell Resolution. Cell stem cell 28, 568–580 e564. 10.1016/j.stem.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, and Wishart DS (2016). Heatmapper: web-enabled heat mapping for all. Nucleic acids research 44, W147–153. 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome biology 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12. [Google Scholar]
- 78.Amemiya HM, Kundaje A, and Boyle AP (2019). The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Scientific reports 9, 9354. 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stark R, and Brown GD (2011). DiffBind: Differential Binding Analysis of ChIP-Seq Peak Data. Bioconductor, Available online at: http://bioconductor.org/packages/release/bioc/html/DiffBind.html.
- 81.Pinello L, Farouni R, and Yuan GC (2018). Haystack: systematic analysis of the variation of epigenetic states and cell-type specific regulatory elements. Bioinformatics 34, 1930–1933. 10.1093/bioinformatics/bty031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic acids research 44, W160–165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shin H, Liu T, Manrai AK, and Liu XS (2009). CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606. 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 84.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nature biotechnology 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562–578. 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tamayo P, Steinhardt G, Liberzon A, and Mesirov JP (2016). The limitations of simple gene set enrichment analysis assuming gene independence. Statistical methods in medical research 25, 472–487. 10.1177/0962280212460441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mustata RC, Vasile G, Fernandez-Vallone V, Strollo S, Lefort A, Libert F, Monteyne D, Perez-Morga D, Vassart G, and Garcia MI (2013). Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep 5, 421–432. 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Diffbind analysis and de novo motifs of TGFB1 vs. vehicle-enriched chromatin-accessible regions as defined by HOMER, related to Figure 6A–C.
Table S2. The measurement of TGFB1 activity in this study, related to Methods.
Table S3. Sorted Ki67+DAPI- cells from crypts of Mki67tm1.1Cle/J mice 56h post IR and non-IR controls, related to Methods.
Table S4. Gene signature lists used in this study, related to Methods.
Data Availability Statement
Single-cell RNA-seq, RNA-seq and ATAC-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Ki67 antibody | Abcam | Cat#: ab16667, RRID:AB_302459 |
| Anti-OLFM4 antibody | Cell Signaling | Cat#: 39141, RRID:AB_2650511 |
| Anti-CD44 antibody | BD Biosciences | Cat#: 558739, RRID:AB_397098 |
| Anti-BrdU antibody | Bio-Rad | Cat#: MCA2060, RRID:AB_323427 |
| Anti-F4/80 antibody | Cell Signaling | Cat#: 70076, RRID:AB_2799771 |
| Anti-SOX9 antibody | Cell Signaling | Cat#: 82630, RRID:AB_2665492 |
| Anti-YAP antibody | Cell Signaling | Cat#: 4912, RRID:AB_2218911 |
| Anti-p-SMAD3 antibody | Cell Signaling | Cat#: 9520, RRID:AB_2193207 |
| Anti-p-SMAD2/3 antibody | Cell Signaling | Cat#: 8828, RRID:AB_2631089 |
| Anti-β-actin antibody | Abcam | Cat#: ab8227, RRID:AB_2305186 |
| Anti-Ecadherin antibody | BD Transduction Labs | Cat#: 610181 |
| Anti-Rabbit-750 | Sigma Aldrich | Cat#: SAB4600373 |
| Anti-Mouse-594 | Jackson Immuno Labs | Cat#: 715-585-150 |
| TGF beta-1,2,3 Monoclonal Antibody (1D11) | Invitrogen | Cat#: MA5-23795, AB_2609812 |
| Mouse IgG1 Isotype Control | R&D | Cat#: MAB002, RRID:AB_357344 |
| Biological samples | ||
| Human duodenal organoid lines | Michigan Translational Tissue Modeling Laboratory | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Biotium | 40043 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| Human TGF-B1 | Peprotech | 100-21 |
| OCT compound | Tissue-Tek | 4583 |
| Hematoxylin | VWR | 95057-858 |
| Eosin | Sigma Aldrich | HT110180 |
| SB525334 | Selleckchem | S1476 |
| A83-01 | Tocris | 2939 |
| Trizol | Invitrogen | 15596018 |
| SYBR Green PCR Master Mix | Applied Biosystems | 4309155 |
| Dextran Sulfate sodium salt (DSS) | Affymetrix | J14489 |
| Cultrex reduced growth factor basement membrane matrix, Type R1 | R&D | 3433-010-R1 |
| Corning Matrigel | Corning | 356231 |
| Y-27632 2HCl | Selleckchem | S1049 5mg |
| CHIR99021 | Axon Medchem | 1386 |
| Murine Noggin | Peprotech | 250-38 |
| Advanced DMEM/F-12 | Gibco | 12634-010 |
| GlutaMax | Gibco | 35050-061 |
| HEPES | Gibco | 15630-080 |
| Penicillin-Streptomycin | Invitrogen | 15140-122 |
| N-2 supplement | Gibco | 17502048 |
| B-27 supplement | Gibco | 12587-010 |
| Recombinant Human EGF Protein | R&D | 236-EG |
| Recombinant Murine EGF | Peprotech | 315-09 |
| Primocin | InvivoGen | ant-pm-1 |
| TrypLE | Thermo Fisher | 12604-013 |
| Dispase | Stem Cell Technologies | 07913 |
| DNAse I | Sigma Aldrich | D4513 |
| HBSS, no Calcium, no Magnesium | Thermo Fisher | 14170120 |
| Collagenase IV | Worthington Biochemical Corporation | LS004188 |
| Collagenase D | Roche | 11088858001 |
| Dispase II | Gibco | 17105041 |
| Fetal Bovine Serum (FBS) | Gibco | 26140-095 |
| TSA Buffer | ACD | 322809 |
| Clondronate Liposomes | FormuMax Scientific | F70101C-NC-10 |
| DAB | Amresco | 0430 |
| Critical commercial assays | ||
| KAPA Mouse Genotyping Kits | KAPA Biosystems | KK7352 |
| Illumina Tagment DNA Enzyme and Buffer Large Kit | Illumina | 20034198 |
| QIAGEN RNeasy Micro Kit | Qiagen | 74004 |
| SuperScript III First-Strand Synthesis SuperMix | Invitrogen | 18080-400 |
| Pierce BCA Protein Assay kit | Thermo Fisher | 23225 |
| Mouse TGF beta 1 ELISA kit | Abcam | ab119557 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 Assay | ACD | 323110 |
| Anti-F4/80 MicroBeads and MS column-based cell separation kit | Miltenyi Biotec | 130-110-443 |
| Mouse growth factor array C3 kit | RayBiotech | AAM-GF-3-4 |
| PIPseq T2 3’ Single Cell Capture and Lysis Kit v2.1 | Fluent Biosciences | N/A |
| Vectastain ABC HRP Kit | Vector Labs | PK-6101 |
| MinElute PCR purification kit | Qiagen | 28004 |
| Deposited data | ||
| RNA-seq, scRNA-seq and ATAC-seq data | This study | GEO: GSE222505 |
| RNA-seq of crypts upon IR | Qu et al., 2021 | GEO: GSE165157 |
| scRNA-seq of normal crypts and irradiated crypts | Ayyaz et al., 2019 | GEO: GSE117783 |
| scRNA-seq of duodenum/jejunum boundary samples upon IR | N/A | GEO: GSE165318 |
| scRNA-seq of sorted Msi1-GFP positive cells (irradiation-resistant) and their progeny cells upon IR | Sheng et al., 2020 | GEO: GSE145866 |
| SMAD4 ChIP in mouse intestinal epithelium | Chen et al., 2019 | GEO: GSE112946 |
| Experimental models: Organisms/strains | ||
| Mouse: Villin-Cre-ERT2 | el Marjou et al., 2004 | JAX: 020282 |
| Mouse: UBC-Cre-ERT2 | Ruzankina et al., 2007 | JAX: 007001 |
| Mouse: Villin-Cre | Madison et al., 2002 | JAX: 004586 |
| Mouse: Tgfbr2f/f | Leveen et al., 2002 | JAX: 012603 |
| Mouse: Smad4f/f | Yang et al., 2002 | JAX: 017462 |
| Mouse: Sox9f/f | Akiyama et al., 2002 | JAX: 013106 |
| Mouse: Mki67tm1.1Cle/J (Ki-67 RFP) | Basak et al., 2014 | JAX: 029802 |
| Mouse: PDGFRa-H2B-EGFP | Hamilton et al., 2003 | JAX: 007669 |
| Mouse: ACTB-EGFP | Okabe et al., 1997 | JAX: 006567 |
| Mouse: ROSA26mT/mG (ACTB-tdTomato-EGFP) | Muzumdar et al., 2007 | JAX: 007676 |
| Mouse: NOD SCID | Blunt et al., 1995 | JAX: 001303 |
| Mouse: C57BL/6 | The Jackson Laboratory | JAX: 000664 |
| Software and algorithms | ||
| Scanpy | Wolf et al., 2018 | https://scanpy.readthedocs.io |
| Velocyto | La Manno et al., 2018 | http://velocyto.org/ |
| GSEA | Subramanian et al., 2005 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Heatmapper | Babicki et al., 2016 | http://www.heatmapper.ca |
| BEDTools | Quinlan, 2014 | https://bedtools.readthedocs.io/en/latest/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Deeptools | Ramirez et al., 2016 | https://github.com/deeptools/deepTools |
| MACS | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| MACS2 | Zhang et al., 2008 | https://github.com/topics/macs2 |
| CutAdapt | Martin, 2011 | https://cutadapt.readthedocs.org |
| Picard | Broad Institute, 2019 | http://broadinstitute.github.io/picard |
| Haystack | Pinello et al., 2018 | https://github.com/pinellolab/haystack_bio |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/motif |
| IGV | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv |
| DiffBind | Stark and Brown, 2011 | http://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Cufflinks | Trapnell et al., 2012 | http://cufflinks.cbcb.umd.edu |
| Kaluza | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| ImageJ | NIH | https://imagej.nih.gov/ |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


