Abstract
Treatment of wound biofilm infections faces challenges from both pathogens and uncontrolled host immune response. Treating both issues through a single vector would provide enhanced wound healing. Here, we report the use of a potent cationic antimicrobial polymer to generate siRNA polyplexes for dual-mode treatment of wound biofilms in vivo. These polyplexes act both as an antibiofilm agent and a delivery vehicle for siRNA for the knockdown of biofilm-associated pro-inflammatory MMP9 in host macrophages. The resulting polyplexes were effective in vitro, eradicating MRSA biofilms and efficiently delivering siRNA to macrophages in vitro with concomitant knockdown of MMP9. These polyplexes were likewise effective in an in vivo murine wound biofilm model, significantly reducing bacterial load in the wound (~99% bacterial clearance) and reducing MMP9 expression by 80% (qRT-PCR). This combination therapeutic strategy dramatically reduced wound purulence and significantly expedited wound healing. Taken together, these polyplexes provide an effective and translatable strategy for managing biofilm-infected wounds.
Graphical Abstract
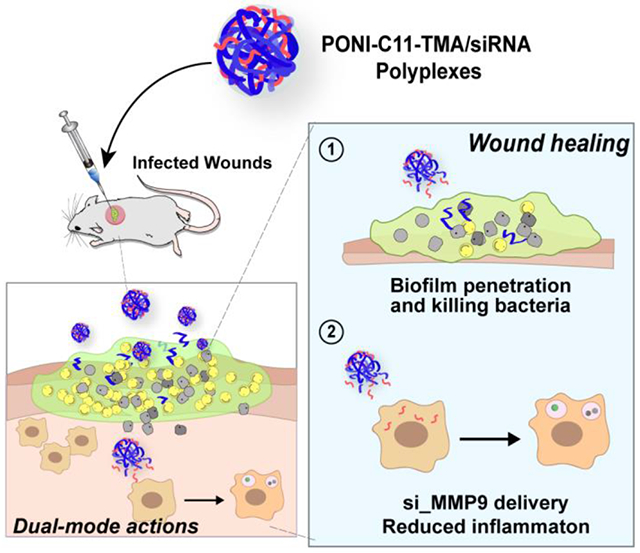
Dual therapeutics use cationic polymers to kill bacteria and deliver siRNA to macrophages to decrease inflammation and enhance wound healing.
Introduction
Chronic non-healing wounds affect 40 million patients worldwide annually, resulting in an estimated cost of USD 100 billion to treat.1 Biofilm infections are the predominant driver of these chronic infections.2 The presence of necrotic tissues and high moisture content in the wound area promote bacterial attachment and proliferation, ultimately leading to the formation of biofilms.3 Once established, these biofilms create a bio-barrier comprised of extracellular polymeric substances (EPS) secreted by bacteria.4,5 The EPS matrix prevents antibiotics and other treatments from reaching the infected wound area.6 This protective microenvironment of biofilm promotes the maturation of embedded bacteria, making antibiotic-based treatment challenging and frequently requiring surgical interventions.7,8
Beyond protecting biofilms from synthetic antimicrobials, the EPS matrix obstructs the access of phagocytic immune cells to the bacteria,9 rendering the host response ineffective in clearing the infection.10 Importantly, the persistent presence of pathogens induces continuous cycles of inflammatory responses, with high levels of inflammatory factors and proteinases within the wound.11 This chronic wound inflammation causes tissue damage, impaired re-epithelialization, and delayed wound healing.6,12,13
Current clinical practice for treatment of wound infections normally focuses on clearing bacteria with long-term and high-dosage treatment topical and/or systemic antibiotics, coupled with frequent wound debridement.14 These strategies have limitations arising from from antibiotic resistance and limited biofilm penetration.15 Besides limited antibiofilm activity, standard biofilm treatment strategies do not address other negative impacts of biofilms on the wound healing process, including prolonged inflammatory state, and tissue damage with consequent delay in reparative cell migration.16,17
Concurrent biofilm killing and modulation of inflammation could offer a promising strategy for accelerated wound healing and treatment of wound biofilm infections.18 The use of a single vector for these two uses would provide an important step in holistic wound treatment. Recently, peptide-loaded hydrogels have been used to target wound infection and inflammation simultaneously.19 An alternative strategy integrating fast-acting antimicrobials with anti-inflammatory siRNA into a single delivery system has the potential for an enhanced wound healing process, a combination that to our knowledge has not been used before. Antimicrobial activity occurs quickly, often in a few hours or less.20,21 In contrast, siRNA knockdown and concomitant anti-inflammatory activity take longer (generally >24 h),22 allowing the host immune system to combat the infection prior to anti-inflammatory action.
Polymeric nanoparticles provide a versatile toolkit to combat wound biofilm infections through antimicrobial action.4, 23 Engineering the physicochemical properties of polymers, including size, hydrophobicity, charge, and surface functionality, enables effective antimicrobial activity and biofilm penetration. 24 , 25 , 26 In our previous work, we reported the antimicrobial activity of poly(oxanorborneneimide) (PONI)-based polymers with long alkyl chains through effective bacterial membrane disruption against a broad spectrum of bacterial strains both Gram-positive and negative.4,27,28 The amphiphilic nature of this antimicrobial polymer is engineered using cationic sidechains, with quaternary ammonium groups appended with a C11 alkyl chain (PONI-C11-TMA, Figure 1).26 In parallel research, structurally related PONI polymers with guanidinium groups were used to provide efficient delivery of siRNA for the treatment of lung inflammation in a mouse model.29
Figure 1.
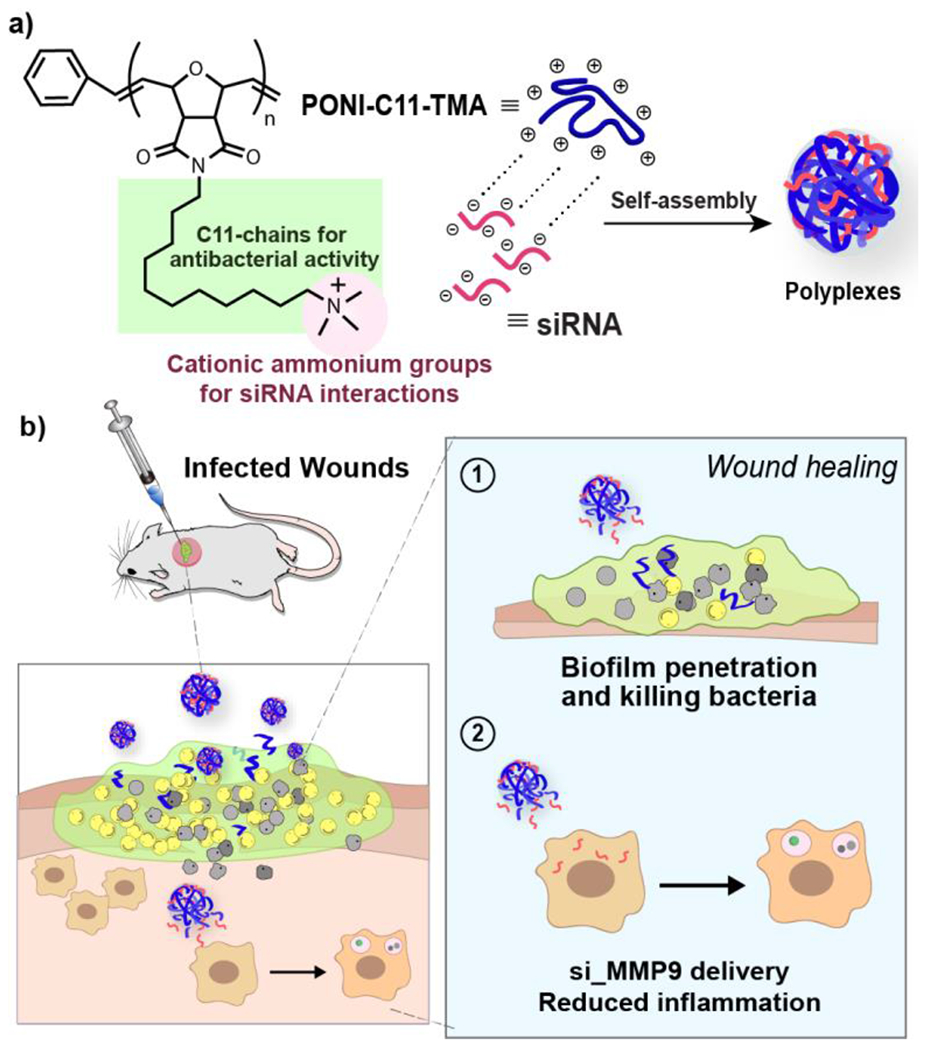
Schematic representation of a) engineering polyplexes via electrostatic interactions of siRNA and PONI-C11-TMA and b) in vivo treatment of polyplexes for infected wounds on mice showing efficient biofilm penetration and eradication of bacteria combined with si_MMP9 delivery strategy induced reduction in inflammation.
Both the antibacterial action and siRNA delivery of PONI polymers build upon key PONI polymer structural features: 1) cationic moieties that interact with negatively charged siRNA while providing the polymers essential interactions with bacterial membrane for antimicrobial activity; 2) the “semi-arthritic” structure of the PONI backbone pre-organized semi-rigid platform that facilitates self-assembly with siRNA cargo30 and provides an efficient presentation of amphiphilic cationic groups for antimicrobial efficacy and cationic polyplexes formation.17
We hypothesized that with appropriate sidechain choice, the PONI polymer scaffold could be used for both antimicrobial activity and siRNA delivery to host immune cells for enhanced wound healing. We report here a dual-mode strategy for the treatment of wound biofilm infections using PONI-C11-TMA/siRNA polyplexes (Figure 1). In this platform, PONI-C11-TMA formed polyplexes through electrostatic complexation with siRNA targeting matrix metalloproteinase 9 (MMP9), a proinflammation-associated protease. The polyplexes demonstrated excellent penetration and eradication of methicillin-resistant Staphylococcus aureus (MRSA) biofilms, and effective knockdown of MMP9 expression in macrophages and fibroblasts in vitro. In vivo efficacy was demonstrated in a MRSA wound biofilm model. The polyplexes exhibited potent antimicrobial activity, effectively clearing 99% (~2 log units) of bacterial load. The polyplexes also achieved an 80% knockdown of MMP9 mRNA expression at the wound site. This dual-mode wound healing strategy significantly improved wound healing, surpassing the results of the PONI polymer alone or clinically indicated vancomycin controls. Overall, the modularity, antimicrobial efficacy and anti-inflammatory modalities of these polymer-siRNA dual-action polyplexes provide a powerful strategy for topical treatment of wound biofilm infections.
Cationic antimicrobial polymer, PONI-C11-TMA was synthesized, characterized, and used for experiments, as it provided the highest antimicrobial efficacy in our previous reports.24,27 Briefly, PONI-C11-TMA (24K MW) polymer was synthesized using ring-opening metathesis polymerization (ROMP) with a third-generation Grubbs catalyst, yielding polymers with low polydispersity (PDI: 1.03). The molecular weight and PDI of the synthesized polymers were measured using gel permeation chromatography (GPC, PMMA calibrated) in tetrahydrofuran (Figure S1).
The synthesized PONI-C11-TMA was then used to form polyplexes with siRNA through simple co-incubation of polymer (N, number of quaternary ammonium groups) and siRNA (P, number of phosphate groups) at varied quaternary ammonium/phosphate (N/P) ratios (details are available in Supporting Information, Experimental Section, Table S1). Stable complexation between PONI-C11-TMA polymer and siRNA was confirmed by gel mobility shift assay showing that PONI-C11-TMA could completely retard the migration of siRNA at over N/P 10 (Figure 2a). Dynamic light scattering (DLS) measurements showed that the polyplexes exhibited discrete particles with an average diameter of ~100 nm and low PDI (< 0.1) (Figure 2b). PONI-C11-TMA alone self-assembles into nanoparticles with an average size of 15 nm. The observed increase in the size upon complexation with siRNA is consistent with successful formation of polyplexes with narrow size distribution. Moreover, the overall surface charge of the formulated polyplexes shifted to a less positive value (+6 to + 9 mV, Figure 2c) compared to that of PONI-C11-TMA only (+25 mV), as measured by zeta potential.15 These results demonstrated that the average surface charge of the resulting polyplexes increased with N/P ratio due to the increased charge contribution by cationic PONI-C11-TMA polymers. Transmission electron microscopy (TEM) images further confirmed the sizes of PONI-C11-TMA/siRNA polyplexes and showed that the system has spherical morphology (Figure 2d, Figure S2).
Figure 2.
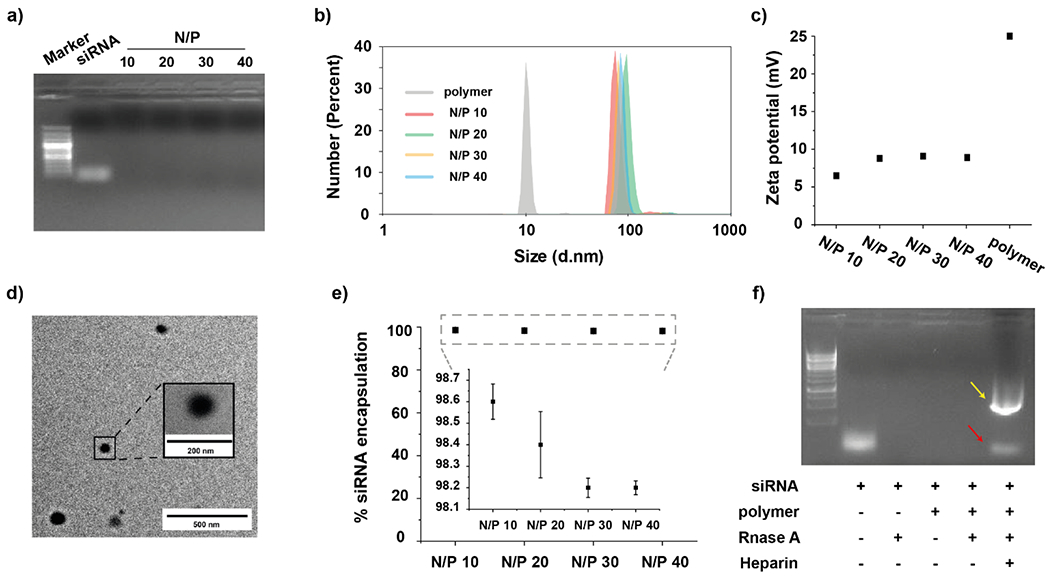
Characterization of PONI-C11-TMA/siRNA polyplexes. a) Agarose gel electrophoresis of PONI-C11-TAM/siRNA polyplexes at varied quaternary ammonium/phosphate (N/P) ratios. b) Hydrodynamic diameters by number and c) zeta potential of PONI-C11-TMA/siRNA polyplexes at varied N/P ratios. d) Representative transmission light microscopy (TEM) micrographs of polyplexes at N/P 40. e) Encapsulation efficiency of siRNA in polyplexes at varied N/P ratios. f) Nuclease protection of PONI-C11-TMA/siRNA polyplexes at N/P 40 incubated with Rnase A. Yellow arrow and red arrow indicate SDS and release siRNA respectively.
Next, the encapsulation efficiency of siRNA was verified using RiboGreen assay (Figure 2e). All formulations yielded siRNA encapsulation efficiency values of more than 96%, indicating efficient encapsulation siRNA. Additionally, polyplexes were prepared with serum media and incubated with RNase A to evaluate the stability of siRNA when exposed to physiological conditions (Figure 2f). Polyplexes efficiently protected siRNA from enzymatic degradation by RNase A, while free siRNA was fully degraded.
Having confirmed the formation of the polyplexes, we next verified the biofilm penetration ability of the formulated polyplexes. The EPS matrix of biofilms acts as a physical barrier impeding the entry of therapeutics. Therefore, we first evaluated the polyplex penetration into the biofilm using confocal laser scanning microscopy (CLSM). The penetration profiles of polyplexes were monitored using RFP-expressing MRSA biofilms and Cy5.5-labeled polymers. As shown in Figure 3a, polyplexes successfully penetrated the biofilm matrix and co-localized with bacterial cells. Next, minimum inhibitory concentrations (MIC) and minimum bactericidal concentrations (MBC) of PONI-C11-TMA polymers and PONI-C11-TMA_siRNA polyplex were determined against clinically isolated MRSA (IDRL-6169). Interestingly, the MIC of both systems was observed at 160 nM (Table S2), but MBC of polyplexes was 2-fold higher than polymer alone possibly due to a less positive charge with negatively charged siRNA presented in the system (Figure S3). Then, we screened polyplexes formulated with varied N/P ratios for antimicrobial activity against biofilms. Notably, the polyplexes exhibited high antimicrobial activity by showing a similar 4-log reduction of colony forming units (CFU) as compared to polymer alone control (Figure 3b).
Figure 3.
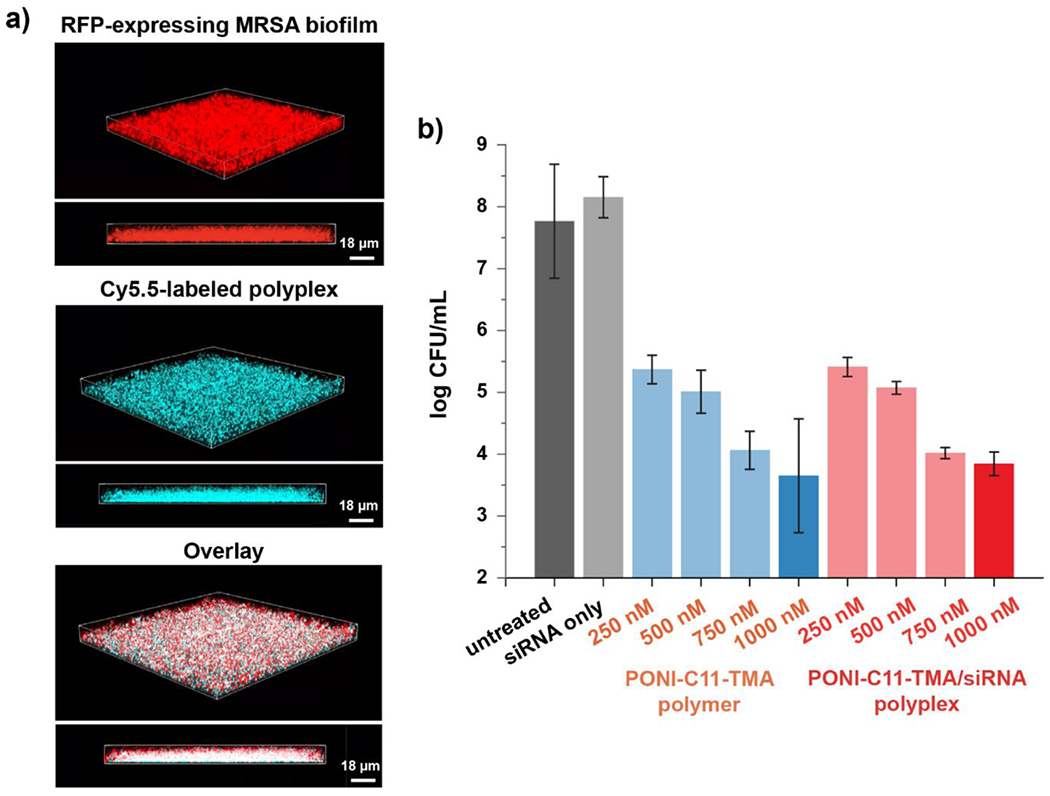
Effects of polyplexes on methicillin-resistant S. aureus (MRSA) biofilm. a) Representative 3D views of confocal image stacks of red fluorescent protein (RFP)-expressing MRSA biofilm after 1 h incubation with Cy5.5-labelled polyplexes (Cyan). Overlay images show Cy5.5-polyplexes completely penetrate the entire biofilm, interacting with MRSA cells. Biofilm thickness is ~ 18μm. b) Screening the polymer and the polyplexes formulated with different N/P ratios via their antimicrobial activity against MRSA IDRL-6169 for biocompatibility.
The host cell toxicity of all the formulations was tested using Alamar Blue cell viability and hemolysis assays. The results showed minimal cytotoxicity towards macrophages and absence of hemolytic activities at concentration levels capable of killing the biofilms, supporting lack of host toxicity of the polyplexes (Figure S4). Given the observed efficacy and safety of the library of polyplexes, we moved forward for the subsequent studies with the formulation (N/P ratio 40) that showed the highest antimicrobial effects.
Next, gene knockdown by the PONI-C11-TMA/siRNA polyplexes was investigated.31 We investigated the efficacy of gene knockdown in macrophages. Stably transfected eGFP-expressing macrophages (RAW 264.7:eGFP) were incubated with polyplexes targeting eGFP (100 nM siRNA) at N/P 40 for 24 h. Then, eGFP expression profile was monitored using CLSM (Figure 4a). The eGFP expression was confirmed and quantified by flow cytometry and data analyses revealed that PONI-C11-TMA/si_eGFP polyplex showed effective knockdown of eGFP expression (~90%) (Figure 4b). As expected, no eGFP silencing was observed when the cells were treated with PONI-C11-TMA polymer only.
Figure 4.
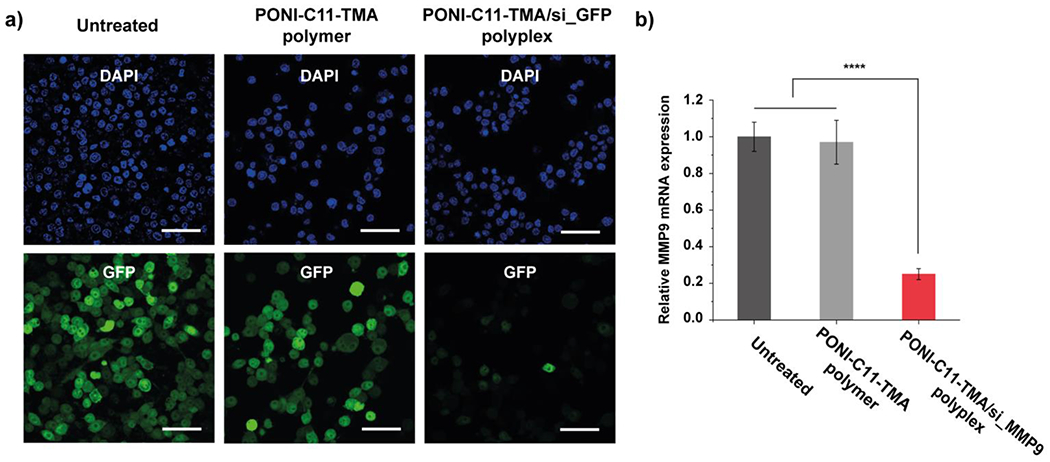
Fluorescent reporter gene silencing and evaluation of siRNA activity. a) Representative confocal microscopy images of cells after treatment with PONI-C11-TMA/si_eGFP polyplexes. Cell nuclei stained with DAPI (Blue). Deliveries were performed with polyplexes formulated at N/P 40 ratio with 100 nM of siRNA. Scale bar: 50 μm. b) Evaluating siRNA activity through MMP9 knockdown in RAW 264.7 macrophages quantified by qRT-PCR. Error bars represent the standard deviation (SD) of three experimental replicates (Data are presented as mean ± SD, one-way ANOVA, and Tukey multiple comparisons, ****p< 0.001).
Next, we evaluated the delivery of therapeutic matrix metalloproteinase (MMP9) siRNA by PONI-C11-TMA/si_MMP9 polyplexes. MMP9 was selected as a therapeutic target given MMP9 the crucial role the protein plays in tissue remodeling and inflammation in chronic wounds.32 MMP9 mRNA expression after the treatment with the polyplexes was verified using qRT-PCR demonstrated significant (~80%) knockdown in macrophages (Figure S5). The polymer alone did not silence the gene as expected due to the lack of cytotoxicity of the polymer.
MMP9 knockdown and subsequent anti-inflammatory effect occur over a longer timeline compared to the rapid-onset antimicrobial activity of the polymer. Notably, the PONI-C11-TMA/si_MMP9 polyplexes disrupt biofilm quickly (3 hours), comparable to polymer alone. This rapid antimicrobial action allows the antimicrobial to work in concert with the host immune system to combat the infection prior to anti-inflammatory action following MMP9 gene silencing.
Encouraged by in vitro efficacy of the polyplexes, we evaluated the system in vivo as a topical wound biofilm therapeutic. We used a murine model of wound biofilm infection with bioluminescent MRSA USA300 NRS384 strain (SAP-231) as the infecting pathogen (Figure 5a). The wound was created by making a 5-mm defect at the dorsum of the mice, infected with 107 CFU MRSA/animal then incubated for 24 h to develop the MRSA biofilm, as confirmed by scanning electron microscopy (SEM) image (Figure 5b). Stable bioluminescent signals on the wound area confirmed established biofilm formation; mice that did not exhibit bioluminescence were not used for further studies. The mice were then randomly grouped into four groups (n=4) to receive one of the following: 1) phosphate-buffered saline (PBS) only, 2) PONI-C11-TMA polymer only 3) PONI-C11-TMA/si_MMP9 polyplex or 4) vancomycin only. Treatments were topically administered once daily for four days. Bioluminescence was monitored to qualitatively assess the activity of the treatment groups. As shown in Figure 5c, mice treated with polymer or polyplex showed no bioluminescent signal on the last day of treatment while mice treated with PBS or vancomycin showed minimal to no change in bioluminescence. After the last day of treatment, mice were sacrificed via CO2 asphyxiation, and then 3 mm skin samples were collected from the inner wound area for quantitative bacterial load determination. Quantitative colony counting using tryptic soy agar plates was used to quantify the remaining bacterial load at the infection site. Results showed that the polyplexes retained their potent antibiofilm activity, with a bacterial reduction of approximately ~2 log units (~99 % reduction), which is similar to that of the polymer only (Figure 5d). Moreover, polymer or polyplex treatments showed better efficacy than the standard treatment, vancomycin, which exhibited bacteria reduction of only ~0.5 log units.
Figure 5.
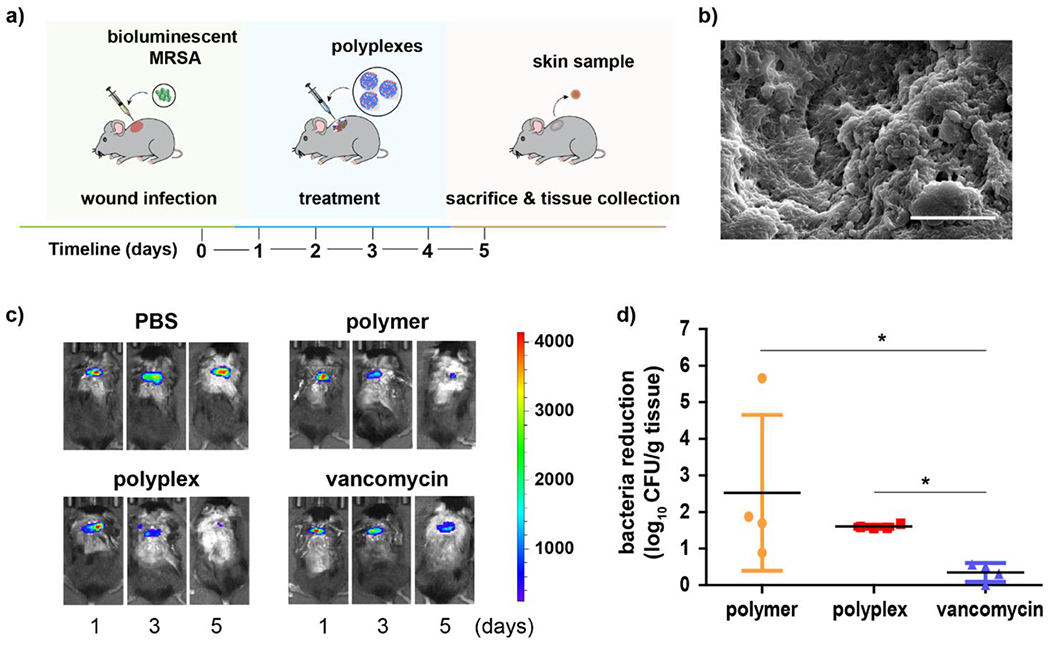
In vivo therapeutic efficacy of PONI-C11-TMA/siRNA polyplexes for severe wound biofilm infections. a) Schematic representation of the murine model of wound biofilm infection. b) SEM image of mice skin sample confirming biofilm formation at the wound site. Scale bar: 5 μm. c) Bioluminescence signals from the wound area of representative mice on the different days of treatment. d) Extent of bacterial reduction relative to negative control, PBS only [bacteria reduction = (log CFU count)PBS - (log CFU count)treatment group]. Error bars represent the mean ± the standard error of the mean (SEM) (n=4, one-way ANOVA and Tukey multiple comparisons, *p < 0.05).
Inflammation is an essential step in the wound healing process at the infected area, however, the prolonged presence of biofilm on the wound can extend the inflammatory state, slowing down or inhibiting other processes in the wound healing cascade.33 Given pus formation is an indicator of infection and inflammation, we monitored the degree of purulence of the wounds using a reported scoring system to assess the effects of the treatment groups on the inflammatory phase of the wound healing process of the infected mice (Figure 6a).34,35 Mice treated with PONI-C11-TMA/si_MMP9 polyplexes obtained the lowest scores, ranging from 1-2, indicating a low level of inflammation. This result is consistent with effective knockdown of MMP9, an inflammatory protease. Mice treated with PONI-C11-TMA polymer had scores averaging at 3, a reduced level of inflammation relative to the PBS control or vancomycin. Effective knockdown of MMP9 in wound samples was using verified qRT-PCR (Figure 6b). Quantifying the MMP9 mRNA levels in the collected tissue samples revealed that treatment with polyplexes resulted in an 80% knockdown of MMP9, while polymer controls have MMP9 mRNA levels comparable to the negative control, PBS group.
Figure 6.
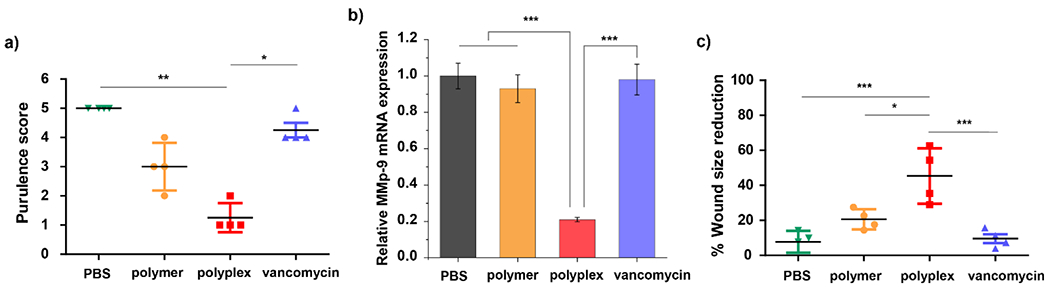
Therapeutic efficacy of PONI-C11-TMA/siRNA polyplexes mediated gene knockdown to induce wound healing in vivo. a) Purulence scores of the mice treated with PBS, polymer (PONI-C11-TMA), polyplex (PONI-C11-TMA/si_MMP9 polyplex), or vancomycin on the day of sacrifice. Kruskal-Wallis test and Dunn’s multiple comparisons, *, ** indicate p-value < 0.05 or 0.01, respectively. b) In vivo treatment of PONI-C11-TMA/si_MMP9 decreased MMP9 mRNA levels quantified by qRT-PCR. Error bars represent the mean ± the standard error of the mean (SEM) (n=4, one-way ANOVA and Tukey multiple comparisons, ***p < 0.001. c) Degree of wound size reduction at the day of sacrifice relative to the first day of treatment. One-way ANOVA and Tukey multiple comparisons, *, **, *** indicate p-value < 0.05, 0.01 or 0.001, respectively.
The combination of antimicrobial action and MMP9 knockdown substantially aided wound healing. Mice treated with the polyplexes demonstrated up to 50% wound closure, significantly better than polymer alone (~25%) or vancomycin alone (~15%) (Figure 6c, Figure S6). Histological analysis of the skin in the wound site also indicated enhanced wound healing for groups treated with the polyplexes (Figure S7). Hematoxylin and eosin staining (H&E staining) revealed increased re-epithelialization with polyplex treatment compared to polymers alone. Meanwhile, groups treated with vancomycin and PBS were still at the early stages of the wound healing process, with an abundance of proteinaceous debris and inflammatory cells. Overall, the dual-mode of polyplexes allowed effective siRNA delivery and maintained antimicrobial effects, resulting in reduced inflammation and improved wound healing. These results highlight the potential of PONI-C11-TMA/siRNA polyplex as an effective therapeutic platform for treating severe wound infections.
Conclusions
In summary, we present a dual-mode antimicrobial/anti-inflammatory strategy that provides a single vector system that is promising non-surgical strategy for the treatment of chronic non-healing wounds caused by biofilm infections. Through combined antimicrobial properties and siRNA delivery capabilities, the engineered PONI-C11-TMA/siRNA polyplexes effectively penetrated and eradicated MRSA biofilms, while also reducing MMP9 expression in vitro. In vivo studies demonstrated increased antimicrobial activity, decreased purulence, and significant improvement in wound healing for the polyplexes relative to the polymer alone and clinically-used vancomycin positive control. This combined approach harnesses the strengths of both antimicrobial and anti-inflammatory modalities, leveraging their disparate timeframes to optimize the host immune response and enhance the wound healing process. Overall, the integration of antimicrobial activity with immunomodulation provides new avenues for the treatment of chronic biofilm infections.
Supplementary Material
New Concepts.
Chronic wound biofilm infections affect 40 million patients worldwide. Biofilms in wound beds present a reservoir for pathogens and result in a prolonged inflammatory phase that delays the wound healing cascade. The protective barrier presented by the biofilm limits antimicrobial efficacy and, causes uncontrolled inflammatory responses that inhibit skin repair. Multimodal therapeutics that address killing the pathogen and modulating immune response have the capability of holistic treatment of wound biofilm infections. We used electrostatic complementarity between a poly(oxanorborneneimide) cationic antimicrobial polymer and anionic small interfering RNA (siRNA) to generate self-assembled polyplexes. These systems combine potent antibiofilm activity with efficient delivery of immunomodulatory siRNA to generate dual-mode wound therapeutics. This platform effectively kills bacteria in wounds while reducing inflammation. This combined activity significantly reduces wound purulence and promotes wound healing.
Acknowledgment
This research was supported by the National Institutes of Health under R01 AI134770 and EB022641. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Bacteria samples of methicillin-resistant S. aureus of clinical isolates from the Infectious Diseases Research Laboratory at Mayo Clinic were kindly provided by Dr. Robin Patel. The bioluminescent MRSA USA300 NRS384 strain, SAP-231, was kindly gifted by Dr. Roger Plaut. Microscopy data was obtained at the Light Microscopy Facility and Nikon Center of Excellence at the Institute for Applied Life Sciences (IALS), UMass Amherst with support from the Massachusetts Life Sciences Center. In vivo work was carried out at UMass Animal Care Facility.
Footnotes
Conflicts of Interest
There are no conflicts to declare.
References
- 1.Sen CK, Adv. Wound Care, 2019, 8, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei D, Zhu XM, Chen YY, Li XY, Chen YP, Liu HY and Zhang M, Chin. Med. J. (Engl), 2019, 132, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razdan K, Garcia-Lara J, Sinha VR and Singh KK, Drug Discov. Today, 2022, 27, 2137–2150. [DOI] [PubMed] [Google Scholar]
- 4.Flemming H-C and Wingender J, Nat. Rev. Microbiol, 2010, 8, 623–633. [DOI] [PubMed] [Google Scholar]
- 5.Muhammad MH, Idris AL, Fan X, Guo Y, Yu Y, Jin X, Qiu J, Guan X and Huang T, Front. Microbiol, 2020, 11, 530515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makabenta JMV, Nabawy A, Li C-H, Schmidt-Malan S, Patel R and Rotello VM, Nat. Rev. Microbiol, 2021, 19, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA and Kjelleberg S, Nat. Rev. Microbiol, 2016, 14, 563–575. [DOI] [PubMed] [Google Scholar]
- 8.Arciola CR, Campoccia D and Montanaro L, Nat. Rev. Microbiol, 2018, 16, 397–409 [DOI] [PubMed] [Google Scholar]
- 9.Domenech M, Ramos-Sevillano E, García E, Moscoso M and Yuste J, Infect. Immun, 2013, 81, 2606–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu YK, Cheng NC and Cheng CM, Trends Biotechnol., 2019, 37, 505–517. [DOI] [PubMed] [Google Scholar]
- 11.Zhao G, Usui ML, Lippman SI, James GA, Stewart PS, Fleckman P and Olerud JE, Adv. Wound Care, 2013, 2, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omar A, Wright JB, Schultz G, Burrell R and Nadworny P, Microorganisms, 2017, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnaswamy VR, Mintz D and Sagi I, Biochim. Biophys. Acta - Mol. Cell Res, 2017, 1864, 2220–2227. [DOI] [PubMed] [Google Scholar]
- 14.Dhar Y and Han Y, Eng. Regen, 2020, 1, 64–75. [Google Scholar]
- 15.Roy R, Tiwari M, Donelli G and Tiwari V, Virulence, 2018, 9, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demidova-Rice TN, Hamblin MR and Herman IM, Adv. Ski. Wound, 2013, 25, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzer S, James GA, Goeres D, Bjarnsholt T, Vickery K, Percival SL, Stoodley P, Schultz G, Jensen SO and Malone M, J. Infect, 2020, 80, 261–270. [DOI] [PubMed] [Google Scholar]
- 18.Puthia M, Butrym M, Petrlova J, Strömdahl AC, Andersson M, Kjellström S and Schmidtchen A, Sci. Transl. Med, 2020, 12, 6601. [DOI] [PubMed] [Google Scholar]
- 19.Puthia M, Butrym M, Petrlova J, Strömdahl AC, Andersson M, Kjellström S and Schmidtchen A, Sci. Transl. Med, 2020, 12, 6601. [DOI] [PubMed] [Google Scholar]
- 20.Vogelman B and Craig WA, The Journal of Pediatrics, 1986, 108, 835–840. [DOI] [PubMed] [Google Scholar]
- 21.Corvaisier S, Maire PH, Bouvier d’yvoire MY, Barbaut X, Bleyzac N and Jelliffe RW, Antimicrob. Agents Chemother, 1998, 42, 1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiyong H, Methods Mol. Biol, 2018, 1706, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, Mumtaz S, Li CH, Hussain I and Rotello VM, Chem. Soc. Rev, 2019, 48, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engler AC, Shukla A, Puranam S, Buss HG, Jreige N and Hammond PT, Biomacromolecules, 2011, 12, 1666–1674. [DOI] [PubMed] [Google Scholar]
- 25.Si Z, Zheng W, Prananty D, Li J, Koh CH, Kang E-T, Pethe K and Chan-Park MB, Chem. Sci, 2022, 13, 345–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta A, Landis RF, Li CH, Schnurr M, Das R, Lee YW, Yazdani M, Liu Y, Kozlova A and Rotello VM, J. Am. Chem. Soc, 2018, 140, 12137–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makabenta JMV, Park J, Li CH, Chattopadhyay AN, Nabawy A, Landis RF, Gupta A, Schmidt-Malan S, Patel R and Rotello VM, Molecules, 2021, 26, 4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopalakrishnan S, Gupta A, Makabenta JMV, Park J, Amante JJ, Chattopadhyay AN, Matuwana D, Kearney CJ and Rotello VM, Advanced Healthcare Materials, 2022, 11, 2201060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon T, Luther DC, Goswami R, Bell C, Nagaraj H, Cicek YA, Huang R, Mas-Rosario JA, Elia JL, Im J, Lee YW, Liu Y, Scaletti F, Farkas ME, Mager J and Rotello VM, ACS Nano, 2022, 17, 4315–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YW, Luther DC, Goswami R, Jeon T, Clark V, Elia J, Gopalakrishnan S and Rotello VM, J. Am. Chem. Soc, 2020, 142, 4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead KA, Langer R and Anderson DG, Nat. Rev. Dru. Discov, 2009, 8, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caley MP, Martins VLC and O’Toole EA, Adv. Wound Care, 2015, 4, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cutting KF, Br. J. Community Nurs, 2003, 8, S4–S9.12819589 [Google Scholar]
- 34.Kim CK, Karau MJ, Greenwood-Quaintance KE, Tilahun AY, Krogman A, David CS, Pritt BS, Patel R and Rajagopalan G, Toxins (Basel)., 2015, 7, 5308–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C-H, Landis RF, Makabenta JM, Nabawy A, Tronchet T, Archambault D, Liu Y, Huang R, Golan M, Cui W, Mager J, Gupta A, Schmidt-Malan S, Patel R and Rotello VM, Mater. Horizons, 2021, 8, 1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


