Abstract
In this study, we have identified a novel Nedd4-like ubiquitin ligase, BUL1, as the host factor involved in budding of type D retrovirus Mason-Pfizer monkey virus (M-PMV). Overexpression of BUL1 enhanced virus particle release, while a BUL1 mutant in which a W to G substitution was introduced into a WW domain, W791G, lost the ability to bind to the viral Gag protein and abolished its ability to mediate virus budding. In addition, a fragment of BUL1 containing only the WW domains inhibited virus budding in a dominant negative manner. These results, together with previous findings, indicate that the M-PMV Gag L domain interacts with the BUL1 WW domain and that this interaction is essential for virus budding. Our observations provide new insights into the mechanism of virus budding, and could be useful in establishing new antiviral strategies targeted at progeny virus release from a host cell.
INTRODUCTION
We previously showed that a PPPY sequence within the Mason-Pfizer monkey virus (M-PMV) Gag polyprotein plays a critical role in a late step in M-PMV virion budding and release (Yasuda and Hunter, 1998). This proline-rich (PPxY) motif is conserved among most retroviruses and has been identified in rhabdoviruses and filoviruses. It has been defined as the L domain essential for viral budding (Wills et al., 1994; Parent et al., 1995; Xiang et al., 1996; Yuan et al., 1999; Harty et al., 1999, 2000; Jayakar et al., 2000), suggesting that budding of these viruses proceeds via an analogous mechanism.
The PPxY motif has also been determined as the core sequence binding to WW domains, a sequence of 38 amino acids containing two widely spaced tryptophans, which is involved in protein–protein interactions (Sudol, 1996). Thus, it is conceivable that the function required for virus budding involves an interaction between the PPxY motif-containing viral proteins and a WW domain-containing cellular protein (Garnier et al., 1996; Xiang et al., 1996). At present, of those cellular proteins containing a WW domain, Nedd4 or Nedd4-like proteins are considered the most likely potential candidates. This is because ubiquitylation of retroviral Gag proteins or some associated cellular component appears to be required for virus budding, and Nedd4 and its yeast homologue Rsp5 possess the ability to ubiquitylate target proteins containing a PPxY motif as a E3 ubiquitin ligase (Huibregtse et al., 1995; Anan et al., 1998; Wang et al., 1999; Ott et al., 2000; Patnaik et al., 2000; Strack et al., 2000).
It was shown recently that the VSV M and Ebola virus VP40 proteins could bind to the yeast Rsp5 containing WW domains in vitro (Harty et al., 1999, 2000, 2001). In addition, Kikonyogo et al. (2001) reported that the Nedd4-related protein, LDI-1, interacts with the L domain of RSV Gag in cells cotransfected with both RSV Gag and LDI-1 expression plasmids, and that the WW domain of this protein inhibited RSV Gag budding in a dominant negative manner. However, it is not clear whether LDI-1 is the host factor that is functionally involved in the RSV budding process, since this protein is not intact and does not positively regulate RSV budding. Thus, the identities of cellular proteins that play critical roles in non-lentiviral budding and facilitate or promote virus budding have not been established.
Here, we demonstrate that a Nedd4-like protein, BUL1, interacts with the L domain of M-PMV Gag and facilitates viral budding, while a BUL1 fragment containing just the WW domains inhibits virus budding. This is the first report to demonstrate that a member of the ubiquitin ligase family, BUL1, is functionally involved in viral budding.
RESULTS AND DISCUSSION
Cellular factors interacting with the M-PMV L domain
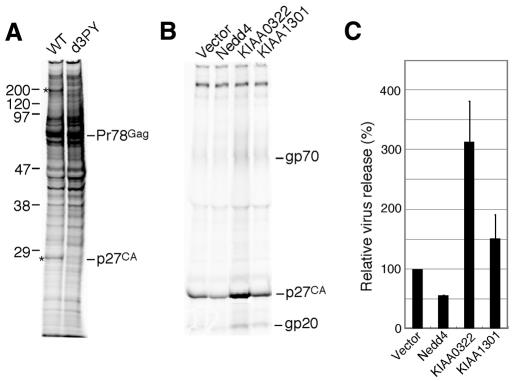
To identify the host factor involved in M-PMV budding, we transfected pSHRM15, which encodes a wild-type M-PMV genome, or pd3PY, which encodes a mutant M-PMV genome with a deletion of the PPPY-encoding sequence in the Gag region (Yasuda and Hunter, 1998), into COS-7 cells and analysed the proteins that coimmunoprecipitated with wild-type or mutant M-PMV Gag.
Two specific bands, at 27 and 200 kDa, were detected only in wild-type M-PMV-expressing cells, but not in those expressing d3PY (Figure 1A). The 27 kDa protein is presumably the viral capsid protein p27, since we previously showed that the mutant Gag precursor Pr78 expressed from pd3PY was stable and very little p27 could be detected even after an 8 h chase (Yasuda and Hunter, 1998). Therefore, the 200 kDa protein was considered to be a candidate for the budding-associated host factor.
Fig. 1. Identification of the cellular factors involved in M-PMV budding. (A) Proteins that interact with the PPPY sequence within the L domain of M-PMV. COS-7 cells were transfected with either pSHRM15 (WT) or pd3PY and labelled for 6 h at 48 h after transfection. The cells were lysed with TNE buffer, and then the lysate was subjected to immunoprecipitation with rabbit anti-Pr78gag antiserum. The proteins coimmunoprecipitated with Gag were resolved by SDS–PAGE. Asterisks on the left indicate differences between wild-type and d3PY. The apparent molecular masses of these components are shown on the left (in kDa). (B) Effects of the expression of Nedd4, KIAA0322 (BUL1) or KIAA1301 on virus production. COS-7 cells were cotransfected with pSHRM15 and expression plasmids for Nedd4, KIAA0322 or KIAA1301. Extracellular virions were pelleted from the culture fluids of the labelled cells and then subjected to immunoprecipitation with rabbit anti-M-PMV antiserum. The migration positions of viral proteins are indicated on the right. These results were representative of three independent experiments. (C) Intensity of the band for viral capsid protein p27 in (B) was quantitated as described in Methods. The extent of p27 released from wild-type proviral vector-transfected cells was set to 100%. The data represent averages and SDs of three independent experiments.
To search for Nedd4-like proteins that have a molecular mass of ∼200 kDa, we performed a BLAST analysis of nucleotide and protein sequence databases for genes that encode a protein larger than 200 kDa and contain the HECT domain. Two clones, KIAA0322 and KIAA1301 (DDBJ/EMBL/GenBank accession Nos AB002320 and AB037722, respectively), matched this search, and encoded deduced ORFs of 1562 and 1580 amino acids, respectively. The clones are very similar and contain a C2 domain at the N-terminus, two WW domains located in the middle part of the protein, and a HECT domain at the C-terminus (Nagase et al., 1997, 2000; Harvey and Kumar, 1999).
Effect of the overexpression of Nedd4-like clones on M-PMV budding
To examine the effect of Nedd4, KIAA0322 and KIAA1301 expression on the virus production by M-PMV, we next cotransfected the expression plasmid for each Nedd4-like molecule with pSHRM15 into COS-7 cells, and compared the virus releases from these transfected cells into the culture media. As shown in Figure 1B and C, overexpression of KIAA0322 significantly enhanced virus release, while Nedd4 appeared to inhibit virus production. KIAA1301 slightly facilitated M-PMV particle release from cells.
These results suggest that the KIAA0322 gene product interacts with the L domain of M-PMV Gag and plays an important role in virus budding. Although Nedd4 may also bind to the L domain of M-PMV Gag, Nedd4 appears to negatively regulate virus budding in a dominant negative manner because of its lack of an additional unknown activity required for budding.
We named the product translated from KIAA0322 BUL1 as Budding-associated Ubiquitin Ligase 1.
Interaction of Gag with BUL1 in cells
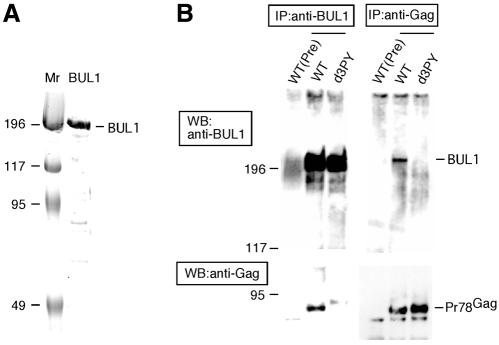
To confirm whether M-PMV Gag interacted with BUL1 in vivo, we investigated the direct interaction between M-PMV Gag and endogenous BUL1 in cells expressing M-PMV proteins by coimmunoprecipitation. Prior to this analysis, we purified the recombinant BUL1 from Escherichia coli expressed BUL1 (Figure 2A) and generated anti-BUL1 antisera by immunizing rabbits using the purified BUL1. The extracts of COS-7 cells transfected with pSHRM15 or pd3PY were immunoprecipitated with either anti-BUL1 or anti-Gag antibodies and then the immunoprecipitates were analysed by western blotting using either anti-Gag or anti-BUL1 antibodies, respectively. As shown in Figure 2B, immunoprecipitation with anti-BUL1 or anti-Gag antibodies for cell extracts expressing wild-type Gag (WT) coimmunoprecipitated Gag or endogenous BUL1, respectively, while immunoprecipitation for the cell extracts expressed the mutant Gag deleted PPPY motif (d3PY) did not show any coimmunoprecipitates of Gag and BUL1. As a control, preimmune sera precipitated neither BUL1 nor Gag from the extract of cells transfected with pSHRM15. These results indicated that M-PMV Gag interacts with endogenous BUL1 through the PPxY motif in vivo.
Fig. 2. Interaction of M-PMV Gag with BUL1. (A) The recombinant BUL1 protein purified from the bacterial extracts expressed BUL1 was resolved by SDS/7.5% PAGE and the gel was stained with Coomassie Brilliant Blue. (B) COS-7 cells transfected with pSHRM15 (WT) or pd3PY (d3PY) were lysed and processed to prepare cell extracts as described in Methods. IP, cell extracts were immunoprecipitated with anti-BUL1, anti-Gag or pre-immune (Pre) sera; WB, immunoprecipitated proteins were separated on a 7.5% resolving gel by SDS–PAGE and then subjected to western blotting with anti-Gag or anti-BUL1 antibodies.
Analyses of the BUL1 mutants
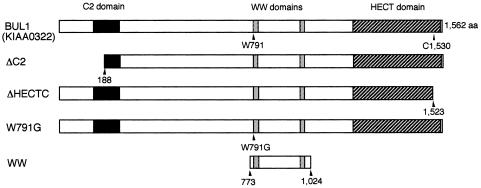
To clarify the functional involvement of BUL1 on M-PMV budding, we constructed BUL1 mutants with specific deletions or point mutations (Figure 3) and examined the effect of BUL1 mutant expression on M-PMV production (Figure 4).
Fig. 3. Schematic representation of the BUL1 mutants. BUL1 contains an N-terminal C2 domain (black boxes), two WW domains (grey boxes) and a C-terminal HECT domain (dashed boxes). All constructs were expressed as proteins containing a Myc-tag at the N-terminus. The cysteine residue at position 1530 is critical for the ubiquitin ligase activity of HECT domain proteins (Anan et al., 1998; Goulet et al., 1998; Harvey and Kumar, 1999).
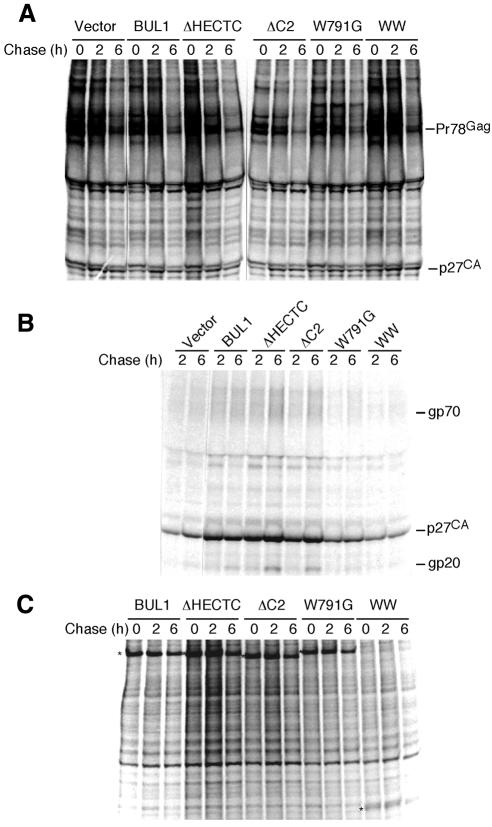
Fig. 4. Pulse–chase analysis of COS-7 cells coexpressing M-PMV and various BUL1 mutants. At 48 h after transfection of each BUL1 mutant DNA with pSHRM15 into COS-7 cells, cells were pulse-labelled and then chased for 2 or 6 h. Extracellular virions were pelleted from the culture fluids of chased cells. M-PMV-specific cell-associated (A) or virion-associated (B) proteins were immunoprecipitated with anti-M-PMV antiserum. The positions of M-PMV proteins are indicated on the right. (C) Expression of BUL1 mutants was also examined by immunoprecipitation with anti-c-Myc antiserum. Asterisks on the left indicate the migration positions of the wild-type or BUL1 mutants.
After transfection of each BUL1 mutant DNA with pSHRM15 into COS-7 cells, pulse–chase experiments were carried out. In COS-7 cells coexpressing M-PMV and either wild-type or mutant BUL1 molecules, the Gag protein precursors were processed into mature proteins at similar rates, as evidenced by the appearance of p27 (CA) (Figure 4A). This indicates that expressions of wild-type and mutant BUL1s have no effect on Gag stability or processing in M-PMV-producing cells.
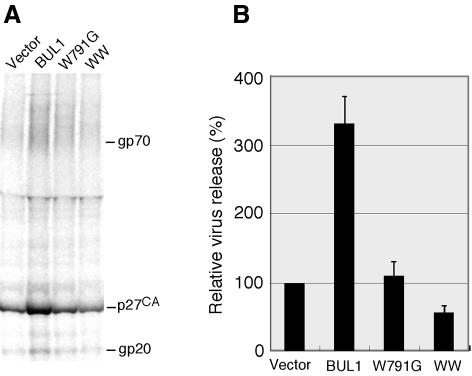
As shown in Figure 4B, two of the deletion mutants, ΔC2, which deleted the putative C2 domain, and ΔHECTC, which deleted 39 amino acids from the C-terminal region containing the active site cysteine in the HECT domain, enhanced viral budding to an extent similar to that of wild-type BUL1. This suggests that neither the putative C2 region nor the ubiquitin ligase activity are required for the function of BUL1 in viral budding, although we have not confirmed that the deletions within each mutant inactivate the putative C2 or HECT functions. In contrast, the W791G mutant, which disrupts the first WW motif, did not enhance virus release, and the amounts of extracellular viral proteins were similar to those seen in pSHRM15-transfected cells. Thus, the ability of BUL1 to enhance virus budding was completely abolished by the W to G substitution, suggesting, as predicted, that BUL1 associated with the L domain of Gag via the WW domain. In the cells cotransfected with the WW construct, which expresses only the region containing the two WW domains, the culture supernatant contained significantly less virion-associated protein than that seen with the wild-type genomic plasmid, indicating that this construct efficiently inhibited M-PMV budding in a dominant negative manner despite its low level of expression (Figure 4B and C).
A quantitative analysis of the effect of BUL1 and the dominant negative construct expression in virus budding is shown in Figure 5A and B. Cotransfection of the BUL1 expression plasmid enhanced M-PMV budding more than 3-fold. In contrast, the coexpression of the WW mutant inhibited virus budding by >50%. In this experiment, the expression level of the WW mutant was again lower than that of BUL1 (data not shown), consistent with the result from the pulse–chase experiment (Figure 4C). It seems likely that modification of the WW construct to yield high levels of expression could induce a much greater reduction in virus budding.
Fig. 5. Quantitative analysis for the effect of BUL1 and its mutants on virus budding. COS-7 cells were transfected with pSHRM15, wild-type or mutant BUL1 DNA, and pCDMβ-gal and labelled for 7 h with a [35S]methionine/cysteine mixture. (A) Radiolabelled virus particles released from cells into the culture medium were prepared as described previously (Yasuda and Hunter, 1998) and subjected to immunoprecipitation with rabbit anti-M-PMV antiserum. (B) Intensity of the band for viral capsid protein p27 in (A) was quantitated as conducted in Figure 1C. The data represent averages and SDs of three independent experiments.
Taken together, our results show that BUL1 is functionally involved in M-PMV budding. Furthermore, the first WW domain of BUL1 is critical for the interaction with the L domain of M-PMV, and a small fragment of BUL1 containing only the WW domains functions as a dominant negative mutant, suggesting that our observations could be useful for the development of antiviral agents targeted the viral budding step.
Recently, Garrus et al. (2001) reported that Tsg101, which functions in vacuolar protein sorting (Vps), binds to the PTAP motif identified as the L domain within HIV Gag, and is required for HIV-1 budding. A dominant negative mutant of Vps4, which functions in Tsg101 cycling and endosomal trafficking in the Vps pathway, arrested virus budding of not only of HIV-1 but also of MLV, which contains a PPPY-based L domain, indicating that the Vps pathway is required for viral budding through both the PTAP and PPPY motifs. BUL1 is a Nedd4-like E3 ubiquitin ligase and could bind to E2 or UEV-containing proteins such as Tsg101. Therefore, BUL1 may recruit M-PMV Gag to the Vps pathway through interaction with UEV or E2. Identification of the putative E2-binding domain of BUL1, the UEV or E2 protein interacting with BUL1, and the physiological targets of BUL1 will be important for understanding the functional role of BUL1 in virus budding and development of antiviral strategies.
METHODS
Plasmids. KIAA0322 and KIAA1301 clones (Nagase et al., 1997, 2000) were gifts from Dr T. Nagase (Kazusa DNA Research Institute). BamHI sites were introduced by standard PCR into KIAA1301 and all of the KIAA0322 (BUL1) constructs at both 5′ and 3′ ends, and subcloned into the pBj-Myc-hNedd4, which was constructed for Nedd4 expression (Anan et al., 1998), by replacing Nedd4 with each DNA fragment using BamHI. For the W791G mutant, a tryptophan to glycine substitution at position 791 was introduced by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The accuracy of all constructs was confirmed by DNA sequencing analysis.
Radiolabelling and coimmunoprecipitation. COS-7 cells were maintained at 37°C in a 5% CO2 incubator in DMEM supplemented with 10% fetal bovine serum. Either pSHRM15 or pd3PY was transfected into COS-7 cells by using Lipofectamine plus reagent (Gibco-BRL). At 48 h after transfection, cells were pulse-labelled for 3 h with a [35S]methionine/cysteine mixture (ICN) and then lysed with TNE buffer (10 mM Tris–HCl pH 7.8, 0.1% NP-40, 0.15 M NaCl, 1 mM EDTA, 10 µg/ml aprotinin). Lysates were used for immunoprecipitation with rabbit anti-M-PMV Pr78gag antiserum. Coimmunoprecipitated proteins with wild-type or mutant Gag were separated on a 10–20% resolving gel by SDS–PAGE. For the detection of the interaction between Gag and endogenous BUL1 in Figure 2, cell lysates were prepared by different methods. Briefly, cells transfected with pSHRM15 were washed with cold PBS at 48 h post-transfection and then collected from dishes by a cell scraper after addition of 0.8 ml of Hypotonic buffer (10 mM Tris–HCl pH 7.5/1 mM MgCl2). Cells were then lysed by homogenizing and subjected to centrifugation. The soluble fraction was analysed by immunoprecipitation followed by western blotting.
Detection and quantification of extracellular virion. At 48 h after transfection, COS-7 cells were labelled for 7 h with a [35S]methionine/cysteine mixture. Radiolabelled virus particles released from cells into the culture medium were prepared as described previously (Yasuda and Hunter, 1998) and subjected to immunoprecipitation with rabbit anti-M-PMV antiserum. Immunoprecipitated proteins were electrophoresed on an SDS–polyacrylamide gel (10–20%). The intensity of the band for viral capsid protein p27 was measured by a FLA-3000 (Fujifilm). Transfection efficiency in each plate was normalized to the activity of β-galactosidase that synthesizes from the cotransfected pCDMβ-gal.
Pulse–chase experiment. At 48 h after transfection, COS-7 cells were pulse-labelled for 1 h with a [35S]methionine/cysteine mixture and chased for various periods of time in complete medium. As described previously (Yasuda and Hunter, 1998), the cells were lysed, and then the lysate was subjected to immunoprecipitation with rabbit anti-c-Myc (A-14) antiserum (Santa Cruz Biotechnology) or rabbit anti-M-PMV antiserum. Radiolabelled virus particles, which were released from the chased cells into the culture medium, were also prepared as described above and subjected to immunoprecipitation with rabbit anti-M-PMV antiserum. Immunoprecipitated proteins were separated on a 10–20% resolving gel by SDS–PAGE.
Production of anti-BUL1 antibody. Full-length BUL1 was expressed in E. coli BL21 (DE3) as a GST fusion (from pGEX6P-2; Amersham Biosciences), and purified from the bacterial extracts according to the manufacturer’s instructions. As the GST fusion proteins attached to the glutathione–Sepharose 4B beads were digested with PreScission protease, the purified BUL1 protein does not contain GST. The purified BUL1 was used then to immunize rabbits.
Supplementary data. Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Kantou Nakajima, Fumiko Kokusen, Aki Okuhara and Ayako Kanayama for their excellent technical assistance. We also thank Dr Marius Sudol and Dr Masami Yamada for providing valuable information. This work was supported by grants from the Japan Society for the Promotion of Science (JSPS), the Japan Health Sciences Foundation (JHSF) and the Naito Foundation.
REFERENCES
- Anan T. et al. (1998) Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells, 3, 751–763. [DOI] [PubMed] [Google Scholar]
- Garnier L., Wills, J.W., Verderame, M.F. and Sudol, M. (1996) WW domains and retrovirus budding. Nature, 381, 744–745. [DOI] [PubMed] [Google Scholar]
- Garrus J.E. et al. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell, 107, 55–65. [DOI] [PubMed] [Google Scholar]
- Goulet C.C., Volk, K.A., Adams, C.M., Prince, L.S., Stokes, J.B. and Snyder, P.M. (1998) Inhibition of the epithelial Na+ channel by interaction of Nedd4 with a PY motif deleted in Liddle’s syndrome. J. Biol. Chem., 273, 30012–30017. [DOI] [PubMed] [Google Scholar]
- Harty R.N., Paragas, J., Sudol, M. and Palese, P. (1999) A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol., 73, 2921–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty R.N., Brown, M.E., Wang, G., Huibregtse, J. and Hayes, F.P. (2000) A PPxY motif within the VP40 protein of ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl Acad. Sci. USA, 97, 13871–13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty R.N., Brown, M.E., Mcgettigan, J.P., Wang, G., Jayakar, H.R., Huibregtse, J., Whitt, M.A. and Schnell, M.J. (2001) Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol., 75, 10623–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K.F. and Kumar, S. (1999) Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol., 9, 166–169. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M., Scheffner, M., Beaudenon, S. and Howley, P.M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl Acad. Sci. USA, 92, 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar H.R., Murti, K.G. and Whitt, M.A. (2000) Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol., 74, 9818–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikonyogo A., Bouamr, F., Vana, M.L., Xiang, Y., Aiyar, A., Carter, C. and Leis, J. (2001) Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl Acad. Sci. USA, 98, 11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T., Ishikawa, K., Nakajima, D., Ohira, M., Seki, N., Miyajima, N., Tanaka, A., Kotani, H. and Ohara, O. (1997) Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res., 4, 141–150. [DOI] [PubMed] [Google Scholar]
- Nagase T., Kikuno, R., Ishikawa, K.I., Hirosawa, M. and Ohara, O. (2000) Prediction of the coding sequences of unidentified human genes. XVI. The complete sequences of 150 new cDNA clones from brain which code for large proteins in vitro. DNA Res., 7, 65–73. [DOI] [PubMed] [Google Scholar]
- Ott D.E., Coren, L.V., Chertova, E.N., Gagliardi, T.D. and Schubert, U. (2000) Ubiquitination of HIV-1 and MuLV Gag. Virology, 278, 111–121. [DOI] [PubMed] [Google Scholar]
- Parent L.J. et al. (1995) Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol., 69, 5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A., Chau, V. and Wills, J.W. (2000) Ubiquitin is part of the retrovirus budding machinery. Proc. Natl Acad. Sci. USA, 97, 13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B., Calistri, A., Accola, M.A., Palu, G. and Gottlimger, H.G. (2000) A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl Acad. Sci. USA, 97, 13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M. (1996) Structure and function of the WW domain. Prog. Biophys. Mol. Biol., 65, 113–132. [DOI] [PubMed] [Google Scholar]
- Wang G., Yang, J. and Huibregtse, J.M. (1999) Functional domains of the Rsp5 ubiquitin-protein ligase. Mol. Cell. Biol., 19, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J.W., Cameron, C.E., Wilson, C.B., Xiang, Y., Bennett, R.P. and Leis, J. (1994) An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol., 68, 6605–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Cameron, C.E., Wills, J.W. and Leis, J. (1996) Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol., 70, 5695–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J. and Hunter, E. (1998) A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol., 72, 4095–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B., Li, X. and Goff, S.P. (1999) Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J., 18, 4700–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.