Abstract
Background:
FOXI3 is a forkhead family transcription factor that is expressed in the progenitors of craniofacial placodes, epidermal placodes, and the ectoderm and endoderm of the pharyngeal arch region. Loss of Foxi3 in mice and pathogenic Foxi3 variants in dogs and humans cause a variety of craniofacial defects including absence of the inner ear, severe truncations of the jaw, loss or reduction in external and middle ear structures, and defects in teeth and hair.
Results:
To allow for the identification, isolation, and lineage tracing of Foxi3-expressing cells in developing mice, we targeted the Foxi3 locus to create Foxi3GFP and Foxi3CreER mice. We show that Foxi3GFP mice faithfully recapitulate the expression pattern of Foxi3 mRNA at all ages examined, and Foxi3CreER mice can trace the derivatives of pharyngeal arch ectoderm and endoderm, the pharyngeal pouches and clefts that separate each arch, and the derivatives of hair and tooth placodes.
Conclusions:
Foxi3GFP and Foxi3CreER mice are new tools that will be of use in identifying and manipulating pharyngeal arch ectoderm and endoderm and hair and tooth placodes.
Keywords: Pharyngeal arch, pharyngeal pouch, pharyngeal cleft, tooth, hair follicle, CreER and reporter mice
INTRODUCTION
Pharyngeal arches are transient embryonic structures that give rise to many craniofacial elements such as the lower jaw, middle and external ears, and endodermal structures such as the thymus, thyroid and parathyroid 1–3. Each arch is lined on its outside by ectoderm, on the inside by endoderm, and is populated by neural crest cells and endothelial precursors. Each pharyngeal arch is separated from its neighbor by constrictions of the ectoderm and endoderm that comprise the pharyngeal clefts and pouches respectively. In addition to demarcating each arch, the pouch and cleft regions can serve as signaling centers that sculpt arch development 3. A number of different human syndromes involving pharyngeal arch defects include DiGeorge syndrome, Pierre Robin Syndrome, Goldenhar syndrome and Treacher-Collins Syndrome 4–7, as well as cleft lip/palate.
Many previous studies report that signaling molecules or the transcription factors present in pharyngeal arch ectoderm or endoderm are essential for pharyngeal arch patterning 8–16. FOXI3, a member of the Forkhead transcription factor class I (Foxi) family plays a crucial role in early craniofacial development. Foxi3 mRNA is expressed in the ectoderm and endoderm of the developing pharyngeal arches, gradually localizing to the pouch/cleft regions between the arches before being down-regulated as arch derivatives differentiate 17–19. Loss of Foxi3 resulted in craniofacial abnormalities, including absence of the outer, middle and inner ears, a misshapen Meckel’s cartilage, a shortened mandible fused with the maxilla, and thymic, parathyroid and aortic arch defects 18,20. Although Foxi3 is not expressed in neural crest cells migrating into the pharyngeal arches, Foxi3 is nevertheless required for the survival of migrating crest. Loss of Foxi3 causes migrating neural crest cells entering the pharyngeal arches to undergo significant cell death, due in part from the loss of FGF8 from arch ectoderm 18. Foxi3 is also expressed in ectodermal appendage progenitors such as hair and whisker follicles, and tooth and mammary buds during embryonic development 21,22. Loss of Foxi3 in these structures leads to defects in tooth and hair development 21,23, and a semidominant neomorphic mutation in dog Foxi3 leads to partial or complete loss of hair and tooth defects in three hairless dog breeds 24–26. Variants or deletions of the FOXI3 locus in humans can also lead to craniofacial abnormalities such as microtia and craniofacial microsomia 27–29.
The specific and transient expression of Foxi3 in pharyngeal arches and the progenitors of ectodermal appendages suggests it may be a useful gene to direct the expression of reporter proteins or Cre recombinase for developmental studies of these tissues. In this study, we introduce two mouse models that can be used to alter the specific genes in ectodermal or endodermal derived tissues in the pharyngeal arches, teeth, and hair/whisker follicles. We developed Foxi3GFP mice in which Venus fluorescent protein is targeted downstream of the Foxi3 coding region. GFP labeling in these mice reveals a very similar expression pattern to Foxi3 mRNA expression previously reported in pharyngeal arches and the progenitors of ectodermal appendages 18,22,24. We created a second line of mice in which the Foxi3 coding region was replaced by a CreER construct encoding a Cre recombinase fused to a modified estrogen receptor. We used these mice to trace the lineage of Foxi3-expressing cells and labeled cells in cranial epidermis, the ear pinna and ear canal (ectodermal derivatives), and in the Eustachian tube, oropharynx, glottis, epiglottis, thyroid, parathyroid and thymus (endodermal derivatives), as well as hair/whisker follicles and tooth placodes. No Foxi3 lineage-labeled cells were present in neural crest- or mesodermal-derived tissues such as cartilages and muscles. Both mouse lines will be useful tools for investigators seeking to identify, purify and manipulate progenitors of the pharyngeal arches, their derivatives and the progenitors of teeth and hair follicles.
RESULTS AND DISCUSSION
Foxi3GFP mice show expression in the pharyngeal arches along anterior to posterior axis
We targeted the Foxi3 locus to insert a Venus fluorescent protein downstream of the Foxi3 coding region, separated by a GSG-P2A sequence to allow bicistronic expression of FOXI3 and the fluorescent reporter (Figure 1A). To validate the expression of our Foxi3GFP mice, we analyzed GFP expression in Foxi3GFP embryos at three consecutive stages E8.5, E9.5 and E10.5 (Figure 2). We previously reported the expression of Foxi3 mRNA in both ectoderm and endodermal regions of the pharyngeal arches at E8.5, with the expression becoming restricted to the clefts and pouches in an anterior-posterior direction 18,19. At E8.5 and E9.5, GFP is expressed continuously along the surface ectoderm of maxillary and mandibular regions of the first arch (Figure 2A, B, E, arrowheads). The first and second pharyngeal clefts and pharyngeal pouches at these stages show very strong GFP expression (Figure 2B, C, F G, arrowheads). By E9.5, GFP could be detected in the third pharyngeal cleft and pharyngeal pouch (Figure 2H, arrowheads). At E10.5, GFP expression along the surface ectoderm decreased in the first pharyngeal arch (Figure 2J), its expression becoming restricted to the pharyngeal pouches and clefts (Figure 2K, L). The duct arising from the third pharyngeal pouch is the primordia of thymus and parathyroid tissue and strongly expresses GFP (Figure 2M). We used light sheet microscopy to visualize these expression patterns in whole mounts of the embryonic head at E10.5 (Supplementary Movies 1–3). Together, these results show that our Foxi3GFP mice faithfully recapitulate the expression of Foxi3 mRNA at all stages of arch development and can thus be used as a reporter and as a tool to isolate Foxi3-expressing progenitors.
Figure 1: Generation of Foxi3GFP and Foxi3CreER mice.
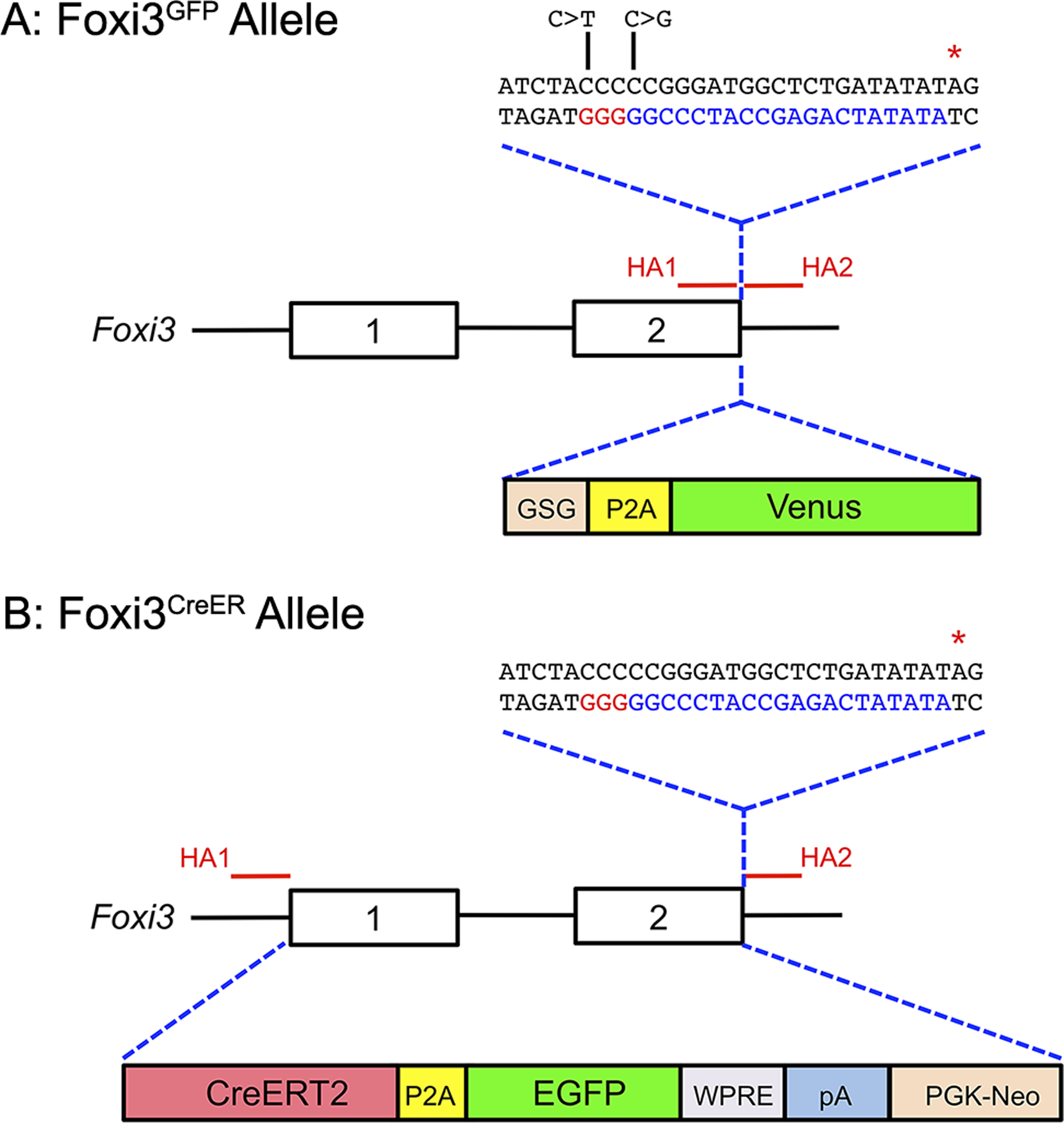
(A) Diagram showing the Foxi3 locus, including the position of the two homology arms (HA1 and HA2) used to target the locus by CRISPR-mediated homology-directed repair. The homology arms enclosed a DNA sequence coding for a GSG-P2A-Venus fluorescent protein, followed by a terminating stop codon (red asterisk). The sequence corresponding to the gRNA targeting the 3’ end of Foxi3 is shown in blue letters, with the PAM sequence shown in red letters. The DNA used for homologous recombination contained two nucleotide changes (C>T and C>G; indicated in the diagram) to prevent the inserted sequence from being targeted by the Foxi3 gRNA.
(B) Diagram showing the Foxi3 locus, including the position of the two homology arms (HA1 and HA2) used to target either end of the Foxi3 gene and insert CreER by homologous recombination in ES cells. Exons 1 and 2 and the intervening intron were replaced by an insert containing a CreERT2 fusion cDNA, a P2A sequence followed by EGFP, a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), a polyA signal, and a PGK-neo resistance cassette. Homologous recombination in ES cells was enhanced by CRISPR-mediated cleavage of the 3’ end of the Foxi3 locus using the same gRNA as in (A) above.
Figure 2: Activity of Foxi3GFP and Foxi3CreER mice in the first and second pharyngeal arches.
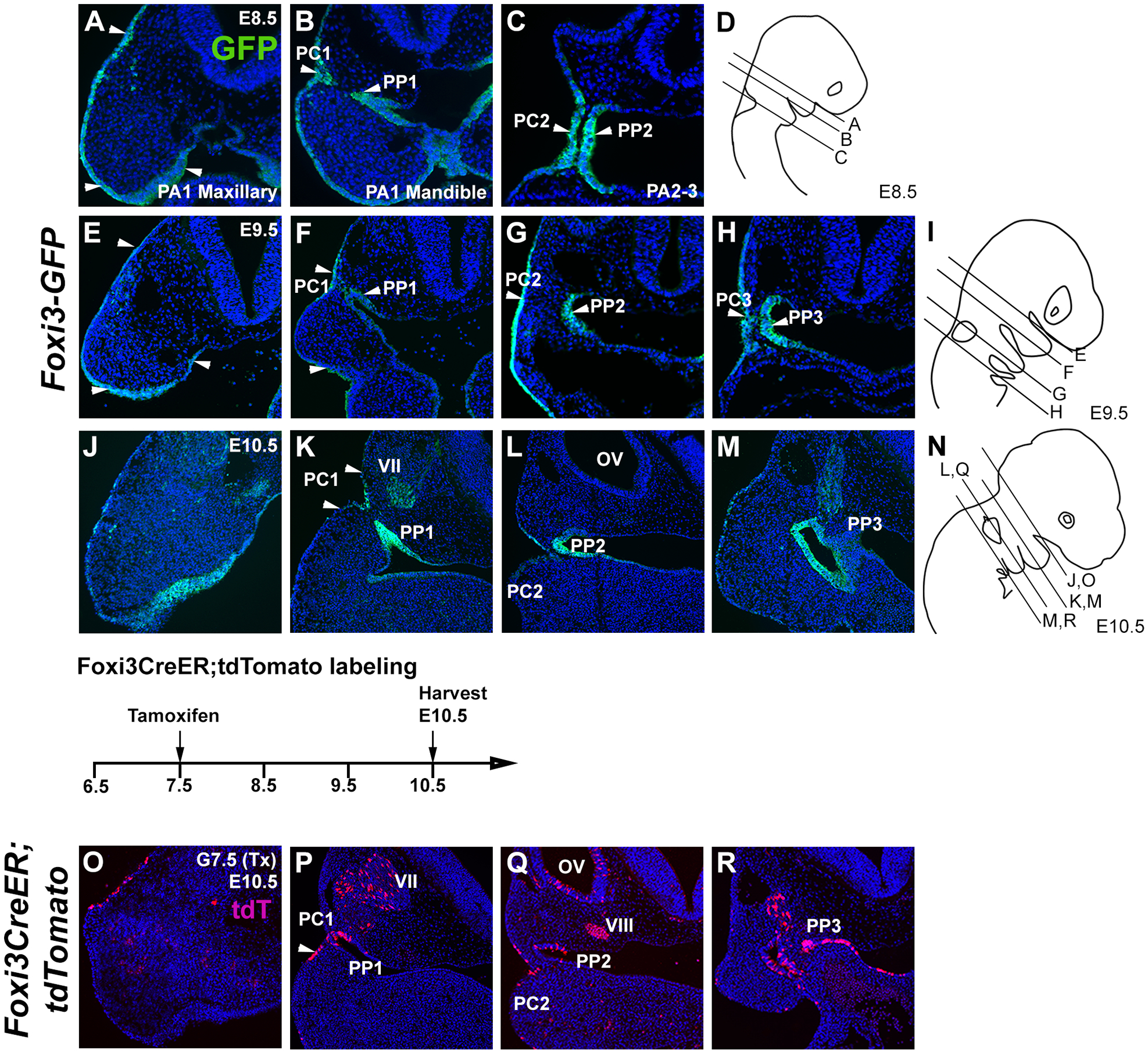
(A-N): Expression of the Foxi3GFP reporter in the first and second pharyngeal arches at E8.5, E9.5 and 10.5. Panels D, I and N illustrate the approximate planes of section for each panel at the three ages. (A-C) Foxi3GFP is expressed in the ectoderm of the mandible and maxilla of the first pharyngeal arch (PA1) at E8.5, and in the pharyngeal cleft and pharyngeal pouch between the first and second arches (PC1 and PP1). (E-H) Foxi3GFP expression begins to be down-regulated from the first arch ectoderm and endoderm by E9.5 but remains expressed in the cleft and pouch regions. Expression is also seen in the second and third cleft and pouch regions (PC2, PC3, PP2, PP3). (J-M) Foxi3GFP continues to be down-regulated from the ectoderm of the first and second pharyngeal arches but persists in the first three cleft and pouch regions. (O-R). Foxi3CreER mice were mated with ROSA-Ai9 Cre reporter mice and the pregnant females received a single dose of tamoxifen at 7.5 dpc. At E10.5, tdTomato-expressing cells can be observed in the ectoderm and endoderm of the first three cleft and pouch regions. Labeled cells can also be seen in the geniculate ganglion (VII, panel P), the otic vesicle (OV) and vestibulo-acoustic ganglion (VIII; panel Q) which are derived from Foxi3 lineage-labeled progenitors in the pre-placodal region.
Fate mapping reveals contribution of Foxi3-expressing cells to the pharyngeal arches
To map the fates of Foxi3-expressing cells, we targeted the Foxi3 locus and replaced the both Foxi3 coding exons and the intervening intron with a construct containing a tamoxifen-inducible CreER fusion construct, a GSG-P2A sequence and an EGFP sequence (Figure 1B). We employed the EGFP sequence to visualize Foxi3-expressing cells; however, we found that although the CreER protein was expressed and active, the low level of expression from the construct did not allow detection of the EGFP protein, either by native fluorescence or by immunostaining with GFP antibodies (data not shown). To determine the contribution of Foxi3-expressing cells to the pharyngeal arch region, we crossed Foxi3CreER mice to ROSA-Ai9 Cre reporter mice, administered a single moderate dose of tamoxifen (75μg/g body weight) at 7.5 days post-coitum (dpc) and harvested the embryos at E10.5. We observed a line of Foxi3 lineage-labeled cells, revealed by expression of the tdTomato reporter, in the surface ectoderm of the maxillary portion of the first pharyngeal arch (Figure 2O). Foxi3 lineage-labeled cells were also located specifically in the first, second and third pharyngeal clefts and pouches (Figure 2P–R). Additionally, we observed labeled derivatives in the cranial ganglia, otocyst and the endodermally-derived thymus-parathyroid primordia (Figure 2P–R). These results demonstrate the contribution of Foxi3 lineage-labeled cells to the ectoderm- and endodermally derived pharyngeal pouches and clefts. Foxi3 is also expressed in the pre-placodal region that gives rise to all craniofacial placodes17,19, and we also observed labeled cells in some placodal derivatives such as the trigeminal ganglion (Figure 2P) and the otic vesicle and its associated vestibulo-acoustic ganglion (Figure 2Q).
To assess the contribution of Foxi3-expressing progenitors to pharyngeal arch derivatives more fully, we analyzed Foxi3CreER embryos at later developmental stages. To examine the presence of Foxi3 lineage-labeled cells in the first two pharyngeal arches at later stages, we administered a single dose of tamoxifen at 7.5 dpc and harvested embryos at E14.5. We identified developing cartilage in the cranial region with immunostaining for Sox9. Foxi3 lineage-labeled cells were found in the ectodermal regions of the ear pinna, groove, and ear canal regions of the outer ear (Figure 3A–C). However, no Foxi3 lineage-labeled cells were seen in the regions of six auricular hillock cartilages of the pinna (Figure 3D). In the middle ear region, no Foxi3 lineage-labeled cells were found in the malleus or incus (the middle ear ossicles derived from the first arch), although small numbers of endodermally-derived Foxi3 lineage-labeled descendants could be seen in the region of the Eustachian tube (Figure 3F–I). Similarly, in the second pharyngeal arch region, the stapes (the third middle ear ossicle; Figure 3I) did not contain any Foxi3CreER labeled cells. These results suggest that Foxi3-expressing cells give rise to tissues in the first two pharyngeal arches derived from ectodermal and endodermal origins, but not to other derivatives derived from migrating neural crest cells or mesodermal cells, which do not express Foxi3 during development. Previous studies have suggested a link between some forms of craniofacial microsomia and defects of the stapedial artery leading to hemorrhage 4,30. We did not observe Foxi3 lineage-labeled cells in the stapedial artery or endothelium any other vessels of the head (not shown), suggesting that cases of craniofacial microsomia with a vascular etiology may not be directly due to FOXI3 pathogenic variants. In sum, Foxi3CreER mice can be used to follow the fate of pharyngeal ectoderm and endoderm derivatives, and to manipulate gene expression in these structures at different ages.
Figure 3: Foxi3-expressing cells contribute to the external and middle ear, but not the middle ear ossicles.
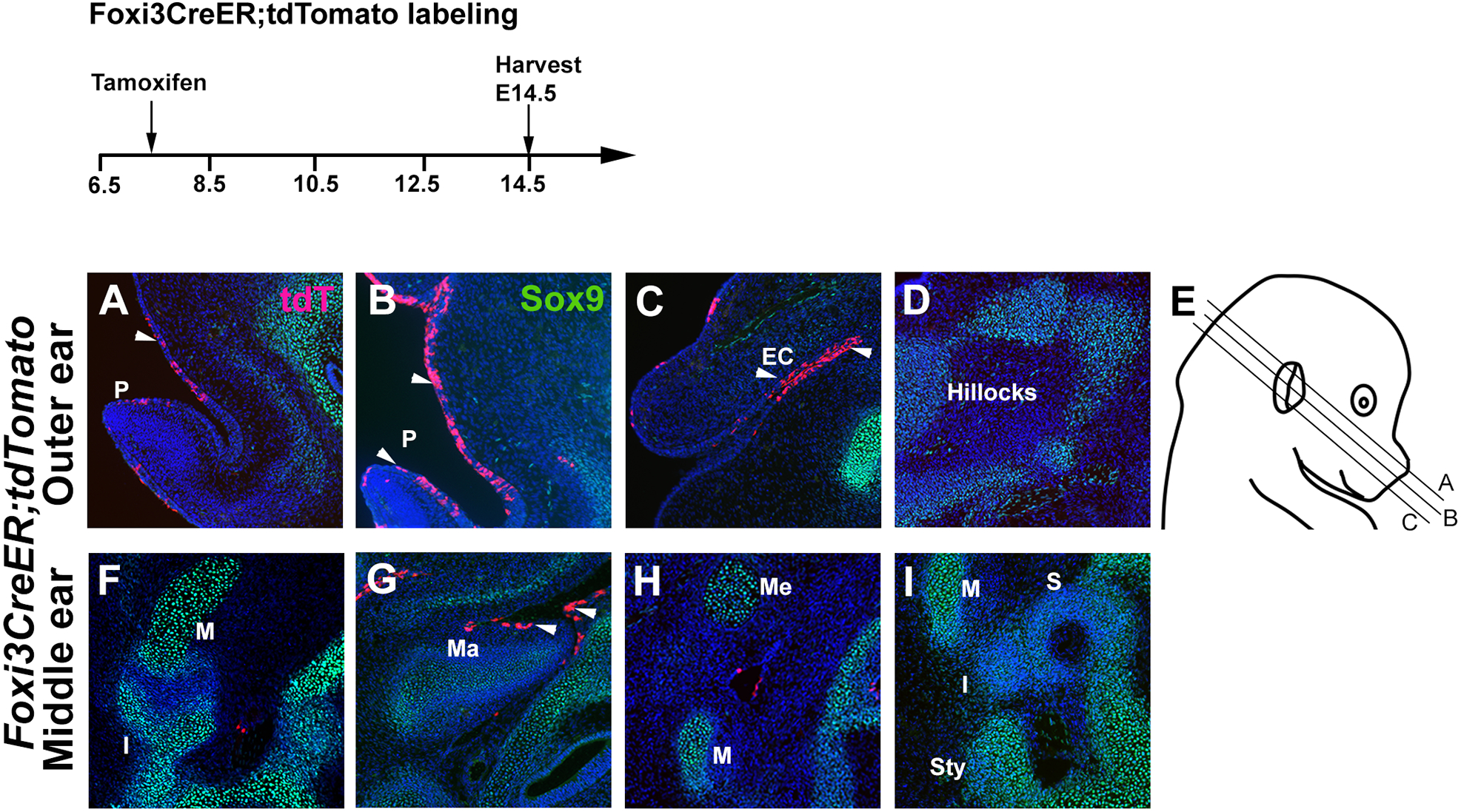
Foxi3CreER mice were mated with ROSA-Ai9 Cre reporter mice and the pregnant females received a single dose of tamoxifen at 7.5 dpc. Mice were analyzed for tdTomato expression at E14.5, together with Sox9 to show developing cartilage. (A-C) Foxi3 lineage-labeled derivatives can be seen in the ectoderm of the external ear pinna (P) and ear canal (EC), but not the mesenchymal auricular hillocks that label with Sox9 (D). (F-I) Although some cells can be observed in the endoderm of the Eustachian tube, the middle ear ossicles – the malleus (M), incus (I) and stapes (S) – are all unlabeled, as are Meckel’s cartilage (Me) and the styloid process (Sty)
Foxi3-expressing cells contribute to the posterior pharyngeal arches
Tissues derived from the posterior pharyngeal arches include elements of the cricoid, arytenoid, and thyroid cartilages which are located surrounding the glottis region, the trachea (which is of mesodermal origin) and the endodermally-derived thymus, parathyroid and thyroid tissues. We mapped the fates of Foxi3-expressing cells by administering tamoxifen at 7.5 dpc and analyzing at E14.5. Foxi3 lineage-labeled cells are present in all endodermally-derived tissues in this region including the oropharynx, glottis, epiglottis, thyroid, parathyroid and thymus tissues (Figure 4A-G). However, no Foxi3 lineage-labeled cells were found in any neural crest-derived, Sox9 -expressing cartilage elements. Thus, our results show that Foxi3 lineage-labeled cells contribute mainly to endodermal derivatives in posterior pharyngeal arches, and that Foxi3CreER mice can be used to label and isolate or manipulate the derivatives, thyroid, parathyroid and thymus tissues.
Figure 4: Foxi3-expressing cells contribute to endodermal derivatives of the third and fourth pharyngeal arches.
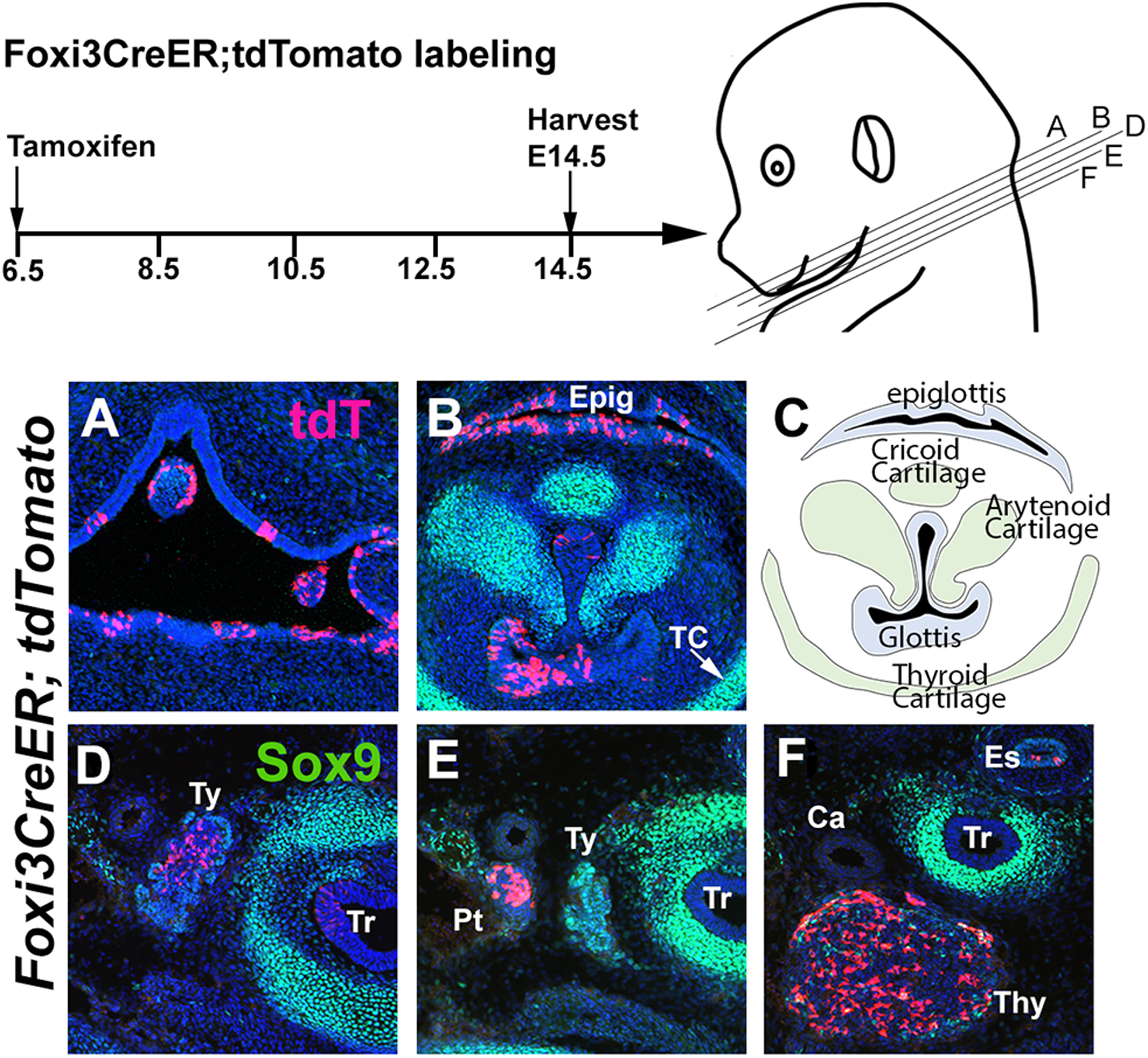
Foxi3CreER mice were mated with ROSA-Ai9 Cre reporter mice and the pregnant females received a single dose of tamoxifen at 6.5 dpc. Mice were analyzed for tdTomato expression at E14.5, together with Sox9 to show developing cartilage. TdTomato cells can be observed in the oropharynx (A), glottis and epiglottis (B), but not the cricoid, arytenoid, or thyroid cartilages (TC; B, C). (D-F) Labeled cells can also be seen in the developing thyroid (Ty), parathyroid (Pt) and thymus (Thy) tissues, as well as the esophagus (Es) and trachea (Tr).
Foxi3GFP and Foxi3CreER mice reveal the fates of Foxi3- expressing progenitors in ectodermal appendages
Previous studies reported that Foxi3 is expressed in the epithelium of all hair and whisker follicles, and in dental epithelium throughout tooth morphogenesis 22,24. We showed that our Foxi3GFP reporter was strongly expressed in ectodermal appendages including whisker and hair follicles and tooth placodes at E12.5 and E14.5. Specifically, GFP expression was strongly observed in the tooth and hair placodes at E12.5, and the epithelial compartments of invaginating whisker and hair follicles and in the tooth germs at E14.5 (Figure 5A–E) similar to what has been reported for Foxi3 mRNA in previous studies 22. GFP is also expressed in the oral epithelium near the tooth placodes (Figure 5D and E). We performed lineage analysis to locate Foxi3 lineage-labeled cells during hair follicle and tooth development. For these experiments, we administered tamoxifen at 11.5 dpc and harvested the mice at E14.5 (for hair follicle and tooth derivatives) or E18.5 (for hair follicle derivatives). Consistent with a role for Foxi3 in tooth and hair development21,23–26, we found Foxi3 lineage-labeled cells in the dental epithelium (Figure 5G, H) and the inner root sheet of hair and whisker follicles (Figure 5F, I). Foxi3GFP and Foxi3CreER mice are thus likely to be useful and specific tools to label, isolate and manipulate hair and tooth progenitors in late embryonic and postnatal animals.
Figure 5: Foxi3-expressing cells contribute to hair and tooth placodes.
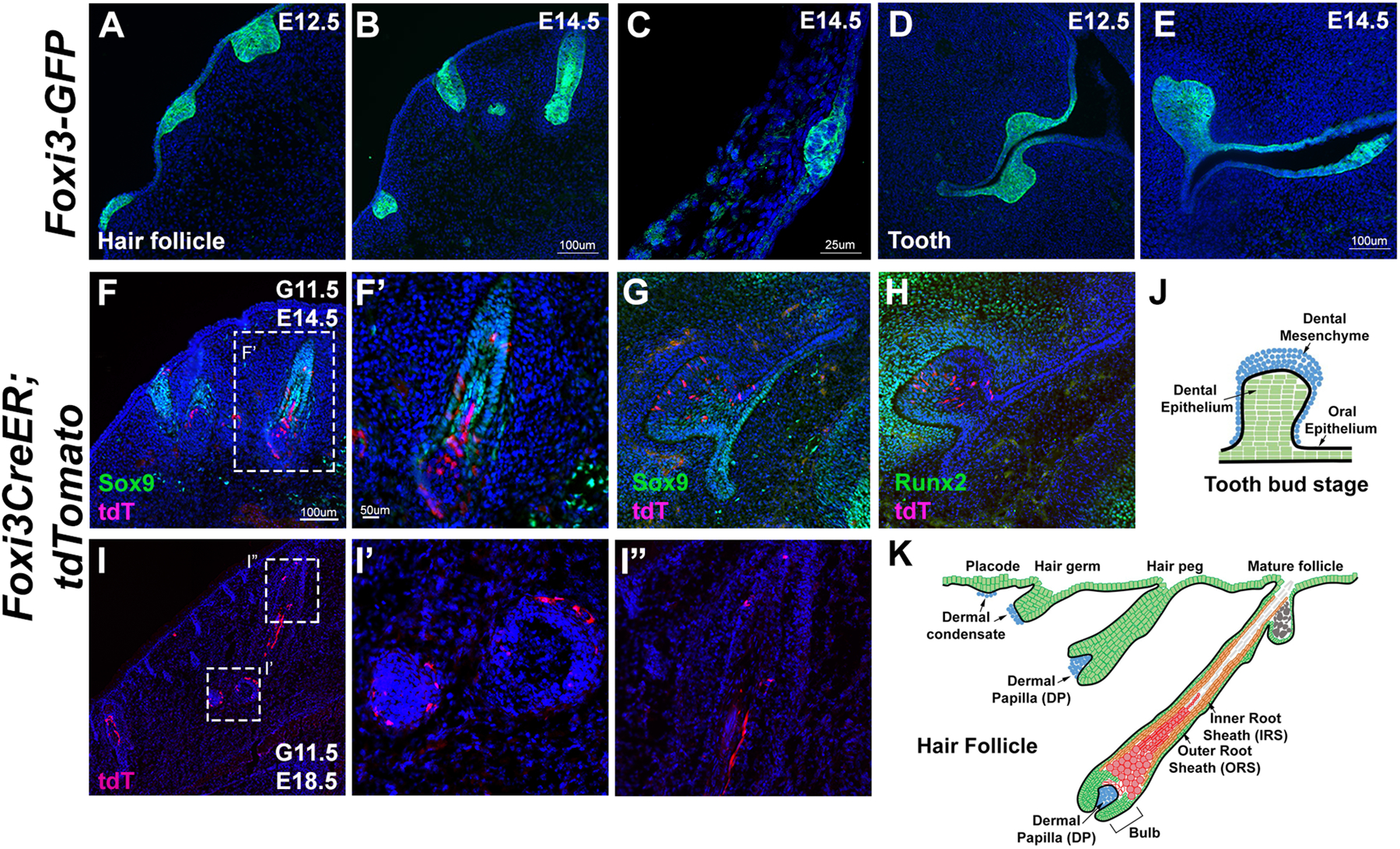
(A-E) Foxi3GFP reporter expression is seen in the progenitors of whisker follicles at E12.5 and 14.5 (A, B), hair follicles from dissected rump skin at E14.5 (C) and tooth placodes at E12.5 and 14.5 (D,E). (F-K) To label the descendants of these populations, Foxi3CreER mice were mated with ROSA-Ai9 Cre reporter mice and the pregnant females received a single dose of tamoxifen by gavage at 11.5 dpc (G11.5). Mice were analyzed for tdTomato expression at E14.5 and E18.5. Cells could be observed in the outer root sheath, cortex, and dermal papilla of the whisker follicle (F, F’, I, I’, I”, K) and in the dental and oral epithelium (G, H, J)
EXPERIMENTAL PROCEDURES
Generation of Foxi3GFP mice
Foxi3GFP reporter mice were created by the Genetically Engineered Rodent Models core at Baylor College of Medicine. The targeting strategy is shown in Figure 1A. Mouse zygotes were injected with 100ng/μl of Cas9 mRNA, 20ng/μl of a guide RNA targeting the end of the Foxi3 coding region (CCC CCGGGATGGCTCTGATATA) and 50ng/μl of a targeting ssDNA sequence. This single stranded DNA molecule contained 150 bases of the 3’ end of the Foxi3 coding region (upstream homology arm) containing two base pair alterations to prevent cleavage by the gRNA (Figure 1A), 66 bases comprising a GSG-P2A sequence, the coding region for mVenus fluorescent protein and a final 115 bases of the 3’ end of the Foxi3 gene downstream from the stop codon (downstream homology arm). Founder mice were genotyped using a forward primer (5’-TTC TCC ACA CTC CAT GCA GC-3’) and a reverse primer (5’-CCC TTT CCA AGA CGC TAA GCT-3’). A correctly targeted band was 1069bp in length; the wild type band was 285bp in length. Positive founders were outcrossed to wild type mice for five generations before the characterization described in this paper.
Generation of Foxi3CreER mice
Foxi3CreER mice were created by the Genetically Engineered Rodent Models core at Baylor College of Medicine. The targeting strategy is shown in Figure 1B. We developed a targeting construct for the Foxi3 locus containing 1456bp upstream of the Foxi3 start codon, a CreER fusion gene, a GSG-P2A sequence, an EGFP sequence, a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), polyA signal, a PGK-neo resistance cassette, and 1200bp downstream from the Foxi3 stop codon. This was targeted to mouse ES cells by homologous recombination. To enhance targeting, AB2.2 ES cells were electroporated with the linearized targeting construct (2μg) were electroporated with 20μg of a pX330 plasmid (Addgene # 42230) expressing Cas9 and a sgRNA sequence to target the 3’ end of the Foxi3 gene (CCC CCGGGATGGCTCTGATATA). Neomycin-resistant colonies were expanded and screened for correct targeting, and three correctly targeted clones were expanded and injected into 129 blastocysts to create chimeras, which were then bred to C57Bl6 mice to establish germline founders. Genotyping was performed using a forward primer (5’-AAA GCC GCT GCC GCT CTG CA-3’) and a reverse primer (5’-TTG GTC GTG GCA GCC CGG AC-3’).
Maintenance and breeding of mouse lines and tamoxifen administration
All the mice were maintained according to IACUC policies (Baylor College of Medicine). We bred Foxi3CreER mice with homozygous Rosa-Ai9 mice 31 that express tdTomato after Cre recombination. Foxi3GFP mice were maintained as homozygotes and with ICR female mice to generate embryos carrying the Foxi3GFP allele. For Foxi3CreER mouse lineage tracing, a single dose of 3mg tamoxifen and 3mg progesterone per 40gm body weight (75μg/g) was administered to pregnant mice by oral gavage at 6.5, 7.5 or 11.5 days post-coitum (dpc). Embryos from both lines were harvested at embryonic day (E) 8.5, 9.5, 10.5, 12.5, 14.5 and 18.5 respectively. For both lines, at least 10 embryos were analyzed at each embryonic age described in the Results section.
Preparation of embryos
Embryos at were harvested and fixed in 4% paraformaldehyde for overnight at 4°C, dehydrated in 30% sucrose in PBS overnight at 4°C, and finally embedded in cryomolds (Tissue-Tek, 4565) using OCT compound (Tissue-Tek, 4583) by freezing in liquid nitrogen. The tissues were sectioned at 12μm thickness using a Leica CM 1850 cryostat with sections collected on Superfrost Plus slides (Fisher scientific, 12-550-15). Sections were stored at −20°C.
Immunohistochemistry
The primary antibodies used in study were rabbit anti-GFP (1:1000, Thermofisher, A-11122), rabbit anti-Sox9 (1:1000, Millipore, AB5535), and rabbit anti-Runx2 (1:1000, Cell Signaling Technology, 12556). The secondary antibodies used for this study were: goat anti-rabbit Alexa 488 (1: 1000, Invitrogen, A11008), and goat anti-mouse Alexa 488 (1:1000, Invitrogen, A11029). Immunohistochemistry was performed based on standard protocols. Briefly, sections were dried at room temperature for 10 minutes and washed with 1X PBS to dissolve the OCT compound. The sections were blocked with blocking solution containing 1X PBS, 10% normal goat serum and 0.1% Triton X-100 at room temperature, then treated with diluted primary antibodies overnight at 4°C. After rinsing with 1% Triton X-100 in PBS at room temperature three times, they were treated with diluted secondary antibodies at room temperature for at least 1 hour, washed thoroughly with 1% Triton X-100 in PBS, followed by a 1X PBS wash, and incubation with DAPI for 5–10 minutes at room temperature. The slides were mounted with Fluoromount-G (Southern Biotech, 0100–01). The slides were imaged with a Zeiss Axio Observer microscope with an Apotome structured illumination attachment and an Axiocam using Zen 3.0 software (Blue edition).
Light Sheet Microscopy
Embryos were harvested at E10.5 from ICR females crossed with Foxi3GFP homozygous mice and were fixed in 4% paraformaldehyde overnight at 4°C. Samples were washed with 1X PBS and incubated in blocking solution (1% Triton-X 100, 1% BSA, 10% Horse serum in 1X PBS) overnight at room temperature, treated with primary GFP antibody diluted at 1:500 in antibody solution (0.1% Triton-X 100, 1% BSA, 5% Horse serum in 1X PBS) for at least three days at room temperature, washed with 0.1% Triton-X100 in 1X PBS, treated with secondary antibody (Goat anti-rabbit Alexa 488) diluted at 1:1000 in antibody solution for at least three days, washed with 0.1% Triton-X100 in 1X PBS, incubated in DAPI for another overnight at room temperature. The samples were washed with 1X PBS for three times and processed for tissue clearing using a modified EZ Clear protocol32. Embryos were incubated in EZ Clear solution (~80% Nycodenz - Accurate Chemical & Scientific #100334–594, 0.05% sodium azide prepared in 0.02 M sodium phosphate; refractive index = ~1.46) for 5+ days and then mounted in 1% agarose using a 1ml syringe to make an agarose cylinder with the sample suspended within. The sample in the syringe was partially ejected into the EZ Clear solution (to be used as imaging buffer) for 3+ days. The samples were imaged using a Zeiss Lightsheet Z.1 microscope. ZEN software (Zeiss), Stitchy (Translucence Biosystems), and Imaris software (Oxford Instruments) were used for data processing to render videos.
Supplementary Material
Supplementary Movie 1: Light sheet 3D rotation video of a whole mount E10.5 Foxi3GFP mouse embryo stained with anti-GFP antibodies.
Supplementary Movie 2: Optical sectioning in the anterior-posterior plane from the light sheet data of an E10.5 Foxi3GFP mouse embryo stained with anti-GFP antibodies.
Supplementary Movie 3: Optical sectioning in the dorso-ventral plane from the light sheet data of an E10.5 Foxi3GFP mouse embryo stained with anti-GFP antibodies.
Grant Sponsor:
National Institute on Deafness and Other Communication Disorders
Grant Number:
R01 DC013072
REFERENCES:
- 1.Edlund RK, Birol O, Groves AK. The role of foxi family transcription factors in the development of the ear and jaw. Curr Top Dev Biol. 2015;111:461–95. 10.1016/bs.ctdb.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcucio R, Hallgrimsson B, Young NM. Facial Morphogenesis: Physical and Molecular Interactions Between the Brain and the Face. Curr Top Dev Biol. 2015;115:299–320. 10.1016/bs.ctdb.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo-Rogers HL, Smithers LE, Yakob W, Liu KJ. New directions in craniofacial morphogenesis. Dev Biol. May 1 2010;341(1):84–94. https://doi.orgS0012-1606(09)01376-1 [pii] 10.1016/j.ydbio.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, Zhao Y, Shen G, Dai J. Etiology and Pathogenesis of Hemifacial Microsomia. J Dent Res. Nov 2018;97(12):1297–1305. 10.1177/0022034518795609. [DOI] [PubMed] [Google Scholar]
- 5.Hartzell LD, Chinnadurai S. Microtia and Related Facial Anomalies. Clin Perinatol. Dec 2018;45(4):679–697. 10.1016/j.clp.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Marszalek-Kruk BA, Wojcicki P, Dowgierd K, Smigiel R. Treacher Collins Syndrome: Genetics, Clinical Features and Management. Genes (Basel). Sep 9 2021;12(9). 10.3390/genes12091392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmetz A, Amiel J, Wieczorek D. Genetics of craniofacial malformations. Semin Fetal Neonatal Med. Dec 2021;26(6):101290. 10.1016/j.siny.2021.101290. [DOI] [PubMed] [Google Scholar]
- 8.Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. Oct 2002;129(19):4613–25. [DOI] [PubMed] [Google Scholar]
- 9.Ankamreddy H, Min H, Kim JY, et al. Region-specific endodermal signals direct neural crest cells to form the three middle ear ossicles. Development. Jan 22 2019;146(2). 10.1242/dev.167965. [DOI] [PubMed] [Google Scholar]
- 10.Billmyre KK, Klingensmith J. Sonic hedgehog from pharyngeal arch 1 epithelium is necessary for early mandibular arch cell survival and later cartilage condensation differentiation. Dev Dyn. Apr 2015;244(4):564–76. 10.1002/dvdy.24256. [DOI] [PubMed] [Google Scholar]
- 11.Clouthier DE, Garcia E, Schilling TF. Regulation of facial morphogenesis by endothelin signaling: insights from mice and fish. American journal of medical genetics Part A. Dec 2010;152A(12):2962–73. 10.1002/ajmg.a.33568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development. Feb 2002;129(4):1061–73. [DOI] [PubMed] [Google Scholar]
- 13.Depew MJ, Compagnucci C. Tweaking the hinge and caps: testing a model of the organization of jaws. J Exp Zool B Mol Dev Evol. Jun 15 2008;310(4):315–35. 10.1002/jez.b.21205. [DOI] [PubMed] [Google Scholar]
- 14.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. Nov 2005;207(5):501–61. 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. Aug 2010;137(16):2605–21. 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- 16.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. Dec 1 1999;13(23):3136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birol O, Ohyama T, Edlund RK, Drakou K, Georgiades P, Groves AK. The mouse Foxi3 transcription factor is necessary for the development of posterior placodes. Dev Biol. Jan 1 2016;409(1):139–151. 10.1016/j.ydbio.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edlund RK, Ohyama T, Kantarci H, Riley BB, Groves AK. Foxi transcription factors promote pharyngeal arch development by regulating formation of FGF signaling centers. Dev Biol. Jun 1 2014;390(1):1–13. 10.1016/j.ydbio.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. Nov 2004;231(3):640–6. 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- 20.Hasten E, Morrow BE. Tbx1 and Foxi3 genetically interact in the pharyngeal pouch endoderm in a mouse model for 22q11.2 deletion syndrome. PLoS Genet. Aug 2019;15(8):e1008301. 10.1371/journal.pgen.1008301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jussila M, Aalto AJ, Sanz Navarro M, et al. Suppression of epithelial differentiation by Foxi3 is essential for molar crown patterning. Development. Nov 15 2015;142(22):3954–63. 10.1242/dev.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirokova V, Jussila M, Hytonen MK, et al. Expression of Foxi3 is regulated by ectodysplasin in skin appendage placodes. Dev Dyn. Jun 2013;242(6):593–603. 10.1002/dvdy.23952. [DOI] [PubMed] [Google Scholar]
- 23.Shirokova V, Biggs LC, Jussila M, Ohyama T, Groves AK, Mikkola ML. Foxi3 Deficiency Compromises Hair Follicle Stem Cell Specification and Activation. Stem cells. Jul 2016;34(7):1896–908. 10.1002/stem.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drogemuller C, Karlsson EK, Hytonen MK, et al. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science. Sep 12 2008;321(5895):1462. 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- 25.Kupczik K, Cagan A, Brauer S, Fischer MS. The dental phenotype of hairless dogs with FOXI3 haploinsufficiency. Sci Rep. Jul 14 2017;7(1):5459. 10.1038/s41598-017-05764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiener DJ, Gurtner C, Panakova L, et al. Clinical and histological characterization of hair coat and glandular tissue of Chinese crested dogs. Vet Dermatol. Apr 2013;24(2):274–e62. 10.1111/vde.12008. [DOI] [PubMed] [Google Scholar]
- 27.Mao K, Borel C, Ansar M, et al. FOXI3 pathogenic variants cause one form of craniofacial microsomia. Nat Commun. Apr 11 2023;14(1):2026. 10.1038/s41467-023-37703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiat D, Timberlake AT, Curran JJ, et al. Damaging variants in FOXI3 cause microtia and craniofacial microsomia. Genet Med. Oct 19 2022. 10.1016/j.gim.2022.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tassano E, Jagannathan V, Drogemuller C, et al. Congenital aural atresia associated with agenesis of internal carotid artery in a girl with a FOXI3 deletion. American journal of medical genetics Part A. Mar 2015;167A(3):537–44. 10.1002/ajmg.a.36895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poswillo D The pathogenesis of the first and second branchial arch syndrome. Oral Surg Oral Med Oral Pathol. Mar 1973;35(3):302–28. 10.1016/0030-4220(73)90070-4. [DOI] [PubMed] [Google Scholar]
- 31.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. Jan 2010;13(1):133–40. https://doi.orgnn.2467 [pii]10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu CW, Cerda J 3rd, Kirk JM, et al. EZ Clear for simple, rapid, and robust mouse whole organ clearing. Elife. Oct 11 2022;11. 10.7554/eLife.77419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1: Light sheet 3D rotation video of a whole mount E10.5 Foxi3GFP mouse embryo stained with anti-GFP antibodies.
Supplementary Movie 2: Optical sectioning in the anterior-posterior plane from the light sheet data of an E10.5 Foxi3GFP mouse embryo stained with anti-GFP antibodies.
Supplementary Movie 3: Optical sectioning in the dorso-ventral plane from the light sheet data of an E10.5 Foxi3GFP mouse embryo stained with anti-GFP antibodies.


