Abstract
Humans and other mammals inhabit hypoxic high-altitude locales. In many of these species, genes under positive selection include ones in the Hypoxia Inducible Factor (HIF) pathway. One is PHD2 (EGLN1), which encodes for a key oxygen sensor. Another is HIF2A (EPAS1), which encodes for a PHD2-regulated transcription factor. Recent studies have provided insights into mechanisms for these high-altitude alleles. These studies have (1) shown that selection can occur on nonconserved, unstructured regions of proteins, (2) revealed that high altitude-associated amino acid substitutions can have differential effects on protein:protein interactions, (3) provided evidence for convergent evolution by different molecular mechanisms, and (4) suggested that mutations in different genes can complement one another to produce a set of adaptive phenotypes.
Keywords: Hypoxia Inducible Factor (HIF), PHD2, EGLN1, EPAS1, genetic adaptation, convergent evolution
High altitude and hypoxia
A salient characteristic of high altitude (see Glossary) is hypobaric hypoxia. At 12,000 ft (3,700 m), which is the elevation of the cities of La Paz (Andean Altiplano) and Lhasa (Tibetan plateau), the oxygen concentration is only about 63% of that at sea level. Yet humans and other mammals have lived for millennia on these plateaus and other high-altitude locales such as the Ethiopian Simien plateau. Hypoxia presents a major challenge for mammals living at high altitude primarily because oxygen is the terminal electron acceptor for mitochondrial oxidative phosphorylation, the bioenergetic process that efficiently produces ATP in cells. In the absence of oxygen, cells rely on substantially less efficient means of generating ATP, such as glycolysis.
In multiple high-altitude mammalian species, such as Tibetans, Andeans, high-altitude North American deer mice, Andean horses, and Tibetan dogs [1–6], signals of natural selection have been identified in genes of the Hypoxia Inducible Factor (HIF) pathway, the central transcriptional pathway by which metazoans respond to hypoxia [7–13]. A major gap in our knowledge regarding high-altitude adaptation is the functional characterization of adaptive alleles and their encoded proteins. While other reviews have discussed the identification of high-altitude genes and physiologic aspects of high-altitude adaptation [7,13], this review will focus on recent and perhaps unexpected observations that have begun to emerge from functional studies of mammalian high-altitude HIF pathway alleles, particularly those of Tibetans, Andeans, and high-altitude North American deer mice and especially those that produce amino acid changes.
The HIF pathway
HIF is a heterodimeric transcription factor that consists of an α subunit, of which HIF-1α and HIF-2α are the major paralogues, and a common β subunit, Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT). Each subunit contains structured N-terminal basic Helix-Loop-Helix (bHLH) and Per-Arnt-Sim (PAS) domains that contact the other subunit and DNA [14]. The remainder of each subunit is largely unstructured. HIF-1α and HIF-2α have overlapping and distinct gene targets [15]. HIF-1α is the key isoform that upregulates genes of glycolysis, thereby promoting the shift from aerobic to anaerobic metabolism. HIF-2α plays a central role in regulating the ERYTHROPOIETIN gene (and therefore red cell mass), the EDN-1 gene (which encodes for the potent vasoconstrictor Endothelin-1 that regulates regional blood pressure in organs such as the lung), and the hypoxic ventilatory response (HVR). Humans and mice with gain of function mutations in the HIF2A gene display erythrocytosis and elevated pulmonary arterial pressure (PAP), while inducible knockout of Hif2a in mice blunts the erythrocytosis and the increase in HVR following exposure to chronic hypoxia (also known as the ventilatory acclimatization to chronic hypoxia, or VAH) [16–20].
In an oxygen-dependent manner, the enzyme Prolyl Hydroxylase Domain protein 2 (PHD2, also known as EGLN1) prolyl hydroxylates HIF-α in its oxygen-dependent degradation (ODD) domain [9] (Figure 1A, left). This modification promotes the binding of the von Hippel Lindau (VHL) protein, which targets HIF-α for degradation [21]. Under hypoxia, this modification is arrested, leading to the stabilization of HIF-α, its dimerization with ARNT, and the activation of target genes (Figure 1A, right). Humans with partial loss of function PHD2 mutations display erythrocytosis, and mice with acute global ablation of Phd2 display augmented HVR as well as erythrocytosis [18,20,22,23].
Figure 1. The HIF pathway.
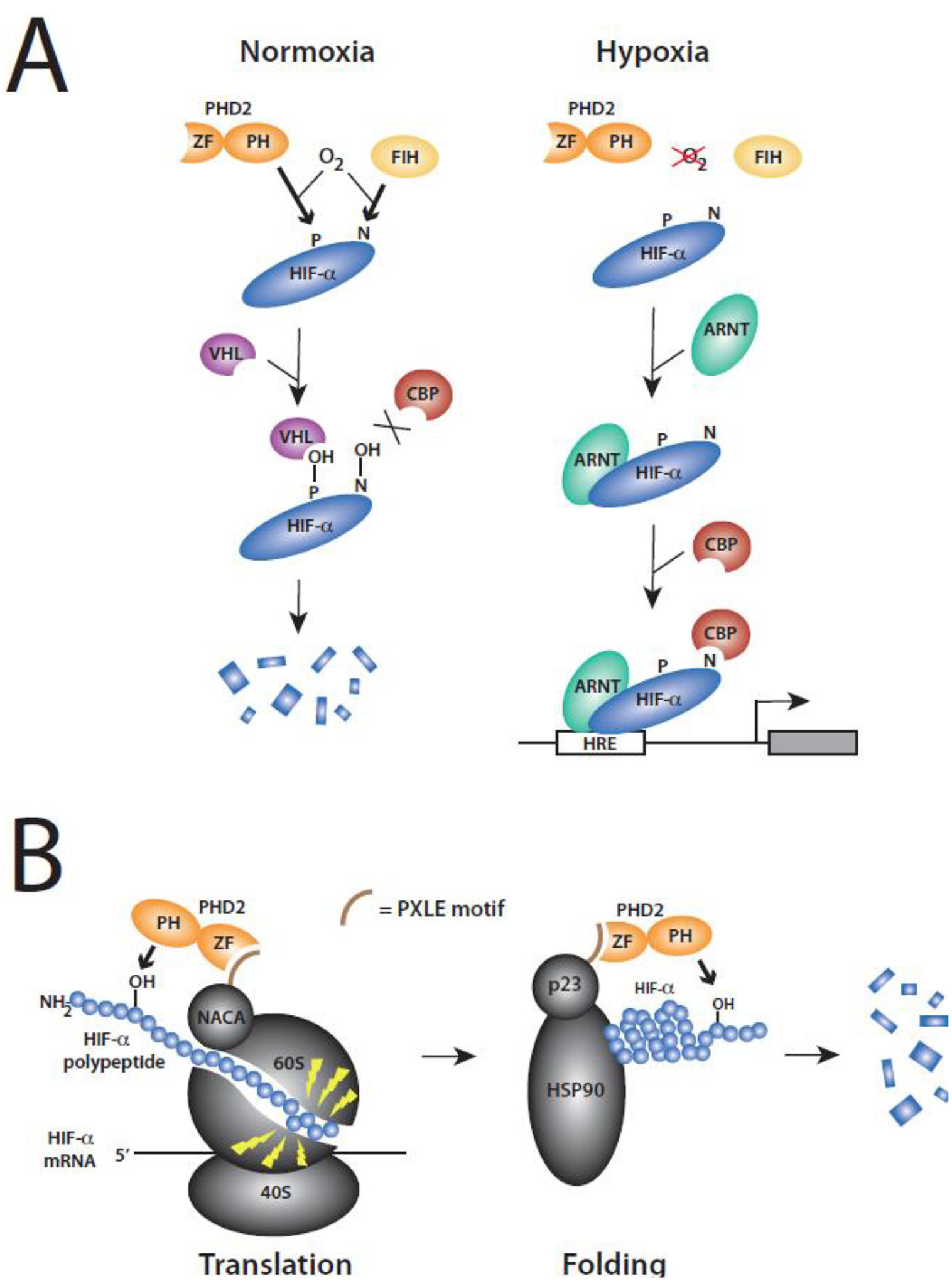
(A) Core components. Under normoxia, PHD2 [which contains zinc finger (ZF) and prolyl hydroxylase (PH) domains] and FIH site-specifically hydroxylate prolyl (P) and asparaginyl (N) residues, respectively, on HIF-α in an oxygen-dependent manner. In human HIF-1α, these residues are Pro-564 and Asn-803, respectively. In human HIF-2α, they are Pro-531 and Asn-847, respectively. Prolyl hydroxylated HIF-α is recognized by VHL, promoting degradation of the former. Asparaginyl hydroxylation blocks binding of CBP/p300 (only CBP shown). Under hypoxia, both hydroxylation reactions are inhibited, allowing HIF-α heterodimerization with ARNT, recruitment of CBP/p300, and activation of HIF target genes. (B) Coupling of HIF prolyl hydroxylation to translation and protein folding. Left, PHD2 employs its zinc finger to allow binding to NACA and recruitment to ribosome, where it cotranslationally hydroxylates HIF-α in a manner enhanced by translational pausing (indicated by lightning bolts). For further details, see [28]. 60S and 40S refer to ribosomal subunits. Right, PHD2 employs its zinc finger to allow binding to p23 and recruitment to the HSP90 complex, where it postranslationally hydroxylates HIF-α, which is a HSP90 client.
In a parallel pathway, Factor Inhibiting HIF (FIH) asparaginyl hydroxylates HIF-α in its C-terminal transcriptional activation domain (CTAD) (Figure 1A, left) [24,25]. This modification blocks the binding of the transcriptional coactivator Creb Binding Protein (CBP, or its paralogue p300). Under hypoxia, this modification is arrested, leading to the recruitment of CBP/p300 to promote the transcription of HIF target genes [26] (Figure 1A, right). HIF-α also recruits, via its PAS domain, the H3K4 methyltransferase SET1B to promote transcription [27].
This core HIF pathway is integrated with two ancient pathways that enhance the efficiency of prolyl hydroxylation. In the first, PHD2 employs a zinc finger domain located at its N-terminus to bind a Pro-Xaa-Leu-Glu (PXLE) motif in the ribosomal chaperone NACA [28] (Figure 1B, left). NACA, in turn, is positioned at the ribosomal exit tunnel [29], thereby allowing the recruitment of PHD2 to cotranslationally modify HIF-α in a manner enhanced by a translational pause sequence in HIF-α. A knockin mouse mutation in the PXLE motif of Naca that ablates its interaction with Phd2 leads to erythrocytosis [28]. PHD2 also employs this same zinc finger to bind a PXLE motif in the HSP90 cochaperone p23 [30] (Figure 1B, right). The p23 protein is a component of the HSP90 complex that folds proteins, and HIF-α is a client of the HSP90 pathway [31]. Thus, recruitment of PHD2 by p23 promotes posttranslational modification of HIF-α. A knockin mouse with a mutation in the PXLE motif of p23 that ablates its interaction with Phd2 leads to augmented HVR [32]. In summary, these zinc finger-guided interactions with NACA and p23 increase the efficiency by which PHD2 can cotranslationally and then postranslationally prolyl hydroxylate HIF-α, respectively, and they play important roles in the control by PHD2 of red cell mass and respiration, respectively.
As noted above, the phenotype of knockin mice with mutations in the PXLE motif of Naca differs from that of mice with mutations in the PXLE motif of p23 [28,32]. The basis for this is not clear. One possibility is that there is a difference in the relative protein levels of NACA and p23 in the renal erythropoietin (EPO)-producing cells that regulate red cell mass and the Type I cells of the carotid body that regulate respiration, leading to a cell-dependent differential reliance of PHD2 on NACA versus p23. Alternatively, the ability of NACA to facilitate PHD2-induced hydroxylation of HIF-2α is itself dependent on a translational pause site in HIF-2α [28], and the strength of this pause site might differ in cell-type specific manner.
Mammalian high-altitude phenotypes
The three most extensively studied human high-altitude populations are the Tibetans, Andeans, and Ethiopians [33–38]. In addition to humans, a multitude of mammalian species have adapted to high altitude (Table 1) [13,39,40]. Phenotypes are available for a number of these species. For example, Tibetans at altitude display (1) normal hemoglobin levels, in contrast to the elevated hemoglobin levels that are seen in lowlanders who ascend to and live at high altitude; (2) persistently augmented HVR, in contrast to the attenuated HVR that is seen in lowlanders who have lived long term at high altitude; and (3) lower right ventricular pressures and lower incidence of pulmonary hypertension than lowlanders who ascend to and live at high altitude [7,41]. High-altitude North American deer mice (Peromyscus maniculatus) display different phenotypes, including (1) a variant hemoglobin that has increased affinity for oxygen and (2) blunted VAH [42,43].
Table 1.
Mammalian high-altitude HIF pathway alleles
| Speciesa | Gene | Amino acid changes | Evidence of selection | Associations | Functional characterization | References |
|---|---|---|---|---|---|---|
| H. sapiens (T) | HIF2A | NDb | yes | ↓ Hb ↓ PAP ↓ expression of HIF2A mRNA |
↓ expression of reporter gene | [1,2,44–46,68,70,78–80] |
| H. sapiens (A) | HIF2A | H194R | yes | ↑ FeNO | ↓ heterodimerization with ARNT ↓ expression of reporter gene Attenuated hypoxia-induced PH in mice |
[48,51] |
| P. maniculatus (N) | Hif2a | T755M | yes | ↑ HR under hypoxia ↓ VAH |
↓ binding to Cbp in vitro | [4,52,54] |
| C. familiaris (T) | Hif2a | G305S, D494E, V500M, P750S | yes | ↓ blood flow resistance | [6,55,56] | |
| B. taurus (N) | Hif2a | E270Q, P362L, A606T, G610S, A671G, L701F | no | ↑ PAP | [61,62] | |
| C. hircus (T) | Hif2a | Q579L | yes | ↑ MCHC | [57] | |
| E. caballus (T) | Hif2a | R144C, E263D | yes | ↓ Hb | ↑ stability of Hif-2α protein | [58] |
| E. caballus (A) | Hif2a | NR | yes | [5] | ||
| S. scrofa (T) | Hif2a | ND | yes | ↓ expression of reporter gene | [59] | |
| O. curzoniae (T) | Hif2a | 24 aa insertion | NR | ↑ stability of Hif-2α protein | [60] | |
| H. sapiens (T) | PHD2 | D4E/C127S | yes | * | ↓ binding to p23 in vitro Preserved binding to NACA in vitro ↑ HVR and VAH in mice ↑ affinity for oxygen in vitro |
[3,32,44,45,65–67,71,73,80] |
| H. sapiens (A) | PHD2 | ND | yes | ↑ VO2maxc | [65,75] | |
| H. sapiens (I) | PHD2 | NR | NR | ↑ pulmonary edemad
↑ expression of PHD2 mRNAd |
[76] | |
| B. taurus (T) | Phd2 | NR | NR | ↓ Hb, ↓ rbc | [81,82] | |
| P. uncia (T) | Phd2 | K228Me | NR | [83] | ||
| H. sapiens (E) | ARNT2 | NR | yes | [63] |
The locales are indicated in parentheses and are as follows: T, Tibet; A, Andes; N, North America; I, India; E, Ethiopia
Abbreviations are as follows: ND, not detected; NR, not reported; Hb, hemoglobin concentration; PAP, pulmonary arterial pressure; FeNO, fractional concentration of exhaled nitric oxide; PH, pulmonary hypertension; VAH, ventilatory acclimatization to chronic hypoxia; MHCH, mean corpuscular hemoglobin concentration; VO2max, maximal oxygen consumption; rbc, red blood cell concentration.
TT genotype of rs1769793
TT genotype of rs479200
Annotation according to full length protein (XP_049500971.1)
One study looking specifically at the D4E/C127S haplotype did not report any association with Hb in highlanders [71]. Another study examining D4E or C127S individually have reported associations with decreased Hb [67]. Additional studies examining other SNPs have reported either no association or an association with decreased Hb [3,68–70].
Signatures of natural selection on the mammalian HIF2A gene
The gene under strongest selection in Tibetans is HIF2A (also known as EPAS1) [1,2,44,45]. This high-altitude HIF2A allele does not encode for any amino acid changes. Rather, it encompasses a haplotype that is introgressed from Denisovans, an archaic human species, and consists of intronic sequence changes [46]. This haplotype is also present in high-altitude Sherpas who reside in the eastern Himalayas [47]. Available evidence suggests that Tibetan HIF2A allele is a partial loss of function allele (reviewed in [48]). It is possible that this allele may result in decreased HIF2A transcription, although there might be other potential effects, such as ones on splicing [49].
In Andean Colla and Quechua populations, signatures of natural selection have also been observed in the HIF2A gene [48,50,51]. In this case, there is enrichment for a HIF2A allele that encodes for a H194R substitution in the PAS domain of HIF-2α [51]. This amino acid substitution impairs heterodimerization with ARNT, leading to decreased transcriptional activity [48]. In a mouse model with a knockin H194R Hif2a mutation, it was observed to result in blunting of hypoxia-induced increases in right ventricular pressure [48]. Together, these observations indicate that the Andean HIF2A is a partial loss of function allele.
Exome sequencing of North American high- and low-altitude deer mice revealed evidence for altitude-dependent natural selection acting on the Hif2a locus [4]. One of the single nucleotide polymorphisms (SNPs) under strongest selection encodes for a T755M substitution. Thr-755 resides between the ODD and CTAD (Figure 2A). In vitro, the T755M mutation impairs binding of Hif-2α to the transcriptional coactivator CBP but not p300 [52]. Because the HIF target genes affected by loss of CBP versus p300 are not identical [53], this would be predicted to impair transcription of some but not all HIF target genes. In vivo, the high-altitude Hif2a haplotype is associated with increased heart rate under hypoxia and impaired VAH [4,54]. Given that impaired VAH is observed in mice with inducible Hif2a knockout [18], these findings are consistent with high altitude North American deer mouse Hif2a being a partial loss of function allele.
Figure 2. HIF-2α and PHD2.
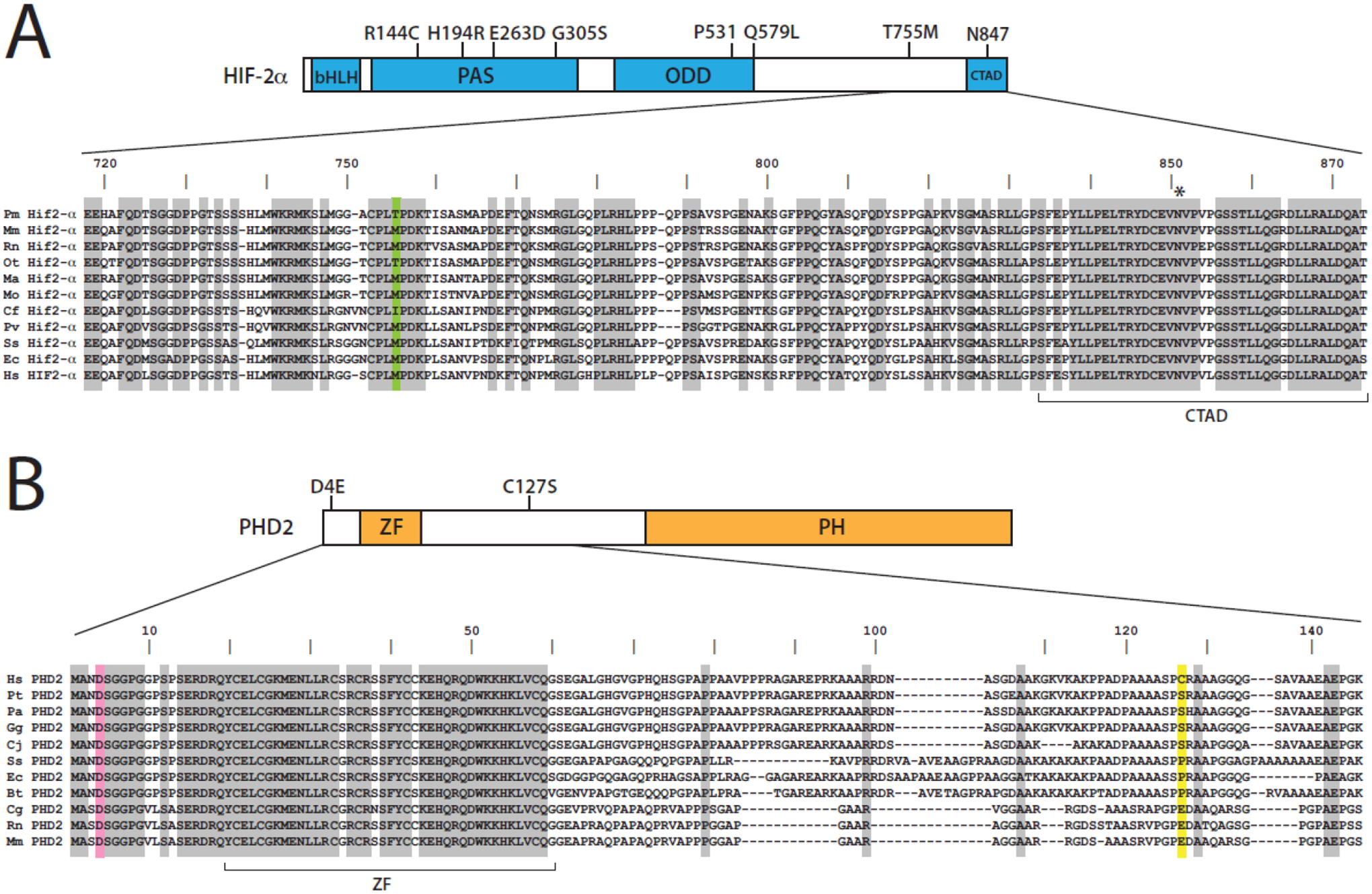
(A) Top, HIF-2α. Select high-altitude mutations shown, including R144C (Tibetan horse), H194R (human Andeans), E263D (Tibetan horse), G305S (Tibetan dogs), Q579L (Tibetan goat), and T755M (high-altitude deer mice). bHLH = basic helix-loop-helix, PAS = Per-Arnt-Sim domain, ODD = oxygen-dependent degradation, CTAD = C-terminal activation domain. Bottom, sequence of residues 718–874 of deer mouse (Peromyscus maniculatus, XP_015860532) Hif-2α, along with corresponding ones from house mouse (Mus musculus, NP_034267), rat (Rattus norvegicus, Q9JHS1), Southern grasshopper mouse (Onychomys torridus, XP_036026399), golden hamster (Mesocricetus auratus, XP_040593547), creeping vole (Microtus oregoni, XP_041486961), dog (Canis lupus familiaris, XP_005626137), harbor seal (Phoca vitulina, XP_032266944), pig (Sus scrofa, NP_001090889), horse (Equus caballus, XP_005600062), and human (Homo sapiens, NP_001421). Green = Thr-755 in deer mouse Hif-2α and the corresponding residues in orthologues. Asterisk, Asn-851. (B) Top, PHD2. Positions of the Tibetan D4E and C127S substitutions shown. ZF = zinc finger, PH = prolyl hydroxylase domain. Bottom, sequence of residues 1–146 of human (Homo sapiens, Q9GZT9) PHD2 is shown, along with corresponding ones from chimpanzee (Pan troglodytes, JAA34701), orangutan (Pongo abelii, XP_002809347), gorilla (Gorilla gorilla gorilla, XP_004028630), marmoset (Callithrix jacchus, XP_003735339), pig (Sus scrofa, XP_020929239), horse (Equus caballus, XP_014588968), cow (Bos taurus, NP_001192975), Chinese hamster (Cricetulus griseus, XP_007651724), rat (Rattus norvegicus, NP_848017), and mouse (Mus musculus, Q91YE3). Pink = Asp-4 and yellow = Cys-127 in human PHD2, and the corresponding residues in orthologues. In (A) and (B), alignment produced using the Clustal W method (Lasergene MegAlign v15.3.0), and shading indicates residues conserved in all species.
Whole genome sequencing or exome sequencing has been performed on a whole range of additional high-altitude mammals, including Tibetan dogs, cashmere goats, horses, pigs, and Pikas, as well as on North American cattle, the latter being a species susceptible to high altitude-associated pulmonary hypertension [5,6,55–62]. In many of these species, there is evidence of natural selection on the Hif2a gene. In a number of species, coding sequence changes in the gene have been reported. While functional characterization of the high-altitude allele has been performed in a few cases [58–60], in many cases the functional consequences are still unknown. These studies are summarized in Table 1, and select amino acid changes in the HIF-2α protein identified in these mammals are highlighted in Figure 2A.
Signatures of natural selection on the human ARNT2 gene
The ARNT2 gene has been nominated as a candidate gene under positive selection in high-altitude Ethiopians [63]. This gene encodes for a paralogue of ARNT that has the capacity to heterodimerize with HIF-2α. It is not known whether the high-altitude allele encodes for any amino acid changes. The ARNT gene itself has not yet been reported to be under positive selection in this or other high-altitude mammalian populations. Perhaps the dual role of ARNT in both the hypoxic response (via heterodimerization with HIF-α) and the xenobiotic response (via heterodimerization with Aryl Hydrocarbon Receptor) [64] might constrain the ability of this gene to participate in evolutionary adaptations to high altitude.
Signatures of natural selection on the human PHD2 gene
In Tibetans, there is also natural selection acting on the PHD2 gene locus [3,65]. The Tibetan PHD2 allele consists of a D4E/C127S double amino acid substitution [66]. Several studies have reported an association between the high-altitude PHD2 allele and lower hemoglobin levels [3,67,68]; in some of these studies, the association was seen in males but not in females [67,68]. In other studies, however, no significant association was seen [69–72]. These latter studies include the one with the largest Tibetan sample size (2,849) among them [70]. That study examined SNP-based haplotypes. Another study that specifically examined the combination of the two amino acid substitutions did not observe an association between D4E/C127S PHD2 haplotype and hemoglobin levels when examined in highlanders [71]. Additionally, Tibetan PHD2 is not associated with PAP [68], and Tibetan Phd2 in mice is not associated with either hemoglobin level or right ventricular systolic pressure (RVSP) [32]. In contrast, Tibetan HIF2A is associated with decreased PAP [68]. These studies provide evidence that the phenotypes of Tibetan PHD2 and Tibetan HIF2A are not identical.
In parallel, the functional effect of the D4E/C127S substitution has been investigated. It has been reported, using in vitro assays employing recombinant proteins, that Tibetan PHD2 exhibits increased affinity for oxygen and therefore is a gain of function allele [66]. A gain of function could lead to an association of the Tibetan PHD2 allele with lower hemoglobin level, which as noted above, has been observed in some, but not all, studies of the Tibetan PHD2 allele.
A very different, though not mutually exclusive, model for Tibetan PHD2 has also been reported [73]. The amino acids affected in PHD2, Asp-4 and Cys-127, flank the N-terminal zinc finger of PHD2 (Figure 2B), which promotes cotranslational and posttranslational hydroxylation of HIF-α (Figure 1B) [28,73]. The D4E/C127S substitutions impair interaction of PHD2 with p23 [73]. Since p23 recruits PHD2 to the HSP90 pathway to facilitate HIF-α prolyl hydroxylation, this would be predicted to lead to a partial loss of function. This is supported by a knockin mouse model bearing the D4E/C127S substitution [32]. These mice display increased HVR, which is also observed in mice with Phd2 haploinsufficiency [74]. Moreover, the increased HVR is phenocopied in mice bearing p23 knockin mutations that ablate p23 interaction with Phd2. These findings, in turn, are consistent with the observation that Tibetans display augmented HVR. Importantly and in contrast to its interaction with p23, Tibetan PHD2 maintains its binding to NACA, the ribosomal chaperone important for PHD2 control over red cell mass [28]. In accord with this, the Tibetan Phd2 knockin mice display hemoglobin levels that are similar to wild type mice under either normoxia or hypoxia [32]. This in turn would be consistent with a lack of an association of the Tibetan PHD2 allele with hemoglobin levels, which as discussed above has been observed in a number of the studies examining the PHD2 allele.
In Andeans, signatures of positive selection have also been identified in the PHD2 gene by SNP genotyping [50,65]. The Andean PHD2 allele, which does not contain any coding sequence substitutions, is associated with increased VO2max, a measure of aerobic capacity [75]. Certain SNPs in this high-altitude Andean allele (rs479200, rs480902) are also enriched in high-altitude Southeast Asian Indian populations [76]. The precise mechanism by which these or other SNPs might act remains to be determined.
Convergent evolution
The possibility of convergent evolution is raised by the observation of natural selection acting on the HIF2A locus in multiple species, including humans (Tibetans and Andeans), North American deer mice, Andean horses, and Tibetan dogs, goats, horses, and pigs (Table 1). Likewise, there is evidence for natural selection acting on the PHD2 locus in humans (Tibetans and Andeans) and possibly Tibetan cattle.
Importantly, there is evidence that convergent evolution can act by different molecular mechanisms (Figure 3). For example, high-altitude North American deer mouse T755M Hif-2α shows a defective interaction with Cbp in vitro (Figure 3B) [52], and deer mice with the Hif2a allele shows impaired VAH [54], consistent with a partial loss of function. Andean H194R HIF-2α displays weakened heterodimerization with ARNT in vitro (Figure 3C), and mice with a knockin Andean Hif2a mutation (H194R) display resistance to hypoxia-induced pulmonary hypertension; both observations are consistent with a partial loss of function [48]. Tibetan HIF2A likewise displays phenotypic properties consistent with a partial loss of function (reviewed in [48]). However, in contrast to the situation with high-altitude deer mouse or Andean HIF-2α, there are no coding sequence mutations in Tibetan HIF-2α, and current evidence is consistent with it being due to decreased transcription (or possibly splicing) of the HIF2A gene (Figure 3D).
Figure 3. Convergent evolution acting on the HIF2A gene occurring by different mechanisms.
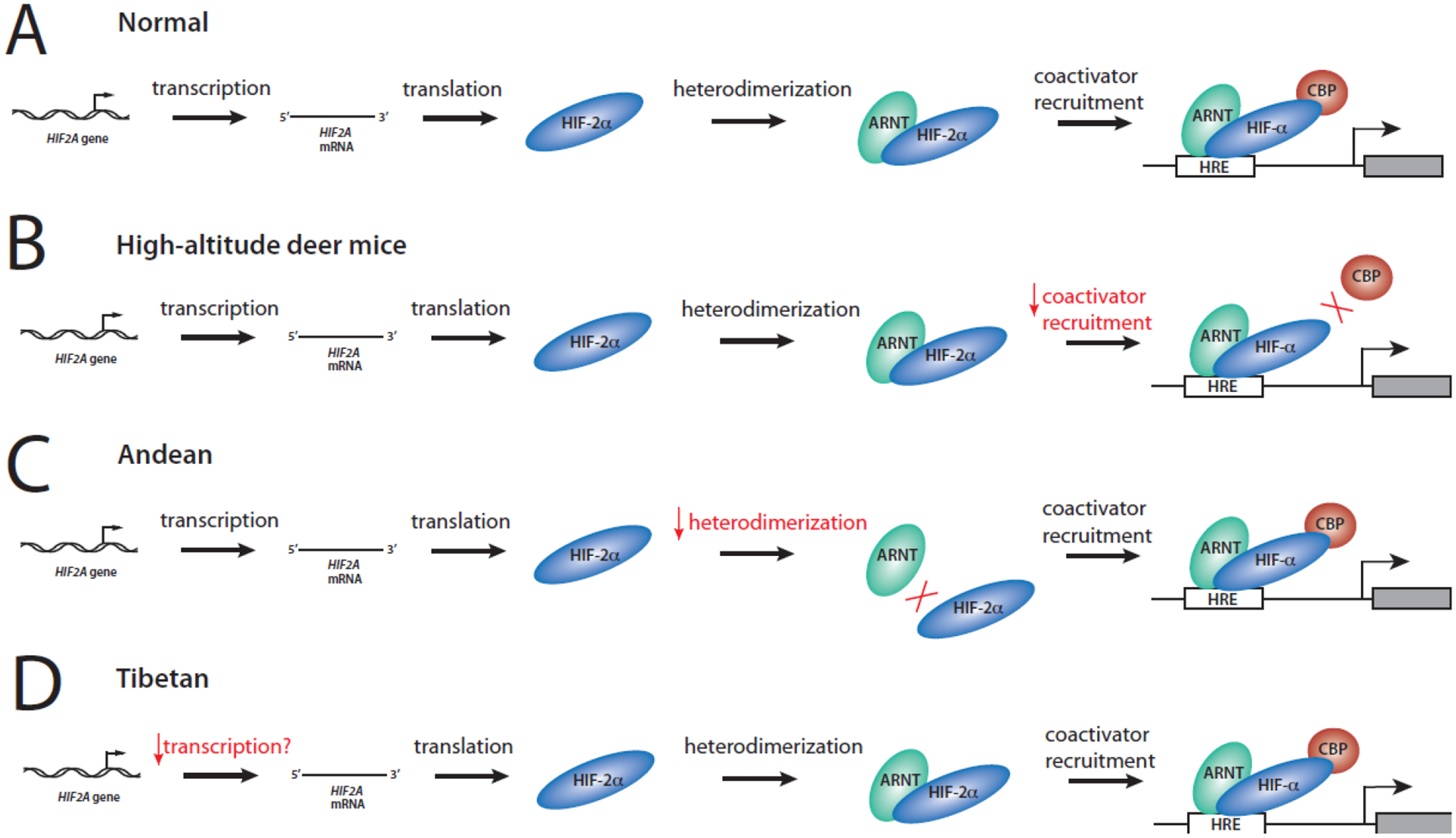
(A) Normal transcription of the HIF2A gene followed by translation, heterodimerization with ARNT, binding to Hypoxia Response Elements (HRE), and recruitment of the coactivator CBP. (B) In high-altitude North American deer mice, there is impaired binding of HIF-2α to the transcriptional coactivator CBP. (C) In Andeans, there is impaired heterodimerization of HIF-2α with ARNT. (D) In Tibetans, there is possibly decreased transcription of the HIF2A gene.
It should be noted that core HIF pathway genes are not invariably targets of natural selection in high-altitude mammals. For example, high-altitude Ethiopian gelada monkeys, like Tibetans, do not exhibit elevated hemoglobin concentrations [77]. However, in contrast to Tibetans, they do not display evidence of natural selection acting on the Hif2a gene.
Selection on nonconserved, unstructured regions of proteins
Natural selection often targets the coding sequences of conserved, structured regions of proteins, resulting in functional changes that can either be readily understood or rationalized based on known three-dimensional structures of the proteins in question. An unexpected finding that has emerged from the recent research on high-altitude HIF pathway alleles is that in some cases, selection has occurred on coding sequences of nonconserved, unstructured regions of proteins. One example of this is Tibetan PHD2. One component of the allele, C127S [66], affects an amino acid that is not conserved (Figure 2B) and is likely to reside in an unstructured region of PHD2 between the N-terminal zinc finger and the C-terminal prolyl hydroxylase domain. The other component of the Tibetan allele, D4E [66,67], affects a conserved amino acid that is just N-terminal to the zinc finger (Figure 2B). It is not known whether Asp-4 resides in a disordered region or alternatively, an ordered part of the protein, perhaps in association with the zinc finger.
A second example is high-altitude North American deer mouse Hif-2α [4]. The T755M mutation affects an amino acid that is not conserved (Figure 2A) and is likely to reside in an unstructured region of Hif-2α between the ODD domain and the CTAD. Interestingly, both the Tibetan PHD2 C127S and the North American deer mouse Hif-2α T755M mutation revert the amino acids to ones present in related species, raising the possibility that the result is reversion to the ancestral amino acid. In the case of PHD2, Ser is present in the position corresponding to Cys-127 in PHD2s from primates such as chimpanzee, orangutan, gorilla, and marmoset (labeled Pt, Pa, Gg, and Cj, respectively, in Figure 2B). Likewise, in the case of Hif-2α, Met is present in the corresponding position in rodent species such as house mouse, rat, and golden hamster (labeled Mm, Rn, and Ma, respectively, in Figure 2A). The basis for this potential reversion to the ancestral amino acid as opposed to some other amino acid is not known. Even more puzzling is how these substitutions can mechanistically produce functional defects since they are predicted to reside in unstructured regions of the protein that are distant from sites of known protein:protein interactions in these proteins. It is noteworthy that the functional effect of specific substitutions can be context dependent. For example, interchanges between Met and Thr at position 755 in Hif-2α have functional effects on Cbp binding in the context of deer mouse Hif-2α but not house mouse Hif-2α [52].
Differential effects of coding sequence mutations on protein:protein interactions
Another notable finding that has emerged from the recent research is that in some cases, the functional result is a differential effect on protein:protein interactions. One example of this is Tibetan PHD2. The D4E/C127S substitution impairs interaction with the HSP90 cochaperone p23 but not the ribosomal chaperone NACA (Figure 4A) [28,73]. The impaired interaction with p23 could account for the augmented HVR seen in Tibetans, since p23 plays an important role in this arm of the HIF pathway in mice [32]. On the other hand, the preserved interaction with NACA, which plays an important role in PHD2 control of red cell mass, could account for the observation that Tibetans are not predisposed to erythrocytosis. This preserved interaction is further supported by the observation that mice with Tibetan Phd2 display normal hemoglobin levels and that, after crossing with mice with a knockin Naca mutation that ablates Phd2 interaction, the resulting mice display erythrocytosis [28,32].
Figure 4. High-altitude mutations in the HIF pathway can differentially affect protein:protein interactions.
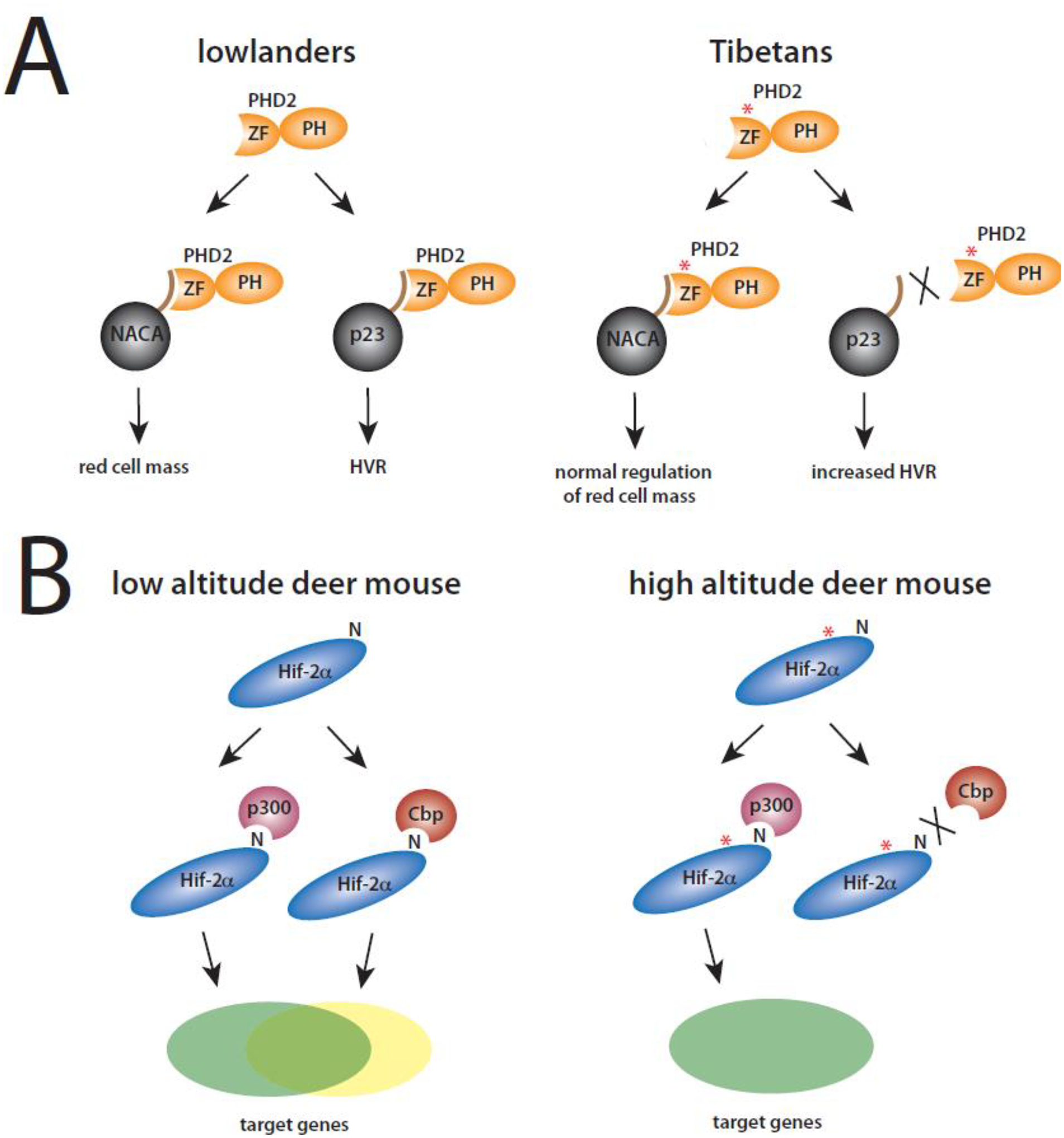
(A) Left, the zinc finger of wild type PHD2 of lowlanders binds to NACA and p23 and therefore maintains appropriate control of red cell mass and HVR, respectively. Right, the zinc finger of Tibetan PHD2 has a defect in interaction with p23 but not with NACA. Hence, there is wild type (normal) control of red cell mass but increased HVR. ZF = zinc finger, PH = prolyl hydroxylase domain, asterisk = D4E/C127S double amino acid substitution. (B) Left, wild type low-altitude deer mouse Hif-2α can interact with both Cbp and p300, leading to activation of both Cbp- and p300-dependent genes (yellow and green, respectively). Right, high-altitude deer mouse Hif-2α has a defect in interaction with Cbp but not with p300. Hence, there is impaired activation of Cbp- but not p300-dependent genes. N = Asn-851 (equivalent to Asn-847 in human), the site of asparaginyl hydroxylation in Hif-2α. Asterisk = T755M amino acid substitution.
A second example of a differential effect on protein:protein interactions is high-altitude North American deer mouse Hif-2α. The T755M substitution impairs interaction with the transcriptional coactivator Cbp but allows maintenance of interaction with its closely related paralogue, p300 (Figure 4B) [52]. While very similar in overall structure, Cbp and p300 serve genetically non-redundant roles in the activation of Hif target genes [53]. It is plausible that this partial loss of function may account for selective effects of the high-altitude North American deer mouse Hif2a allele. For example, this allele is associated with reduced VAH but not hemoglobin levels in this high-altitude species [4,54].
These findings suggest that these alleles with differential protein:protein interaction effects might have been selected for during evolution in order to avoid widespread pleiotropic effects that might otherwise accompany strong loss or gain of function mutations. The observation that they are partial as opposed to complete loss of function is consistent with the notion that these alleles modulate, as opposed to completely ablate or activate, the hypoxic response in adaptive ways.
Complementary effects of apparently antagonistic alleles
Tibetans show evidence of natural selection acting on two key genes of the HIF pathway, PHD2 and HIF2A, with studies supporting both being partial loss of function alleles [1,2,32,68,70,73,78]. The partial loss of function of the zinc finger of Tibetan PHD2 would be predicted to lead to augmented HIF activity. Decreased transcription of the HIF2A allele, in contrast, would be predicted to lead to attenuated HIF-2α activity. On the surface, this is puzzling because these are antagonistic activities.
One resolution to this apparent paradox lies in the differential effect of the Tibetan PHD2 mutation on protein:protein interactions. As noted above, Tibetan PHD2 is defective in its interaction with p23 [73] (Figure 4A). Since the PHD2:p23 interaction is critical for maintenance of HVR (Figure 5A, left), this could account for the augmented HVR observed in Tibetans (Figure 5B, left). At the same time, Tibetan PHD2 maintains its interaction with NACA (Figure 5B, right) [28]. Since the PHD2:NACA interaction is critical for appropriate control of red cell mass (Figure 5A, right), this explains why a PHD2 allele that otherwise shows a defect in p23 interaction (which leads to increased HVR) does not predispose to erythrocytosis. Thus, Tibetan PHD2 behaves very differently than complete loss of function of PHD2. For example, inducible global loss of function of Phd2 in mice is accompanied by both increased HVR and erythrocytosis [18,22,23]. In the setting of a Tibetan PHD2 allele with this selective defect, a hypomorphic HIF2A allele could then be the main factor accounting for the protection against erythrocyosis and pulmonary hypertension that has been observed in Tibetans (Figure 5B, right). Thus, complementary effects of apparently antagonistic alleles can produce a set of phenotypes that neither allele could produce by itself.
Figure 5. Model for HIF pathway alleles in Tibetan adaptation to high altitude.
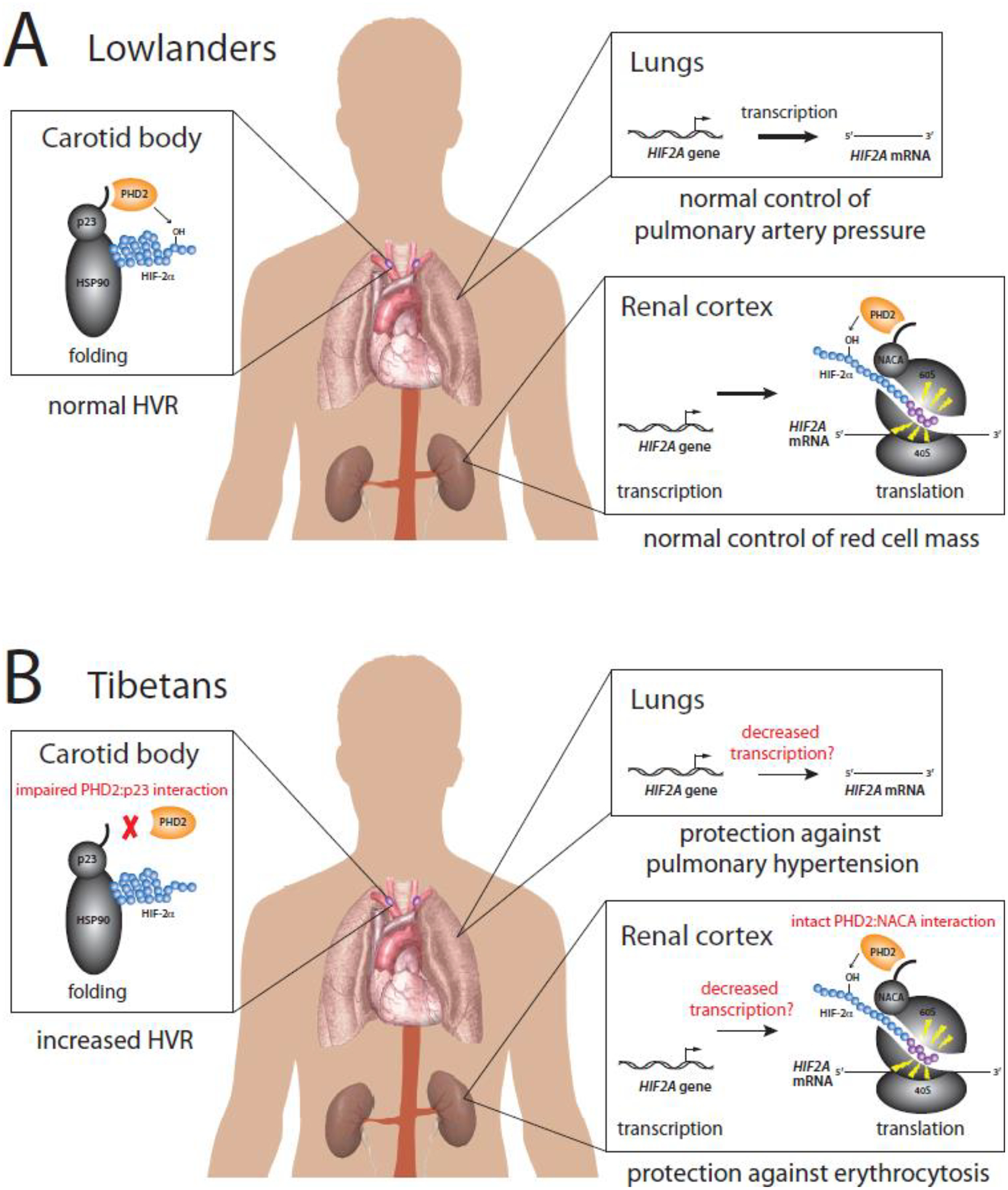
(A) Transcription, translation, and protein folding of HIF-2α in tissues of lowlanders. For simplicity, only select processes are shown in each tissue. Appropriate control of HVR, pulmonary artery pressure, and red cell mass is maintained. (B) In Tibetans at high altitude, impaired interaction of PHD2 with p23 in the carotid body leads to increased HVR (left). In the renal cortex, PHD2 interaction with NACA is preserved in the EPO-producing cells; however, HIF-2α activity is decreased, possibly due to decreased transcription of the HIF2A gene, leading to protection against erythrocytosis (right). Decreased transcription in the lungs may also lead to protection against pulmonary hypertension (right).
Naturally, this now leads to the question of why the presumed loss of function Tibetan HIF2A allele does not depress HVR and act in the opposite direction of the loss of function Tibetan PHD2 allele in the control of ventilation. The Tibetan HIF2A haplotype consists of multiple SNPs which, when tested in vitro using reporter genes and select cell lines, decrease transcription [79]. It is conceivable that these SNPs may have different effects in the EPO-producing interstitial cells of the renal cortex as compared to the Type I cells of the carotid body that play a critical role in oxygen sensing and maintenance of the HVR. In other words, it is plausible that the Tibetan HIF2A allele may exert its effects in a cell type-specific manner.
Concluding remarks
Functional correlations for high-altitude HIF pathway alleles are just beginning to emerge, and further investigations will reveal the extent to which the trends highlighted here extend to other species. Such studies may address other intriguing issues as well, such as whether these alleles select for other phenotypes (see Outstanding questions). The availability of in vitro and in vivo experimental methods to accurately model coding and noncoding nucleotide changes is critical. There are certainly challenges. It may be difficult to predict a priori which nucleotide changes will be functionally important. With coding sequence changes, for example, we now know that they sometimes occur in nonconserved, unstructured regions (Figure 2). Amino acid changes may also have differing effects. Their effects may only be manifest in specific amino acid contexts (e.g., high-altitude deer mouse Hif-2α) [52] or only within specific cells. That being said, a more complete understanding of human high-altitude adaptation may provide the basis for pharmacologically mimicking it. Such an approach could facilitate high altitude adaptation in low altitude residents who sojourn to high altitude, or alternatively ameliorate chronic mountain sickness (Monge’s disease) that can afflict long term high-altitude residents. More broadly, such an approach may also have an impact on more commonplace ischemic cardiovascular diseases, such as heart attack or stroke.
Outstanding questions.
Are all high-altitude HIF2a alleles loss of function alleles?
Can cell-type specific effects of high-altitude HIF pathway alleles be demonstrated?
Why are phenotypic outputs of high-altitude HIF pathway alleles not always identical across species? For example, a presumed loss of function HIF2A allele is associated with low Hb in Tibetans but not in high-altitude deer mice. Conversely, this allele is associated with blunted VAH in high-altitude deer mice but not in Tibetans.
What other phenotypes are selected for by high-altitude HIF pathway alleles?
Can noncoding sequence changes in high-altitude alleles be modeled in mice or other experimental animals?
Can high-altitude alleles such as the hypomorphic PHD2 and HIF2A alleles be mimicked pharmacologically to treat conditions such as chronic mountain sickness?
Highlights.
Many high-altitude mammals, including humans, display signatures of selection on genes of the HIF pathway, particularly HIF2A and PHD2.
HIF2A, encoding the transcription factor HIF-2α, is a recurrent target of selection. Mechanisms of convergent evolution include impaired heterodimerization, defective coactivator recruitment, and possibly decreased transcription of the HIF2A gene.
It has been observed that high altitude-associated amino acid changes in PHD2 and HIF-2α occur in nonconserved, unstructured regions of these proteins, and these changes can selectively alter interactions with other proteins, producing partial as opposed to complete loss of function.
In Tibetans, pairing of a partial loss of function PHD2 allele with a partial loss of function HIF2A allele could produce a panel of adaptive phenotypes that neither allele alone could produce, raising the possibility that this could be mimicked pharmacologically.
Acknowledgments
Work in the author’s laboratory has been supported by NIH grants R01-HL159611 and R33-HL120751, and NSF grant BCS-1638642. I thank Dr. Terence Lappin for valuable comments on the manuscript.
Glossary
- Adaptive allele
a gene variant that confers a survival or fitness benefit in an environment
- Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT)
a transcription factor, also known as HIF-β, that form heterodimeric complexes with a number of other transcription factors, including HIF-α and Aryl Hydrocarbon Receptor
- Basic Helix-Loop-Helix (bHLH)
a protein domain that binds to DNA which structurally consists of a basic region followed by two α helices separated by a loop
- C-terminal transcriptional activation domain (CTAD)
a segment of HIF-α that binds to Cbp/p300 in manner that is regulated by site-specific asparaginyl hydroxylation. It is likely largely unstructured
- Creb binding protein (Cbp)
a transcriptional coactivator with histone acetyl transferase activity that binds to a number of transcription factors, including HIF-α, to facilitate transcription at target genes
- Erythrocytosis
excessive red blood cells, leading to increased circulating red cell mass
- Factor Inhibiting HIF (FIH)
an oxygen-dependent asparaginyl hydroxylase that hydroxylates and thereby downregulates the activity of the HIF-α CTAD
- High altitude
typically defined as altitudes >8,000 ft (>2,500 m), an altitude at which oxygen saturation of hemoglobin in the blood begins to fall
- Hypobaric
low atmospheric pressure, associated with high altitude
- Hypoxia Inducible Factor (HIF)
a heterodimeric transcription factor consisting of α and β subunits that is the main mediator of the transcriptional response to hypoxia in animals
- Hypoxic ventilatory response (HVR)
the increase in breathing that occurs upon acute (seconds to minutes) exposure to hypoxia
- Oxygen-dependent degradation (ODD) domain
a segment of the HIF-α protein that is site-specifically prolyl hydroxylated by PHD2, thereby marking HIF-α for degradation. It is likely largely unstructured
- Per-Arnt-Sim (PAS)
a globular protein domain originally identified in the Period, Arnt, and Sim proteins
- Prolyl Hydroxylase Domain protein 2 (PHD2)
an oxygen-dependent prolyl hydroxylase that is the central oxygen sensor in the HIF pathway
- Pulmonary artery pressure (PAP)
the pressure in the pulmonary arteries that supply the lungs, elevations of which indicate pulmonary hypertension
- Right ventricular systolic pressure (RVSP)
the pressure in the right ventricle during systole. Since the right ventricle pumps blood through the pulmonary arteries, elevations in RVSP can be a reflection of pulmonary hypertension
- Ventilatory acclimatization to chronic hypoxia (VAH)
the augmentation of HVR that occurs upon chronic (hours to weeks) exposure to hypoxia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The author declares no competing interests.
References
- 1.Beall CM et al. (2010) Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences of the United States of America 107, 11459–11464. 10.1073/pnas.1002443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi X et al. (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simonson TS et al. (2010) Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75. 10.1126/science.1189406 [DOI] [PubMed] [Google Scholar]
- 4.Schweizer RM et al. (2019) Physiological and genomic evidence that selection on the transcription factor Epas1 has altered cardiovascular function in high-altitude deer mice. PLoS Genet 15, e1008420. 10.1371/journal.pgen.1008420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickson SL (2013) A genome wide study of genetic adaptation to high altitude in feral Andean Horses of the paramo. BMC Evol Biol 13, 273. 10.1186/1471-2148-13-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y et al. (2014) Population variation revealed high-altitude adaptation of Tibetan mastiffs. Mol Biol Evol 31, 1200–1205. 10.1093/molbev/msu070 [DOI] [PubMed] [Google Scholar]
- 7.Beall CM (2014) Adaptation to High Altitude: Phenotypes and Genotypes. Annual Review of Anthropology, Vol 43 43, 251–272. 10.1146/annurev-anthro-102313-030000 [DOI] [Google Scholar]
- 8.Bigham AW and Lee FS (2014) Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes & development 28, 2189–2204. 10.1101/gad.250167.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaelin WG Jr. and Ratcliffe PJ (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 10.Majmundar AJ et al. (2010) Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40, 294–309. S1097–2765(10)00750–1 [pii] 10.1016/j.molcel.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schodel J and Ratcliffe PJ (2019) Mechanisms of hypoxia signalling: new implications for nephrology. Nat Rev Nephrol 15, 641–659. 10.1038/s41581-019-0182-z [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408. 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storz JF and Cheviron ZA (2021) Physiological Genomics of Adaptation to High-Altitude Hypoxia. Annu Rev Anim Biosci 9, 149–171. 10.1146/annurev-animal-072820-102736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu D et al. (2015) Structural integration in hypoxia-inducible factors. Nature 524, 303–308. 10.1038/nature14883 [DOI] [PubMed] [Google Scholar]
- 15.Keith B et al. (2012) HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews. Cancer 12, 9–22. 10.1038/nrc3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Percy MJ et al. (2008) A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med 358, 162–168. 358/2/162 [pii] 10.1056/NEJMoa073123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale DP et al. (2008) Autosomal dominant erythrocytosis and pulmonary arterial hypertension associated with an activating HIF2 alpha mutation. Blood 112, 919–921 [DOI] [PubMed] [Google Scholar]
- 18.Hodson EJ et al. (2016) Regulation of ventilatory sensitivity and carotid body proliferation in hypoxia by the PHD2/HIF-2 pathway. J Physiol 594, 1179–1195. 10.1113/JP271050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Q et al. (2013) Erythrocytosis and Pulmonary Hypertension in a Mouse Model of Human HIF2A Gain of Function Mutation. The Journal of biological chemistry 288, 17134–17144. 10.1074/jbc.M112.444059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lappin TR and Lee FS (2019) Update on mutations in the HIF: EPO pathway and their role in erythrocytosis. Blood Rev 37, 100590. 10.1016/j.blre.2019.100590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaelin WG Jr. (2022) Von Hippel-Lindau disease: insights into oxygen sensing, protein degradation, and cancer. J Clin Invest 132. 10.1172/JCI162480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda K et al. (2008) Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood 111, 3229–3235. blood-2007–09-114561 [pii] 10.1182/blood-2007-09-114561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minamishima YA et al. (2008) Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood 111, 3236–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lando D et al. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewitson KS et al. (2002) Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem 277, 26351–26355 [DOI] [PubMed] [Google Scholar]
- 26.Arany Z et al. (1996) An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A 93, 12969–12973. 10.1073/pnas.93.23.12969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortmann BM et al. (2021) The HIF complex recruits the histone methyltransferase SET1B to activate specific hypoxia-inducible genes. Nat Genet 53, 1022–1035. 10.1038/s41588-021-00887-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song D et al. (2022) The ribosomal chaperone NACA recruits PHD2 to cotranslationally modify HIF-alpha. EMBO J 41, e112059. 10.15252/embj.2022112059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamerdinger M et al. (2019) Early Scanning of Nascent Polypeptides inside the Ribosomal Tunnel by NAC. Mol Cell 75, 996–1006 e1008. 10.1016/j.molcel.2019.06.030 [DOI] [PubMed] [Google Scholar]
- 30.Song D et al. (2013) Prolyl Hydroxylase Domain Protein 2 (PHD2) Binds a Pro-Xaa-Leu-Glu Motif, Linking It to the Heat Shock Protein 90 Pathway. The Journal of biological chemistry 288, 9662–9674. 10.1074/jbc.M112.440552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isaacs JS et al. (2002) Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem 277, 29936–29944 [DOI] [PubMed] [Google Scholar]
- 32.Song D et al. (2020) Tibetan PHD2, an allele with loss-of-function properties. Proc Natl Acad Sci U S A 117, 12230–12238. 10.1073/pnas.1920546117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azad P et al. (2017) High-altitude adaptation in humans: from genomics to integrative physiology. J Mol Med (Berl) 95, 1269–1282. 10.1007/s00109-017-1584-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beall CM (2006) Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr Comp Biol 46, 18–24. 10.1093/icb/icj004 [DOI] [PubMed] [Google Scholar]
- 35.Julian CG and Moore LG (2019) Human Genetic Adaptation to High Altitude: Evidence from the Andes. Genes (Basel) 10. 10.3390/genes10020150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petousi N and Robbins PA (2014) Human adaptation to the hypoxia of high altitude: the Tibetan paradigm from the pregenomic to the postgenomic era. J Appl Physiol (1985) 116, 875–884. 10.1152/japplphysiol.00605.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheinfeldt LB and Tishkoff SA (2010) Living the high life: high-altitude adaptation. Genome biology 11, 133. 10.1186/gb-2010-11-9-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonson TS (2015) Altitude Adaptation: A Glimpse Through Various Lenses. High Alt Med Biol 16, 125–137. 10.1089/ham.2015.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baik AH and Jain IH (2020) Turning the Oxygen Dial: Balancing the Highs and Lows. Trends Cell Biol 30, 516–536. 10.1016/j.tcb.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witt KE and Huerta-Sanchez E (2019) Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos Trans R Soc Lond B Biol Sci 374, 20180235. 10.1098/rstb.2018.0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beall CM (2007) Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A 104 Suppl 1, 8655–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natarajan C et al. (2013) Epistasis among adaptive mutations in deer mouse hemoglobin. Science 340, 1324–1327. 10.1126/science.1236862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz JF and Scott GR (2019) Life Ascending: Mechanism and Process in Physiological Adaptation to High-Altitude Hypoxia. Annu Rev Ecol Evol Syst 50, 503–526. 10.1146/annurev-ecolsys-110218-025014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng Y et al. (2011) Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol 28, 1075–1081. 10.1093/molbev/msq290 [DOI] [PubMed] [Google Scholar]
- 45.Xu S et al. (2011) A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol 28, 1003–1011. 10.1093/molbev/msq277 [DOI] [PubMed] [Google Scholar]
- 46.Huerta-Sanchez E et al. (2014) Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197. 10.1038/nature13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hackinger S et al. (2016) Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum Genet 135, 393–402. 10.1007/s00439-016-1641-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen K et al. (2023) High-Altitude Andean H194R HIF2A Allele Is a Hypomorphic Allele. Mol Biol Evol 40, msad162. 10.1093/molbev/msad162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu D et al. (2022) How Placenta Promotes the Successful Reproduction in High-Altitude Populations: A Transcriptome Comparison between Adaptation and Acclimatization. Mol Biol Evol 39. 10.1093/molbev/msac120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foll M et al. (2014) Widespread signals of convergent adaptation to high altitude in Asia and america. American journal of human genetics 95, 394–407. 10.1016/j.ajhg.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichstaedt CA et al. (2017) Evidence of Early-Stage Selection on EPAS1 and GPR126 Genes in Andean High Altitude Populations. Sci Rep 7, 13042. 10.1038/s41598-017-13382-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song DS et al. (2021) High-altitude deer mouse hypoxia-inducible factor-2 alpha shows defective interaction with CREB-binding protein. Journal of Biological Chemistry 296. ARTN 100461 10.1016/j.jbc.2021.100461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasper LH et al. (2005) Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J 24, 3846–3858. 10.1038/sj.emboj.7600846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivy CM et al. (2022) Genetic variation in HIF-2alpha attenuates ventilatory sensitivity and carotid body growth in chronic hypoxia in high-altitude deer mice. J Physiol 600, 4207–4225. 10.1113/JP282798 [DOI] [PubMed] [Google Scholar]
- 55.Wang GD et al. (2014) Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the tibetan plateau. Genome Biol Evol 6, 2122–2128. 10.1093/gbe/evu162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gou X et al. (2014) Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res 24, 1308–1315. 10.1101/gr.171876.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song S et al. (2016) Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genomics 17, 122. 10.1186/s12864-016-2449-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X et al. (2019) EPAS1 gain-of-function mutation contributes to high-altitude adaptation in Tibetan horses. Mol Biol Evol 36, 2591–2603. 10.1093/molbev/msz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma YF et al. (2019) Population Genomics Analysis Revealed Origin and High-altitude Adaptation of Tibetan Pigs. Sci Rep 9, 11463. 10.1038/s41598-019-47711-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu N et al. (2022) A highland-adaptation mutation of the Epas1 protein increases its stability and disrupts the circadian clock in the plateau pika. Cell Rep 39, 110816. 10.1016/j.celrep.2022.110816 [DOI] [PubMed] [Google Scholar]
- 61.Newman JH et al. (2015) Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nat Commun 6, 6863. 10.1038/ncomms7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heaton MP et al. (2016) Using diverse U.S. beef cattle genomes to identify missense mutations in EPAS1, a gene associated with pulmonary hypertension. F1000Res 5, 2003. 10.12688/f1000research.9254.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheinfeldt LB et al. (2012) Genetic adaptation to high altitude in the Ethiopian highlands. Genome biology 13, R1. 10.1186/gb-2012-13-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McIntosh BE et al. (2010) Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol 72, 625–645. 10.1146/annurev-physiol-021909-135922 [DOI] [PubMed] [Google Scholar]
- 65.Bigham A et al. (2010) Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet 6. 10.1371/journal.pgen.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorenzo FR et al. (2014) A genetic mechanism for Tibetan high-altitude adaptation. Nature genetics 46, 951–956. 10.1038/ng.3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiang K et al. (2013) Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol Biol Evol 30, 1889–1898. 10.1093/molbev/mst090 [DOI] [PubMed] [Google Scholar]
- 68.Peng Y et al. (2017) Down-Regulation of EPAS1 Transcription and Genetic Adaptation of Tibetans to High-Altitude Hypoxia. Mol Biol Evol 34, 818–830. 10.1093/molbev/msw280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wuren T et al. (2014) Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PLoS One 9, e88252. 10.1371/journal.pone.0088252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J et al. (2017) Genetic signatures of high-altitude adaptation in Tibetans. Proc Natl Acad Sci U S A 114, 4189–4194. 10.1073/pnas.1617042114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tashi T et al. (2017) Gain-of-function EGLN1 prolyl hydroxylase (PHD2 D4E:C127S) in combination with EPAS1 (HIF-2alpha) polymorphism lowers hemoglobin concentration in Tibetan highlanders. J Mol Med (Berl) 95, 665–670. 10.1007/s00109-017-1519-3 [DOI] [PubMed] [Google Scholar]
- 72.Jeong C et al. (2018) Detecting past and ongoing natural selection among ethnically Tibetan women at high altitude in Nepal. PLoS Genet 14, e1007650. 10.1371/journal.pgen.1007650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song D et al. (2014) Defective Tibetan PHD2 Binding to p23 Links High Altitude Adaptation to Altered Oxygen Sensing. The Journal of biological chemistry 289, 14656–14665. 10.1074/jbc.M113.541227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bishop T et al. (2013) Carotid body hyperplasia and enhanced ventilatory responses to hypoxia in mice with heterozygous deficiency of PHD2. The Journal of physiology 591, 3565–3577. 10.1113/jphysiol.2012.247254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brutsaert TD et al. (2019) Association of EGLN1 gene with high aerobic capacity of Peruvian Quechua at high altitude. Proc Natl Acad Sci U S A 116, 24006–24011. 10.1073/pnas.1906171116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aggarwal S et al. (2010) EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. Proceedings of the National Academy of Sciences of the United States of America 107, 18961–18966. 10.1073/pnas.1006108107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chiou KL et al. (2022) Genomic signatures of high-altitude adaptation and chromosomal polymorphism in geladas. Nat Ecol Evol 6, 630–643. 10.1038/s41559-022-01703-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petousi N et al. (2014) Tibetans living at sea level have a hyporesponsive hypoxia-inducible factor system and blunted physiological responses to hypoxia. J Appl Physiol (1985) 116, 893–904. 10.1152/japplphysiol.00535.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gray OA et al. (2022) A pleiotropic hypoxia-sensitive EPAS1 enhancer is disrupted by adaptive alleles in Tibetans. Sci Adv 8, eade1942. 10.1126/sciadv.ade1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu H et al. (2017) Evolutionary history of Tibetans inferred from whole-genome sequencing. PLoS Genet 13, e1006675. 10.1371/journal.pgen.1006675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu DD et al. (2018) Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat Ecol Evol 2, 1139–1145. 10.1038/s41559-018-0562-y [DOI] [PubMed] [Google Scholar]
- 82.Chen N et al. (2018) Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat Commun 9, 2337. 10.1038/s41467-018-04737-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho YS et al. (2013) The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun 4, 2433. 10.1038/ncomms3433 [DOI] [PMC free article] [PubMed] [Google Scholar]


