Abstract
Rhodanese domains are ubiquitous structural modules occurring in the three major evolutionary phyla. They are found as tandem repeats, with the C-terminal domain hosting the properly structured active-site Cys residue, as single domain proteins or in combination with distinct protein domains. An increasing number of reports indicate that rhodanese modules are versatile sulfur carriers that have adapted their function to fulfill the need for reactive sulfane sulfur in distinct metabolic and regulatory pathways. Recent investigations have shown that rhodanese domains are also structurally related to the catalytic subunit of Cdc25 phosphatase enzymes and that the two enzyme families are likely to share a common evolutionary origin. In this review, the rhodanese/Cdc25 phosphatase superfamily is analyzed. Although the identification of their biological substrates has thus far proven elusive, the emerging picture points to a role for the amino-acid composition of the active-site loop in substrate recognition/specificity. Furthermore, the frequently observed association of catalytically inactive rhodanese modules with other protein domains suggests a distinct regulatory role for these inactive domains, possibly in connection with signaling.
Introduction
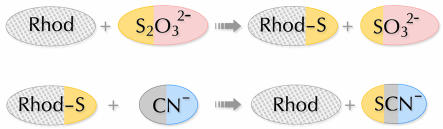
Rhodaneses (thiosulfate:cyanide sulfurtransferases or TSTs; E.C. 2.8.1.1) are widespread enzymes that catalyze the transfer of a sulfane sulfur atom from thiosulfate to cyanide in vitro (Figure 1). 3-mercaptopyruvate:cyanide sulfurtransferases (MSTs; E.C. 2.8.1.2) catalyze a similar reaction, using 3-mercaptopyruvate as the sulfur donor. MSTs display clear sequence homology with rhodaneses, with which they share up to 66% identical residues (Nagahara et al., 1995). The most well characterized sufurtransferase is bovine liver rhodanese, which has been the subject of numerous functional investigations (e.g. Westley et al., 1983; Berni et al., 1991; Luo and Horowitz, 1994; Nandi et al., 2000). The catalysis occurs via a double displacement mechanism involving the transient formation of a persulfide-containing intermediate (Rhod-S), in which the transferring sulfur is bound to the invariant catalytic Cys residue (Figure 1).
Fig. 1. Scheme representing the sulfur-transfer reaction catalyzed by rhodanese.
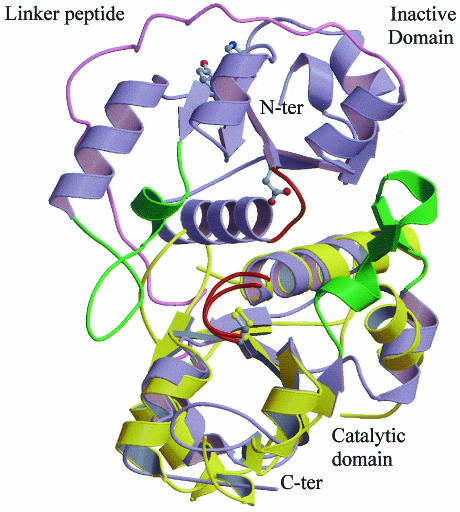
The crystal structures of bovine (Rhobov) and Azotobacter vinelandii (RhdA) rhodaneses are known (Ploegman et al., 1978; Bordo et al., 2000). These two proteins, which exhibit 22% sequence identity, display very similar three-dimensional conformations. They are each composed of two identically folded domains (Figure 2), called hereafter rhodanese domains or modules, each about 120 amino acids long, which display weak sequence similarity to one another (13 and 21% identical residues, respectively). Due to this peculiar tandem domain architecture, rhodanese has been considered the prototype of divergent evolution from a common ancestor protein which, after gene duplication and under the constraint of tertiary structure conservation, led to the almost complete obliteration of sequence similarity between the two domains (Ploegman et al., 1978). Only the C-terminal rhodanese domain (referred to as the catalytic domain) hosts the catalytic Cys residue, which is the first residue of a six-amino-acid active-site loop that folds in a cradle-like structure defining the enzyme catalytic pocket; in the N-terminal domain the same position is occupied by Asp (Figure 2). The recent biochemical and structural characterization of GlpE, a TST from Escherichia coli composed of a single rhodanese domain (Ray et al., 2000; Spallarossa et al., 2001), indicates that the catalytically inactive N-terminal domain found in TSTs and MSTs is not essential for catalysis. Furthermore, the structure of the catalytic domain of two human Cdc25 phosphatases, Cdc25A and Cdc25B, two key enzymes involved in cell cycle control, has revealed that these proteins display a rhodanese-like three-dimensional fold and maintain the same location of the active-site Cys residue (Figure 2; Fauman et al., 1998; Reynolds et al., 1999). In spite of the very weak sequence similarity (17% identical residues), sensitive profile analysis indicated that the two enzyme families stem from a common ancestor (Hofmann et al., 1998). The most relevant structural difference between rhodanese and Cdc25 enzymes is the length of the active-site loop, which in Cdc25 proteins is formed by seven residues instead of the six in sulfurtransferases; this results in a wider catalytic pocket that can accommodate a phosphorous atom, which has a van der Waals radius slightly larger than a sulfur atom (1.9 versus 1.85 Å).
Fig. 2. Three-dimensional structure of A. vinelandii RhdA and of human Cdc25A phosphatase, superimposed on their respective catalytic domains. RhdA and Cdc25A are shown in blue and yellow, respectively. Active-site loops and the structurally equivalent loop on the RhdA N-terminal domain are shown in red. The catalytic Cys and the Asp counterpart on the inactive domain are represented in ball-and-stick. Conserved residues, Tyr37 and His41 (RhdA numbering), putatively involved in regulative function (see text) are also shown. The linker polypeptide connecting the two RhdA domains is shown in pink. The βC–αC loops involved in RhdA in interdomain interaction (Bordo et al., 2000) are shown in green.
The biological role of sulfurtransferases is still largely debated, since the identification of their in vivo substrates has thus far proven unsuccessful, and it appears unlikely that these substrates are those identified by in vitro reactions (Nandi et al., 2000; Ray et al., 2000). Proposed functions include cyanide detoxification (Sorbo, 1957), formation of prosthetic groups in iron–sulfur proteins (Pagani et al., 1984), maintenance of the sulfane pool (Westley, 1989), selenium metabolism (Ogasawara et al., 2001) and thiamin biosynthesis (Palenchar et al., 2000). The involvement of rhodanese enzymes in distinct biological functions is also suggested by the wide variability in the amino acids of the active-site loop. In fact, the amino-acid side chains extending from this structural element define the ridge of the catalytic pocket and are expected, therefore, to play a key role in substrate recognition and catalytic activity.
In this review, the information on rhodanese-like homology domains contained in the protein database is analyzed together with the available structural and biological information to provide a general view of the distinct structural/functional contexts in which these structural domains occur in living organisms.
Rhodanese homology domains encoded in sequenced genomes
The complete sequencing of more than 80 genomes has shown that proteins bearing weak but significant sequence similarity to known rhodaneses or Cdc25 phosphatase enzymes are widely distributed among living organisms. Sensitive homology search programs based on profile analysis or Hidden Markov Models have detected rhodanese homology domains in about 500 genes in the three major evolutionary phyla (see, for example, SMART, http://smart.embl-heidelberg.de; DART, http://www. ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml; Hofmann et al., 1998). Often, distinct proteins containing rhodanese domains are encoded by the same genome. For example, in the human genome, 47 such instances are observed, whereas in the E. coli K12 genome, seven rhodanese-like proteins are found (see, for example, the COG database; Tatusov et al., 2000). From a structural viewpoint, rhodanese-like proteins are either composed of two rhodanese domains, with the C-terminal domain displaying the putative catalytic Cys as observed in Rhobov and RhdA, or composed of a single catalytic rhodanese domain, as found in GlpE. Rhodanese domains, either catalytic or inactive (i.e. where the active-site Cys is replaced by another residue), are also found associated with other protein domains such as MAPK-phosphatases (Keyse and Ginsburg, 1993; Fauman et al., 1998; Hofmann et al., 1998) or ThiI, an E. coli enzyme involved in thiamin and thiouridine biosynthesis (Palenchar et al., 2000).
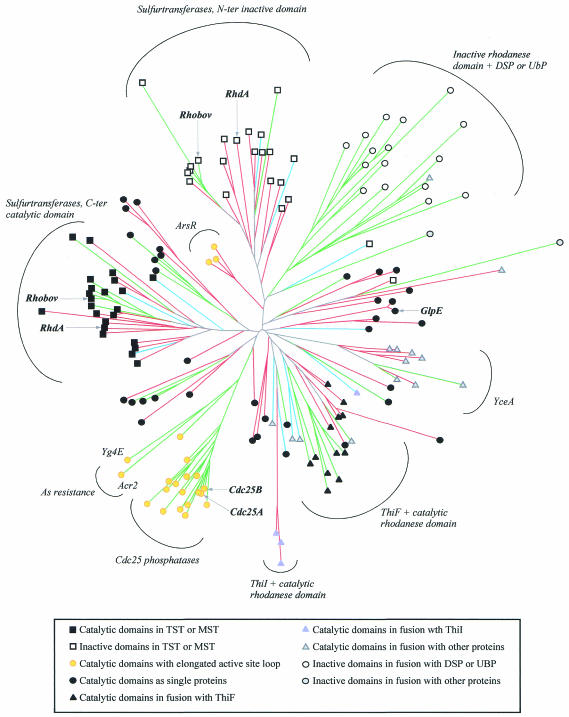
A multiple alignment of the rhodanese homology domain, containing a representative subset of the entries detected in the genomic database, is provided by the SMART resource (Schultz et al., 1998). The deduced neighbor-joining (N-J) tree provides a graphical representation of the mutual similarity among distinct amino-acid sequences (Figure 3). As expected from the wide amino-acid sequence divergence found within this family, bootstrap analysis indicates that the detailed tree structure might be unreliable, especially in its deep branching. Nevertheless, the tree is useful for identifying protein subfamilies that may relate to distinct biological functions, as discussed below.
Fig. 3. Neighbor-joining tree representing the rhodanese superfamily generated by CLUSTALW (correction for multiple substitution was adopted; Thompson et al., 1997). The multiple alignment was derived from that included in the SMART resource, to which four E. coli, two archaea and two ThiI proteins were added, for a total of 155 rhodanese modules. The SMART alignment represents an even sample of the total number of rhodanese modules, as closely related sequences are represented only once. The tree was displayed and validated by bootstrap analysis using programs in the PHYLIP package (Felsenstein, 1989). Archaeal, bacterial and eukaryotic proteins are represented in blue, red and green, respectively. Symbols are used to indicate the distinct structural or functional roles of the rhodanese modules, as described in the inset.
Catalytically active rhodanese and Cdc25 phosphatase domains
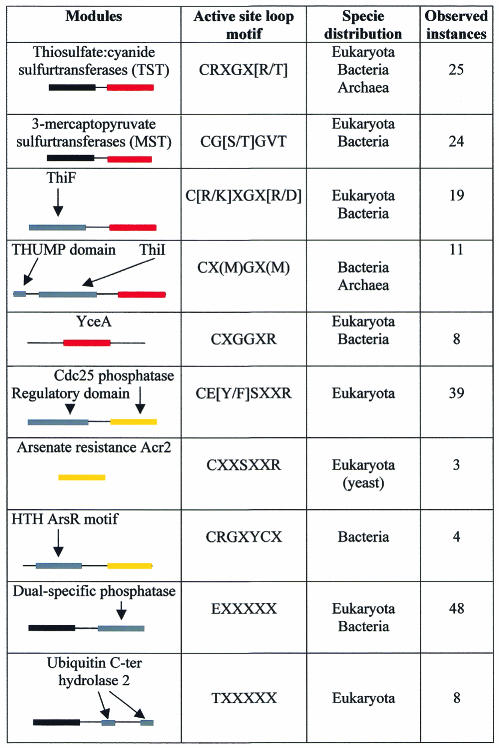
The N- and C-terminal domains of TST enzymes form two distinct subfamilies (Figure 3). Each subfamily also contains previously characterized MST sequences from Rattus norvegicus, Arabidopsis thaliana and E. coli (Nagahara et al., 1995; Nagahara and Nishino, 1996; Papenbrock and Schmidt, 2000; Colnaghi et al., 2001). In TSTs and MSTs, the putative catalytic Cys is invariably found in the C-terminal domain, whereas in the N-terminal domain, this residue is often replaced by Asp or Gly. In three-dimensional structures of RhdA and Rhobov, the side chain of this Asp residue is involved in multiple hydrogen bonds with the main chain amide groups of the active-site loop (Ploegman et al., 1978; Bordo et al., 2000), which makes this side chain unavailable for substrate or ion binding. From an evolutionary viewpoint, as the clusters of both N- and C-terminal domains of TST/MST subfamilies contain eukaryotic, bacterial and archaeal sequences (Figure 3), the gene duplication event that gave rise to the tandem domain architecture found in MSTs and TSTs must have occurred very early during evolution, probably before the diversification of living organisms into the three major evolutionary phyla. Notably, active-site loop amino-acid sequences are distinct in TSTs and in MSTs, which display motifs CRXGX[R/T] and CG[S/T]GVT, respectively (square brackets indicate alternative residues, ‘X’ any amino acid; Figure 4). The active-site loop of known TSTs always contains two basic residues (i.e. if the last loop residue is Thr, a basic residue is observed at one of the X positions), whereas no charged residues are observed in biochemically characterized MSTs; this may relate to the distinct ionic charge of the respective in vitro substrates, thiosulfate (2–) and 3-mercaptopyruvate (1–).
Fig. 4. Co-occurrence of rhodanese modules in association with other protein domains. Domains are schematized as colored boxes. Catalytic rhodanese modules with a six-amino-acid active-site loop are shown in red, and inactive rhodanese modules are displayed in black. Rhodanese modules with a seven-amino-acid active-site loop are displayed in yellow. Other protein domains are shown in gray. Active-site loop positions displaying at least 80% amino-acids conservation within each subfamily are also shown as conserved motifs.
Cdc25 phosphatases are composed of variable N-terminal domains, which are poorly characterized but are suspected to serve regulatory functions (Draetta and Eckstein, 1997; Forrest and Gabrielli, 2001), and conserved catalytic, rhodanese-like, C-terminal domains (Figure 2; Fauman et al., 1998; Reynolds et al., 1999). In the N-J tree, these catalytic domains form a distinct clade (Figure 3). Similarly to MSTs/TSTs, the active-site amino-acid sequences of Cdc25 enzymes display a conserved motif (CE[Y/F]SXXR; Figure 4), which is consistent with the role of this structural element in substrate recognition.
An elongated seven-amino-acid active-site loop, with the putative catalytic Cys at the first position, is also found in the yeast arsenic-resistance protein Acr2 and in the closely related yeast protein Yg4E (Figure 3; Bobrowicz et al., 1997; Mukhopadhyay et al., 2000). Although both proteins lack the regulatory N-terminal domain observed in Cdc25 phosphatase enzymes, the N-J tree suggests a close evolutionary relationship with this family of enzymes. A seven-amino-acid active-site loop bearing the Cys at the first position is also found in a small rhodanese subfamily, formed by eukaryotic proteins from Mycobacterium tuberculosis and Streptomyces peucetis. These rhodanese-like domains are found in association with the helix–turn–helix motif of the arsenic resistance operon repressor ArsR (Figures 3 and 4). This structural association supports the hypothesis that these rhodanese modules are able to bind arsenic, and it is tempting to speculate that they may act as sensory domains modulating ArsR activity.
Taken together, the available biochemical data support the hypothesis that rhodanese domains displaying a seven-amino-acid active-site loop are able to bind substrates containing phosphorous or the chemically similar arsenic, whereas rhodanese-like domains displaying a six-amino-acid loop with Cys at the first position interact with substrates containing reactive sulfur or, in some cases, selenium (Ogasawara et al., 2001). Considering the very distant relationship between bacterial ArsR proteins and eukaryotic Cdc25/Acr2 proteins, it is likely that these two subfamilies result from two independent mutational events, each resulting from a single amino-acid insertion in the active-site loop of sulfur-binding rhodanese modules.
Proteins composed of a single rhodanese-like module, displaying the putative catalytic Cys and a six-amino-acid active-site loop, are widespread, especially among bacteria (Figure 3). These domains display largely divergent amino-acid sequences, being a cluster of uncharacterized proteins similar to E. coli YceA and are the only example of bacterial and eukaryotic proteins forming a well-defined subfamily of closely related sequences (Figure 4). Besides GlpE from E. coli, only the periplasmic sulfide dehydrogenase (Sud) from Wolinella succinogenes has been described as a (polysulfide:cyanide) sulfur- transferase (Krafft et al., 1995; Klimmek et al., 1998). At the genetic level, single rhodanese domain proteins have been associated with specific stress conditions. Examples include the Drosophila melanogaster heat shock protein 67B2, the Vibrio cholerae shock protein Q9KN65 or the E. coli phage shock protein GspE. Single rhodanese domains are also associated with the process of leaf senescence in A. thaliana, Nicotiana tabacum and Raphanus sativus (Sen1, Ntdin and Din1, respectively; Azumi and Watanabe, 1991; Oh et al., 1996).
Putatively catalytic rhodanese domains also frequently associate with proteins involved in thiamin, thiouridine or molybdopterin biosynthesis (Figure 3). In particular, a rhodanese domain is found at the C-terminal end of several ThiF/MoeB proteins (see, for example, the DART resource). In these cases the active-site loop consensus sequence is C[R/K]XGXR and C[R/K]XGXD in bacteria and eukaryota, respectively (Figure 4). ThiF, which is involved in the synthesis of thiamin in bacteria, is deemed responsible for the adenylation of ThiS. Therefore, the C-terminal carboxy-adenylate ThiS might interact with a sulfur donor to proceed in the biosynthesis of thiazole, which is an intermediate compound in thiamin biosynthesis (Taylor et al., 1998; Xi et al., 2001). In this context, the role of the rhodanese module might be to provide the sulfane sulfur required by ThiS.
In bacteria and archaea, a catalytic rhodanese module is also recurrently found in association with ThiI, another enzyme participating in thiouridine and thiamin biosynthesis (Figure 3). The sulfurtransferase activity of this rhodanese domain has been established in vitro using ThiI from E. coli (Palenchar et al., 2000). Also, in this case, the significance of sulfur-delivering proteins in the synthesis of thiouridine and thiamin has been recognized (Begley et al., 1999). In particular, the sulfur transfer between IscS and ThiI has recently been shown and the essential role of the rhodanese Cys residue established (Kambampati and Lauhon, 1999; Mueller et al., 2001).
Catalytically inactive rhodanese domains
Rhodanese domains lacking the active-site Cys, which is often replaced by acidic or non-polar residues, are found as N-terminal domains in about one-third of the known dual-specific phosphatases (DSP) and in several ubiquitinating enzymes (UBP), including yeast UBP4, UBP5, UBP7, human UBP8 and the murine deubiquitinating enzyme UbpY (Fauman et al., 1998; Hofmann et al., 1998). In the N-J tree, these inactive rhodanese domains form a distinct group of sequences that appear to form a clade (Figure 3). The role of the inactive rhodanese domains, either those found in TST and MST enzymes or those in association with DSP or UBP, is still to be elucidated. Although in bovine rhodanese the N-terminal domain is deemed to be involved in substrate recognition mediated by a loop folding over the C-terminal domain in the vicinity of the active site (Ploegman et al., 1979), the absence of a structurally equivalent loop in A. vinelandii RhdA indicates that substrate recognition in sulfurtransferases, if mediated by the N-terminal domain at all, must rely upon more complex mechanisms involving non-contiguous surface patches. In DSP and UBP, the N-terminal inactive rhodanese domain might play a regulatory role, perhaps in connection with signaling. In this regard, the multiple sequence alignment of the rhodanese-like domains associated to DSP and UBP (see, for example, SMART) highlights the conservation of two residues, Tyr37 and His41 (RhdA numbering; Bordo et al., 2000), which occupy neighboring positions at the protein surface (Figure 2). This points to the possibility that either of these two amino acids could be a target for tyrosine or histidine kinases. Obviously, further experimental evidence is needed to prove this hypothesis.
Conclusions
The complete sequencing of a variety of genomes has shown that distinct proteins containing rhodanese-like modules are often encoded by the same genome and now enables the identification of distinct subfamilies of rhodanese-like proteins, possibly associated with different biological functions (paralogs). Each subfamily, displayed in the N-J family tree as a cluster of closely related sequences, harbors conserved motifs in the amino-acid sequence of the (putative) active-site loop. However, despite the increasing amount of experimental information, there remains the open question of the identity of the true biological substrates, either proteins or small molecules, that interact with rhodanese domains. The information now available in the genomic databases should be very helpful in designing appropriate experiments to answer these questions.

Domenico Bordo & Peer Bork

Acknowledgments
Acknowledgements
We wish to thank Toby Gibson for critical reading of the manuscript and Jose Castresana for many helpful suggestions. This research was supported in part by the MIUR grant ‘Sulfotransferasi procariotiche’ and CNR strategic project ‘Molecular genetics’.
References
- Azumi Y. and Watanabe, A. (1991) Evidence for a senescence-associated gene induced by darkness. Plant Physiol., 95, 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley T.P., Xi, J., Kinsland, C., Taylor, S. and McLafferty, F. (1999) The enzymology of sulfur activation during thiamin and biotin biosynthesis. Curr. Opin. Chem. Biol., 3, 623–629. [DOI] [PubMed] [Google Scholar]
- Berni R., Musci, G., Pallini, R. and Cannella, C. (1991) Chemical modification of rhodanese with sulfite. Free Radical Res. Commun., 15, 203–209. [DOI] [PubMed] [Google Scholar]
- Bobrowicz P., Wysocki, R., Owsianik, G., Goffeau, A. and Ulaszewski, S. (1997) Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisae. Yeast, 13, 819–828. [DOI] [PubMed] [Google Scholar]
- Bordo D., Deriu, D., Colnaghi, R., Carpen, A., Pagani, S. and Bolognesi, M. (2000) The crystal structure of a sulfurtransferase from Azotobacter vinelandii highlights the evolutionary relationship between the rhodanese and phosphatase enzyme families. J. Mol. Biol., 298, 691–704. [DOI] [PubMed] [Google Scholar]
- Colnaghi R., Cassinelli, G., Drummond, M., Forlani, F. and Pagani, S. (2001) Properties of the Escherichia coli rhodanese-like protein SseA: contribution of the active-site residue Ser240 to sulfur recognition. FEBS Lett., 500, 153–156. [DOI] [PubMed] [Google Scholar]
- Draetta G. and Eckstein, J. (1997) Cdc25 protein phosphatases in cell proliferation. Biochim. Biophys. Acta, 1332, M53–M63. [DOI] [PubMed] [Google Scholar]
- Fauman E.B., Cogswell, J.P., Lovejoy, B., Rocque, W.J., Holmes, W., Montana, V.G., Piwnica-Worms, H., Rink, M. and Saper, M.A. (1998) Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell, 93, 617–625. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989) PHYLIP. Phylogeny Inference Package (Version 3.2). Cladistics, 5, 164–166. [Google Scholar]
- Forrest A. and Gabrielli, B. (2001) Cdc25B activity is regulated by 14-3-3. Oncogene, 20, 4393–4401. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Bucher, P. and Kajava, A.V. (1998) A model of Cdc25 phosphatase catalytic domain and Cdk-interaction surface based on the presence of a rhodanese homology domain. J. Mol. Biol., 282, 195–208. [DOI] [PubMed] [Google Scholar]
- Kambampati R. and Lauhon, C.T. (1999) IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry, 38, 16561–16568. [DOI] [PubMed] [Google Scholar]
- Keyse S.M. and Ginsburg, M. (1993) Amino acid sequence similarity between CL100, a dual specific MAP kinase phosphatase and Cdc25. Trends Biochem. Sci., 18, 377–378. [DOI] [PubMed] [Google Scholar]
- Klimmek O., Kreis, V., Klein, C., Simon, J., Wittershagen, A. and Kroger, A. (1998) The function of the periplasmic Sud protein in polysulfide respiration of Wolinella succinogenes. Eur. J. Biochem., 253, 263–269. [DOI] [PubMed] [Google Scholar]
- Krafft T., Gross, R. and Kroger, A (1995) The function of Wolinella succinogenenes psr gene in electron transport with polysulfide as terminal electron acceptor. Eur. J. Biochem., 230, 601–606. [DOI] [PubMed] [Google Scholar]
- Luo G.-X. and Horowitz, P.M. (1994) The sulfurtransferase activity and structure of rhodanese are affected by site-directed replacement of Arg186 and Lys249. J. Biol. Chem., 269, 399–408. [PubMed] [Google Scholar]
- Mueller E.G., Palenchar, P.M. and Buck, C.J. (2001) The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J. Biol. Chem., 276, 33588–33595. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R., Shi, J. and Rosen, B. (2000) Purification and characterization of ACR2P, the Saccharomyces cerevisae arsenate reductase. J. Biol. Chem., 275, 21149–21157. [DOI] [PubMed] [Google Scholar]
- Nagahara N. and Nishino, T. (1996) Role of amino acid residues in the active site of rat liver mercaptopyruvate sulfurtransferase. cDNA cloning, overexpression, and site-directed mutagenesis. J. Biol. Chem., 271, 27395–27401. [DOI] [PubMed] [Google Scholar]
- Nagahara N., Okazaki, T. and Nishino, T. (1995) Cytosolic mercaptopyruvate sulfurtransferase is evolutionarily related to mitochondrial rhodanese. J. Biol. Chem., 270, 16230–16235. [DOI] [PubMed] [Google Scholar]
- Nandi D.L., Horowitz, P.M. and Westley, J. (2000) Rhodanese as thioredoxin oxidase. Int. J. Biochem. Cell Biol., 32, 465–473. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y., Lacourciere, G. and Stadtman, T.C. (2001) Formation of a selenium-substituted rhodanese by reaction with selenite and glutathione: possible role of a protein perselenide in a selenium delivery system. Proc. Natl Acad. Sci. USA, 98, 9494–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.A., Lee, S.Y., Chung, I.G., Lee, C.H. and Nam, H.G. (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol., 30, 739–754. [DOI] [PubMed] [Google Scholar]
- Pagani S., Bonomi, F. and Cerletti, P. (1984) Enzymic synthesis of the iron–sulfur cluster of spinach ferredoxin. Eur. J. Biochem., 142, 361–366. [DOI] [PubMed] [Google Scholar]
- Palenchar P.M., Buck, C.J., Cheng, H., Larson, T.J. and Mueller, E.G. (2000) Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J. Biol. Chem., 275, 8283–8286. [DOI] [PubMed] [Google Scholar]
- Papenbrock J. and Schmidt, A. (2000) Characterization of two sulfurtransferase isozymes from Arabidopsis thaliana. Eur. J. Biochem., 267, 5571–5579. [DOI] [PubMed] [Google Scholar]
- Ploegman J.H., Drent, G., Kalk, K.H., Hol, W.G.J., Heinrikson, R.L., Keim, P., Weng, L. and Russel, J. (1978) The covalent and tertiary structure of bovine liver rhodanese. Nature, 273, 124–129. [DOI] [PubMed] [Google Scholar]
- Ploegman J.H., Drent, G., Kalk, K.H. and Hol, W.G.J. (1979) The structure of bovine liver rhodanese. II. The active site in the sulfur-substituted and sulfur-free enzyme. J. Mol. Biol., 127, 149–162. [DOI] [PubMed] [Google Scholar]
- Ray W.K., Zeng, G., Potters, M.B., Mansuri, A.M. and Larson, T.J. (2000) Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J. Bacteriol., 182, 2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R.A., Yem, A.W., Wolfe, C.L., Deibel, M.R., Jr, Chidester, C.G. and Watenpaugh, K.D. (1999) Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle. J. Mol. Biol., 293, 559–568. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz, F., Bork, P. and Ponting, C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbo B.H. (1957) Sulfite and complex-bound cyanide as sulfur acceptors for rhodanese. Acta Chem. Scand., 11, 628–633. [Google Scholar]
- Spallarossa A., Donahue, J., Larson, T.J., Bolognesi, M. and Bordo, D. (2001) Escherichia coli GlpE is a prototype sulfurtransferase for the single-domain rhodanese homology superfamily. Structure, 9, 1117–1125. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Galperin, M.Y., Natale, D.A. and Koonin, E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.V., Kelleher, N.L., Kinsland, C., Chiu, H.-J., Costello, C.A., Backstrom, A.D., McLafferty, F.W. and Begley, T.P. (1998) Thiamin biosynthesis in Escherichia coli. Identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem., 273, 16555–16560. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westley J. (1989) Depletion of the sulphane pool: toxicological implications. In Damiani, L.A. (ed.), Sulphur-Containing Drugs and Related Organic Compounds, Vol. 2B. J. Wiley & Sons, New York, NY, pp. 87–99.
- Westley J., Adler, H., Westley, L. and Nishida, C. (1983) The sulfurtransferases. Fundam. Appl. Toxicol., 3, 377–382. [DOI] [PubMed] [Google Scholar]
- Xi J., Ge, Y., Kinsland, C., McLafferty, F.W. and Begley, T.P. (2001) Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein–protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc. Natl Acad. Sci. USA, 98, 8513–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]