Abstract
Streptococcus pneumoniae causes more than one million deaths every year, mostly of young children in developing countries, due to pneumonia, bacteremia and meningitis. The emergence and dissemination of drug-resistant pneumococcal strains, coupled to changing patterns of virulence and the inadequacy of available vaccines, calls for an aggressive search for novel targets for antibiotic and vaccine development. Microbial genomics techniques allow genetic and biochemical tools to be employed to tackle discovery, design and development of new anti-infective agents based on the identification of hundreds of new targets. In this review, novel approaches employed to identify potential antibiotic and vaccine targets in S. pneumoniae are highlighted. Recently identified virulence factors, as well as molecules essential for bacterial viability, cell wall integrity and infectivity, are discussed.
Introduction
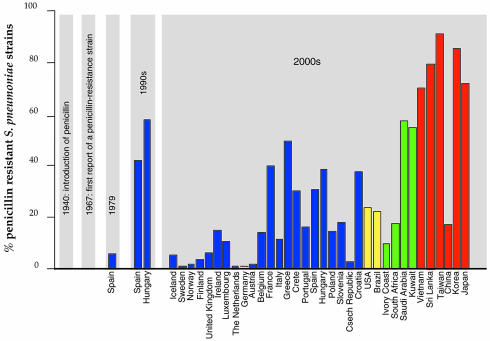
Streptococcus pneumoniae is responsible for a high proportion of cases of pneumonia, acute otitis media, acute sinusitis, bacteremia and meningitis. At present, strains of pneumococci resistant to tetracycline, trimetroprim-sulfamethoxazole, chloramphenicol, erythromycin, vancomycin and cephalosporins have been isolated; 25% of all invasive S. pneumoniae strains are resistant to penicillin (Pallares et al., 2000; Figure 1). In addition, the current adult vaccine includes 23 purified capsular polysaccharides of S. pneumoniae (of the 90 known) and is only moderately effective due to poor immunogenicity in high-risk groups (children under 2 years old and the elderly). The identification of new antibacterial targets, coupled to the development of vaccines based on protein antigens that are common to all virulent serotypes, is therefore of central importance for the treatment of S. pneumoniae infections.
Fig. 1. Each bar represents summed percentages of S. pneumoniae strains that display intermediate (MICs 0.21–1 µg/ml) and high (MICs >1 µg/ml) resistance levels to penicillin. Values for European countries are shown in blue, the American continent in yellow, African and Arabic countries in green and Asian countries in red. European values were obtained from http://www.earss.rium.nl/ (with the exception of Crete).
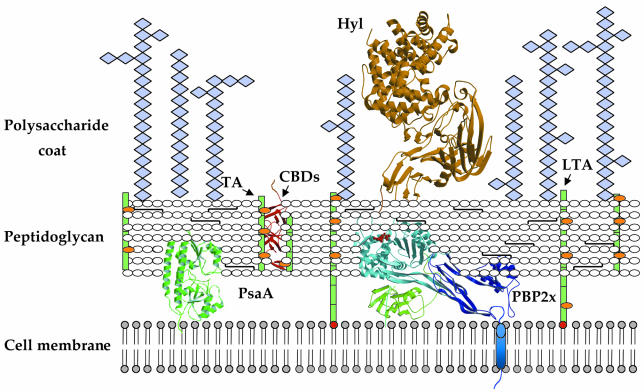
The bacterial cell wall metabolic machinery is the target for a variety of highly successful antibacterials, such as isoniazid (which targets mycobacterial enoyl-ACP reductase), vancomycin (which targets the D-Ala-D-Ala moiety of the peptidoglycan stem peptide) and the β-lactams, such as penicillin and cephalosporin (which bind to penicillin-binding proteins, PBPs). The pneumococcal plasma membrane is surrounded by a layer of peptidoglycan, a three-dimensional, cross-linked mesh that protects the bacterial cell from changes in osmotic pressure and plays key roles in shape determination and daughter cell formation (Höltje, 1998; Figure 2). The peptidoglycan anchors the carboxyl end of a variety of surface proteins, which may be relevant for pathogenicity, and is associated with teichoic and lipoteichoic acids. Both of the latter are polysaccharides of identical repeating units rich in choline, an essential growth factor for pneumococcal survival (Tomasz and Fischer, 2000). Lastly, the polysaccharide capsule, present in virulent streptococcal strains and surrounding the peptidoglycan layer, is the primary virulence determinant. There are 90 known serotypes of S. pneumoniae, each of which produces a structurally different capsular polysaccharide; during infection, these all function by inhibiting host complement-mediated phagocytosis. Problems with the development of polysaccharide-based vaccines arise from the variation in pneumococcal types throughout the world, necessitating distinct formulations, as well from the fact that the types that most often cause disease in children are not those that do so in adults (Klein, 2000).
Fig. 2. Schematic representation of the streptococcal cell wall. Teichoic acids (TAs) and lipoteichoic acids (LTAs) (both in green) are carbohydrate phosphate polymers rich in choline (orange spheres). TAs are linked to the peptidoglycan via a phosphodiester linkage, whereas LTAs are linked to the cell membrane via a C-terminal fatty acyl group (in red). Choline-binding proteins are linked to cell-wall TAs or LTAs via choline-binding domains (CBDs) (red ribbon diagram). Pneumococcal surface antigen (PsaA) is located underneath the peptidoglycan layer and is attached to the cell membrane via an LXXC motif; penicillin-binding proteins (PBPs) are located in the periplasmic space and interact with the peptidoglycan, and display a single, N-terminal transmembrane helix (in blue). Hyaluronate lyase (Hyl) is tethered to the peptidoglycan via an LPXTG motif.
An important barrier to target-based drug and vaccine development has been the limited number of potential macromolecular candidates. The availability of the complete genome sequence of virulent and non-virulent isolates of S. pneumoniae (http://www.tigr.org) has now provided new classes of genes as potential targets. The single circular chromosome of over 2 million base pairs contains slightly more than 2000 predicted protein coding regions; interestingly, ∼30% of open reading frames (ORFs) still remain unannotated, displaying unknown or hypothetical function (Hoskins et al., 2001; Tettelin et al., 2001). Therefore, in vivo and in vitro techniques that complement in silico genomics searches are playing a crucial role in the identification of the genes that provide the greatest promise in terms of novelty and applicability. In addition, such approaches will provide new insight into the mechanisms of streptococcal invasion as well as those of host–pathogen interactions.
Infection
For S. pneumoniae to successfully invade a host, it must first colonize the surface mucosa of the nasopharynx, a step that requires direct interaction between the pneumococcal cell-surface proteins and nasopharyngeal epithelial cells. Subsequently, it may migrate to the alveolar epithelium and invade the bloodstream (causing bacteremia) or traverse the blood–brain barrier (causing meningitis). Tissue damage is a result of stimulation of interleukins 1, 6, 8 and tumor necrosis factor, as well as the activation of the coagulation and complement pathways; intense inflammation is also a consequence of the activation of phospholipase A2 (Musher et al., 2000).
Prior to the genomics era, proteins that were known to be essential for pneumococcal pathogenicity included pneumolysin (Ply), hyaluronate lyase (Hyl), choline binding protein A (CbpA), autolysin (LytA), neuraminidase (NanA/NanB), pneumococcal surface antigen (PsaA) and pneumococcal surface proteins (PspA) (Jedrzejas, 2001). Of these, the most promising targets for vaccine development are Ply, LytA and PspA. Mutations in the corresponding genes reduce pneumococcal virulence, and Ply and PspA have been shown to be protective immunogens (Wu et al., 1997; Berry and Paton, 2000). The importance of each of these genes in pathogenicity was verified by generating single or double knockouts, as well as by examining their effects in animal models. With the availability of novel genomics-based tools, hundreds of genes can now be tested, making it possible to identify a variety of targets that may be associated with different aspects of virulence.
Sequence scanning and genomic analysis
The search of the pneumococcal genome for signature sequence motifs commonly found in cell-surface-exposed or virulence-related proteins in other bacteria has aided the identification of hundreds of potential new vaccine candidates. Choline-binding repeats are of particular interest, since a variety of proteins interact with the cell wall non-covalently through a choline- binding domain (CBD) consisting of 2–10 sequence repeats (Fernandez-Tornero et al., 2001). CbpA is the largest and the most abundant of the choline-binding proteins in streptococcal species, functioning as a surface adhesin and playing an important role in nasopharyngeal colonization (Rosenow et al., 1997). By performing genome-wide searches with the C-terminal choline-binding region of cbpA, Gosink et al. (2000) identified and constructed mutants of six novel genes predicted to encode proteins ranging from 20 to 80 kDa (CbpD, E, F, G, I and J). Mutants defective in the expression of CbpD and CbpE showed significant loss of nasopharyngeal colonization in an infant rat model (i.e. following intranasal inoculation) but did not differ in virulence in a sepsis model (i.e. intraperitoneal inoculation). However, mutants defective in CbpG, which displays sequence similarity to trypsin-like serine proteases, showed a strong loss of adherence to nasopharyngeal cells as well as sharply decreased virulence in the sepsis model. Therefore, CbpG may play an important role in invasion and infection of both the mucosa and the bloodstream (Gosink et al., 2000; Table I), and its possible structural similarity to proteases suggests that it may be an excellent candidate for target-based drug development.
Table I. Potential cell-wall-related antibiotic and vaccine development targets identified by genomics-related approaches in S. pneumoniae for which mutant and/or animal model studies are available.
| Gene and accession No. | Method employed in identification | Possible function | Mutant strain characterization | Animal model results |
|---|---|---|---|---|
| CbpD; AF278686 | Sequence scanning | Protein–protein interactions | Loss of nasopharyngeal colonization | Mutant strain: no reduction in virulence |
| CbpE; AF278687 | Sequence scanning | Loss of nasopharyngeal colonization | Mutant strain: no reduction in virulence | |
| CbpF; AF278688 | Sequence scanning | No effect on nasopharynx colonization | Mutant strain: no reduction in virulence | |
| CbpG; AF278688 | Sequence scanning | Proteolysis of host macromolecules | Loss of adherence to nasopharynx | Mutant strain: 103 fold reduction in virulence |
| CbpI; AF278689 | Sequence scanning; DNA microarray | No effect on nasopharynx colonization | Mutant strain: no reduction in virulence | |
| CbpJ; AF278690 | Sequence scanning; DNA microarray | No effect on nasopharynx colonization | Mutant strain: no reduction in virulence | |
| LytB; CAA09078 | Sequence scanning | Glucosaminidase | Loss of adherence to nasopharynx | Recombinant protein: protection from pneumococcal challenge |
| LytC; CAA08765 | Sequence scanning | Lysozyme | Loss of adherence to nasopharynx | Recombinant protein: protection from pneumococcal challenge |
| PavA; AF181976 | Signature-tagged mutagenesis | Fibronectin binding | No binding to immobilized fibronectin | Mutant strain: 104 fold attenuation in virulence |
| SmcA (PBP1A-like) | Signature-tagged mutagenesis | Peptidoglycan metabolism | Mutant strain: attenuation in virulence | |
| SmcB (YokZ-like) | Signature-tagged mutagenesis | Carboxypeptidation | Mutant strain: attenuation in virulence | |
| SmcC (BacA-like) | Signature-tagged mutagenesis | Undecaprenyl recycling | Mutant strain: attenuation in virulence | |
| PhtA; AF291695 | Sequence scanning | Metal/nucleoside binding | Recombinant protein: limited protection from challenge | |
| PhtB; AF318954 | Sequence scanning | Metal/nucleoside binding | Recombinant protein: limited protection from challenge | |
| PhtD; AF318955 | Sequence scanning | Metal/nucleoside binding | Recombinant protein: protection from challenge with diverse streptococcal subtypes | |
| PhtE; AF318956 | Sequence scanning | Metal/nucleoside binding | Recombinant protein: no protection against challenge |
Proteins displaying high similarity to LytA have also been found to play key roles in infectivity. Streptococcus pneumoniae strains harboring mutations in LytB and LytC were incapable of colonizing the nasopharynx (Gosink et al., 2000). Notably, both LytB (which may act as a glucosaminidase) and LytC (which displays lysozyme-like activity) may play fundamental roles in peptidoglycan remodeling and, in the case of LytB, in cell separation (Lopez et al., 2000). Purified LytB and LytC were evaluated as vaccine candidates in a mouse model of sepsis, and both proteins were shown to confer protection against S. pneumoniae infection (Wizemann et al., 2001).
In Gram-positive pathogens, a variety of surface-exposed proteins are associated with the cell-wall complex via lipid attachment sites (LXXC) or are covalently tethered to the peptidoglycan through a LPXTG-like motif (also LPXAG, LPXTN; Tettelin et al., 2001; Wizemann et al., 2001). Other surface-exposed proteins may harbor signal peptidase I and II sequences, as well as integrin-binding motifs. Genes encoding surface-exposed proteins represent 4% of the S. pneumoniae genome (Wizemann et al., 2001), underlining the large number of candidates from which potential drug development or vaccine targets may be obtained. In addition to the previously identified virulence determinants, proteins which could be involved in host polysaccharide degradation, adherence to host cells, and mucin biosynthesis have been identified by sequence scanning with variations of such motifs (Adamou et al., 2001; Hoskins et al., 2001; Tettelin et al., 2001; Wizemann et al., 2001). Of the several ORFs identified, a few are of note. ORF SP0462 from a virulent, capsulated S. pneumoniae strain (TIGR4) codes for an 893-residue, membrane-associated protein displaying a 230-residue von Willebrand factor (vWF) type A domain (http://smart.embl-heidelberg.de). Although the functions of prokaryotic vWFA homologs are unclear, they are believed to be analogous to those of their eukaryotic counterparts, which range from platelet adhesion to signal transduction. ORF SP1833 (TIGR4), also identified due to an LPXTG cell anchorage motif, is a 708-residue protein whose fold may contain five parallel β-helix repeats (http://smart.embl-heidelberg.de). Such repeats are found in the structures of pectate lyase and carageenases, enzymes that are involved in polysaccharide metabolism, which suggests that this newly identified surface-exposed protein may metabolize host cell polysaccharides. Interestingly, this protein is unique to pneumococci.
The new Pht family of pneumococcal proteins was identified by searching the streptococcal N4 genome with a lipoprotein motif (LXXC; Adamou et al., 2001). The four proteins identified (PhtA, PhtB, PhtD and PhtE) contain between five and six histidine triad motifs (HXXHXH), a proline-rich sequence and a predicted coiled-coil region. Immunization with PhtA, PhtB or PhtD protected mice from subsequent challenges with a variety of streptococcal strains. Since Pht-like proteins are present in different classes of streptococci, they may be good vaccine candidates that can offer broad subtype protection (Adamou et al., 2001).
Signature-tagged mutagenesis
Whole-genome analysis based on signature-tagged mutagenesis (STM) identifies, by negative selection, which genes are important for bacterial viability within an infected host. In this comparative hybridization technique, mutagenesis vectors carrying DNA tags flanked by short constant regions (the latter allowing for PCR amplification and tag detection) are employed in the cloning of random fragments of bacterial genomic DNA. Upon transformation into a pathogenic strain, the tagged plasmids are incorporated into the chromosome by homologous recombination, and the resulting mutants are thus signature-tagged at the site of mutation. Individual clones are then grown separately, pooled and inoculated into an animal host or (less preferably) a cell culture system (the input pool). Subsequently, bacteria are isolated from the animal (output pool), and the presence or absence of mutants within the output is detected by PCR amplification of the tags. As a consequence of the competitive growth disadvantage held by mutants in which an essential gene has been disrupted, the abundance of PCR products amplified from inserts within an essential gene will be substantially diminished, allowing for the ‘negative’ identification of the essential ORFs (Polissi et al., 1998; Shea et al., 2000).
Large numbers of mutants of Gram-positive pathogens screened in different animal models have identified both known and new virulence factors. Polissi et al. (1998) and more recently Lau et al. (2001) tested STM-generated S. pneumoniae mutants by using mouse models for intranasal and intraperitoneal challenge studies. In the Polissi study, in addition to the previously characterized virulence factors LytA and NanA, the IgA1 protease and a choline-binding protein were also identified, as well as a variety of genes with unknown function. Using a pneumonia model of infection, Lau et al. (2001) identified two strains that contained mutations encoding a putative adhesin, PavA. PavA is 71% identical to FbP, a putative fibronectin-binding protein from Streptococcus pyogenes. Both proteins display a coiled-coil region, as is typical of surface-exposed molecules in Gram-positive bacteria, but lack a conventional hydrophobic leader sequence, signal peptidase cleavage site, cell-wall anchorage motif and choline-binding sequence, which explains why they have not been identified in sequence scanning searches. PavA harbors a fibronectin-recognition region within its C-terminal 189 amino acids; interestingly, it lacks the repeated GGX3–4(I/V)DF motif that is common in fibronectin-recognizing proteins in streptococcal and staphylococcal species (Sun et al., 1997). A PavA mutant in a mouse sepsis model displayed highly attenuated virulence (Holmes et al., 2001).
Although STM is a powerful tool in the identification of novel vaccine targets when combined with genomic analysis, it has certain limitations. For example, it is unlikely to identify genes that are essential for growth in vitro, as the mutants would not be viable. In addition, STM has not yet identified secreted toxins or virulence factors (such as pneumolysin), probably because such molecules can be trans-complemented by the other mutants in the input pool (Shea et al., 2000). Thus, the use of other techniques, such as the ones outlined here, is important in order to complement this strategy.
DNA microarrays
DNA microarray technology has only recently become useful in the search for vaccine targets, since it is highly dependent on the availability of the entire genome sequence. In this technique, DNA fragments from a specific genome are spotted onto a porous surface which may be composed of glass, plastic, silicon, gold or even beads at the ends of fiber optical bundles. Gene expression is monitored during specific cellular cycles or under particular growth conditions by preparing RNA from the culture in question, labeling it with fluorescent dyes and then hybridizing it to the DNA fragments on the surface of the chip. Fluorescence signals emitted by each spot upon laser beam excitation are quantified to define the transcriptional activity of all arrayed genes (Lockhart and Winzeler, 2000).
In terms of the study of pathogenic systems, the DNA microarray technique has been employed in the analysis of gene expression in Mycobacterium tuberculosis responding to antibiotic treatment (Wilson et al., 1999), as well as in the analysis of host–pathogen interactions between human cells and Salmonella, Listeria and Bordetella spp. (Belcher et al., 2000; Cohen et al., 2000; Eckmann et al., 2000). Recently, DNA microarray technology was employed as a comparative tool to identify differences between related streptococcal isolates, i.e. infectious and non-infectious strains. A comparative genome hybridization performed with the TIGR4 isolate (capsulated, virulent) and both the R6 (non-capsulated, non-virulent) and D39 (capsulated, virulent) strains revealed that loci which did not hybridize between the TIGR4 isolate and the two other strains included mostly surface-exposed or pathogenesis-related clusters, such as the capsule biosynthesis locus (significantly different in the capsulated strains), an IgA1 protease paralog, sortase motif proteins and CbpI (Tettelin et al., 2001).
Horizontal gene transfer is a powerful strategy used by highly transformable streptococcal species for the acquisition of antibiotic-resistance genes. Employing a high-density oligonucleotide array with over 130 000 oligonucleotide probes, Hakenbeck et al. (2001) investigated genomic variation in 20 S. pneumoniae isolates and identified 470 genes in which variation was detected in at least one of the strains. Sequence variation was identified mainly among the genes coding for choline-binding proteins (PspA, CbpJ, CbpI and PspC). Notably, other genes implicated in virulence, such as NanA/NanB, LytA and Ply, did not show any variation, suggesting that they may be better candidates for vaccine development due to the homogeneity of their sequences in different strains.
From target identification to vaccine elaboration
Once target genes have been identified and selectively tested through the observation of protective effects in animal models and/or loss of virulence in knockout mutants, the challenge consists of analyzing their value as potential vaccines. Problems may arise if the molecule plays only a minor role in pathogenicity, which may not have been revealed due to the inadequacy of the animal model; if the antigen is present in only a limited number of pneumococcal serotypes (in this case, a vaccine that protects only against the homologous strain, but not heterologous ones, may be produced); or if the epitope is not widely accessible to antibodies in the presence of polysaccharide capsules from different strains (leading to a serotype-specific vaccine). In all cases, protection studies should be performed by active and passive immunization approaches. In addition, targets must be expressed and purified, which may be challenging for proteins with a tendency to aggregate or to be harbored in inclusion bodies. An elegant study of the identification of vaccine targets through a genomic approach has been described recently for the capsulated, meningitis-causing bacterium Neisseria meningitidis (Pizza et al., 2000).
Antibiotic resistance and drug development
The classical approach to the discovery of antibacterials has included assaying for compounds that promote bacterial death in a whole-cell system. The possibility of selecting targets through genomics-related methodologies, however, now allows the rationalization of the antibiotic discovery process. Once genes of interest are selected through sequence analysis and/or related technologies as described above, appropriate assays are developed for the purified, characterized protein products. Such assays are employed in high-throughput screens, in which large collections of compounds are tested for inhibition of the new target. Compounds that are selected at this step (‘hits’) are subsequently optimized in structure–activity relationship efforts, which are complemented by tests for potency, solubility, specificity and bacterial permeability. Lead compounds are subsequently tested for pharmacokinetic properties, safety and efficacy in animal models and are eventually included in human clinical trials.
A variety of cell-surface proteins in pneumococci are potential drug-development targets. The recently reported crystal structure of the CBD of LytA reveals a β-hairpin solenoid fold that folds into a left-handed superhelix, with a single choline-binding site requiring residues from three consecutive binding repeats. Since choline molecules are bound in a hydrophobic cavity on the protein surface, small molecules that compete with choline for cleft binding could be explored as potential antibacterials, since they would target all proteins containing CBDs (Fernandez-Tornero et al., 2001). Since choline analogs could act as anti-infectives without causing bacterial death, such compounds would not be subject to elevated selective pressures toward the development of bacterial resistance and could be attractive new tools in the control of bacterial invasion. In addition to the C-terminal CBDs, the N-terminal autolysin activities can also be targeted. Inactivation of LytA, LytB or LytC induces the formation of long chains of streptococci, which are incapable of dividing into normal daughter cells. Since such chain formation inhibits the dissemination of bacteria during infection, autolysins may be an attractive target for the development of drugs that control bacterial division (Lopez et al., 2000).
The two last steps in the biosynthesis of peptidoglycan include polysaccharide elongation (glycosyl transfer) and cross-linking of peptide branches (transpeptidation). These reactions are catalyzed by PBPs, some of which harbor both glycosyl transfer and transpeptidation activities on the same polypeptide. β-lactams (such as penicillin and cephalosporin), which inhibit transpeptidation, affect the maturation of the peptidoglycan and are thus powerful antimicrobials.
Streptococcus pneumoniae acquires β-lactam resistance by horizontal gene transfer, and two primary antibiotic resistance determinants, PBP2x and PBP2b, may contain hundreds of mutations generated by homologous recombination. The crystal structure of PBP2x from a highly resistant clinical isolate reveals an open active site generated by destabilizing mutations, possibly causing poor antibiotic binding as well as suboptimal catalysis (Dessen et al., 2001). High-throughput screening of compound banks using purified proteins from clinical resistant strains, as well as the employment of the crystal structures of such proteins in the design of modified chemotherapeutics, should provide novel methodologies to combat resistance. In addition, to date there is no structural information available for a glycosyl transfer domain of any PBP. However, moenomycin, a non-β-lactam antibiotic approved only for use in poultry owing to its highly toxic effects in humans, is known to stabilize the glycosyl transfer fold (Di Guilmi et al., 1999). The localization of PBPs on the extracellular side of the plasma membrane makes them easily accessible to a variety of small chemotherapeutics (Figure 2). Hence, the glycosyl transfer domain of PBPs is one of the most promising potential drug-development targets. The crystal structure of such a domain, coupled to moenomycin, should shed light on the possibilities for rational drug-design strategies.
Conclusions
Although undeniable progress has been made in the field of pneumococcal disease management, new approaches to infection control are urgently needed for two reasons. First, drug-resistant pneumococci are on the increase, and the common dose of penicillin employed in hospitals worldwide is already 600 times higher than that employed to treat S. pneumoniae infections in the early 1950s. Secondly, the classic polysaccharide-based vaccine is not effective throughout different age and geographic groups and is not employed by the WHO in childhood immunization programs. A more recent, conjugate-based vaccine displays high immunogenicity among children but is marketed at very high cost. The exploration of genomics and structural tools will therefore be essential for the development of novel infection treatment methodologies that employ new mechanisms of action.

Andréa Dessen & Anne Marie Di Guilmi. Andréa Dessen is the recipient of an EMBO Young Investigator Award.
Acknowledgments
Acknowledgements
The authors would like to thank O. Dideberg and T. Vernet for helpful discussions, as well as for their continuous, enthusiastic support. A.D. is the recipient of an ‘ACI Jeunes Chercheurs’ grant from the French Ministry of Research.
References
- Adamou J.E. et al. (2001) Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun., 69, 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher C.E. et al. (2000) The transcriptional responses of respiratory epithelial cells to Bordetella pertussis reveal host defensive and pathogen counter-defensive strategies. Proc. Natl Acad. Sci. USA, 97, 13847–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A.M. and Paton, J.C. (2000) Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun., 68, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. et al. (2000) Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J. Biol. Chem., 275, 11181–11190. [DOI] [PubMed] [Google Scholar]
- Dessen A., Mouz, N., Gordon, E., Hopkins, J. and Dideberg, O. (2001) Crystal structure of PBP2x from a highly penicillin resistant Streptococcus pneumoniae clinical isolate. J. Biol. Chem., 276, 45106–45112. [DOI] [PubMed] [Google Scholar]
- Di Guilmi A.M., Mouz, N., Martin, L., Hoskins, J., Jaskunas, S.R., Dideberg, O. and Vernet, T. (1999) Glycosyltransferase domain of penicillin-binding protein 2a from Streptococcus pneumoniae is membrane associated. J. Bacteriol., 181, 2773–2781.10217767 [Google Scholar]
- Eckmann L., Smith, J.R., Housley, M.P., Dwinell, M.B. and Kagnoff, M.F. (2000) Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J. Biol. Chem., 275, 14084–14094. [DOI] [PubMed] [Google Scholar]
- Fernandez-Tornero C., Lopez, R., Garcia, E., Gimenez-Gallego, G. and Romero, A. (2001) A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol., 8, 1020–1024. [DOI] [PubMed] [Google Scholar]
- Gosink K.K., Mann, E.R., Guglielmo, C., Tuomanen, E.I. and Masure, H.R. (2000) Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun., 68, 5690–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck R., Balmelle, N., Weber, B., Gardes, C., Keck, W. and de Saizieu, A. (2001) Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation in Streptococcus pneumoniae. Infect. Immun., 69, 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A.R. et al. (2001) The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol., 41, 1395–1408. [DOI] [PubMed] [Google Scholar]
- Höltje J.V. (1998) Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev., 62, 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J. et al. (2001) Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol., 183, 5709–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrzejas M.J. (2001) Pneumococcal vaccine factors: structure and function. Microbiol. Mol. Biol. Rev., 65, 187–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.L. (2000) Pneumococcal disease and the role of conjugate vaccines. In Tomasz, A. (ed.), Streptococcus pneumoniae. Mary Ann Liebert Inc. Publishers, New York, NY, pp. 467–477.
- Lau G.W. et al. (2001) A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol., 40, 555–571. [DOI] [PubMed] [Google Scholar]
- Lockhart D.J. and Winzeler, E.A. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. [DOI] [PubMed] [Google Scholar]
- Lopez R., Gonzalez, M.P., Garcia, E., Garcia, J.L. and Garcia, P. (2000) Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol., 151, 437–443. [DOI] [PubMed] [Google Scholar]
- Musher D.M., Breiman, R.F. and Tomasz, A. (2000) Streptococcus pneumoniae: at the threshold of the 21st century. In Tomasz, A. (ed.), Streptococcus pneumoniae. Mary Ann Liebert Inc. Publishers, New York, NY.
- Pallares R., Viladrich, P.F., Linares, J., Cabellos, C. and Gudiol, F. (2000) Impact of antibiotic resistance on chemotherapy for pneumococcal infections. In Tomasz, A. (ed.), Streptococcus pneumoniae. Mary Ann Liebert Inc. Publishers, New York, NY. [DOI] [PubMed]
- Pizza M. et al. (2000) Identification of vaccine candidates against serogroup B meningococcus by whole genome sequencing. Science, 287, 1816–1820. [DOI] [PubMed] [Google Scholar]
- Polissi A., Pontiggia, A., Feger, G., Altieri, M., Mottl, H., Ferrari, L. and Simon, D. (1998) Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun., 66, 5620–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenow C., Ryan, P., Weiser, J.N., Johnson, S., Fontan, P., Ortquist, A. and Masure, H.R. (1997) Contribution of novel choline-binding proteins to adherence, colonization, and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol., 25, 819–825. [DOI] [PubMed] [Google Scholar]
- Shea J.E., Santangelo, J.D. and Feldman, R.G. (2000) Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol., 3, 451–458. [DOI] [PubMed] [Google Scholar]
- Sun Q., Smith, G.M., Zahradka, C. and McGavin, M.J. (1997) Identification of D motif epitopes in Staphylococcus aureus fibronectin-binding protein for the production of antibody inhibitors of fibronectin binding. Infect. Immun., 65, 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H. et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science, 293, 498–506. [DOI] [PubMed] [Google Scholar]
- Tomasz A. and Fischer, W. (2000) The cell wall of Streptococcus pneumoniae. In Fishcetti, V.A., Novick, R.P., Ferretti, J.J., Portnoy, D.A. and Rood, J.I. (eds), Gram-Positive Pathogens. ASM Press, Washington, DC, pp. 191–200.
- Wilson M., DeRisi, J., Kristensen, H.H., Imboden, P., Rane S, Brown, P.O. and Schoolnik, G.K. (1999) Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl Acad. Sci. USA, 96, 12833–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizemann T.M. et al. (2001) Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun., 69, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Nahm, M.H., Guo, Y., Russell, M.W. and Briles, D.E. (1997) Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J. Infect. Dis., 175, 839–846. [DOI] [PubMed] [Google Scholar]