SUMMARY:
The extrahepatic branches of the biliary tree have glands that connect to the surface epithelium through narrow pits. The duct epithelia undergo homeostatic renewal; yet, the identity and multiplicity of cells that maintain this tissue is unknown. Using marker-free and targeted clonal fate mapping in mice, we provide evidence that the extrahepatic bile duct is compartmentalized. Pit cholangiocytes of extramural glands renewed the surface epithelium, whereas basally oriented cholangiocytes maintained the gland itself. In contrast, basally positioned cholangiocytes replenished the surface epithelium in mural glands. Single-cell sequencing identified genes enriched in the base and surface epithelial populations with trajectory analysis showing graded gene expression between these compartments. Epithelia were plastic, changing cellular identity upon fasting and refeeding. Gain of canonical Wnt signaling caused basal cell expansion, gastric chief cell marker expression, and a decrease in surface epithelial markers. Our results identify the cellular hierarchy governing extrahepatic biliary epithelial renewal.
Graphical Abstract
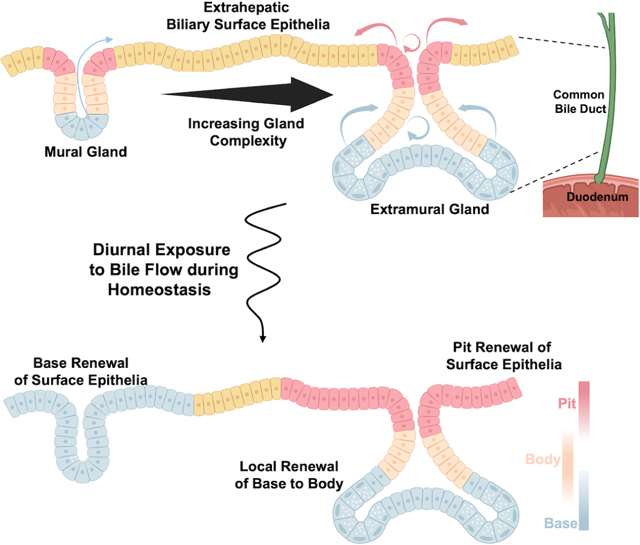
In brief
Singh et al. utilized 3-dimensional genetic lineage tracing to understand homeostatic renewal in the extrahepatic bile duct. They found that the large complex extramural glands show compartmentalized behavior with pits renewing the surface epithelium and the base maintaining the gland itself.
INTRODUCTION
Homeostatic turnover of epithelial cells is critical for proper tissue maintenance. Each organ governs this turnover with distinct mechanisms. Tissues driven by stem cells are typically defined by a stem or progenitor cell within an architecturally demarcated niche1–4. The progeny of these stem cells replaces differentiated cells that are lost through cellular “wear and tear” with tissue specific dynamics5.
The biliary tree forms the interface between the liver and intestinal tract, with the extrahepatic biliary tree having a separate developmental origin6. These ducts are critical for proper function of the liver as therapy for diseases of the biliary tree primarily require liver transplantation. The intrahepatic branches, known for their cuboidal morphology, can be derived from adult hepatocytes through transdifferentiation and have been explored in their ability to generate bi-potential organoids and repopulate the liver7–11.
Conversely, the largest branches of the biliary tree have an architecture in which the lumen is lined with columnar cholangiocytes and the bile duct wall contains an array of glands, heterogenous in their appearance. Glands can be generally subdivided into mural glands that are superficial, simple tubular glands, and extramural glands, which are deeply embedded within the mesenchyme showing a complex tubulo-alveolar structure, often with multiple alveoli12. Prior studies have indicated that epithelia within the peribiliary glands are multipotent and can give rise to hepatic or pancreatic lineages in vitro and can regenerate a damaged epithelium13–15. How the surface epithelium and glandular elements contribute to homeostasis, and whether a stem cell compartment is responsible for homeostatic maintenance of this epithelium, remains unknown.
Using a marker free lineage tracing strategy, we demonstrate that clones anchored in the pit renewed the surface epithelium over the long term; whereas, basally anchored clones contribute exclusively to the base and body region of extramural glands. By contrast, the minority of mural glands showed evidence of a different behavior, with basally located epithelia giving rise to progeny that migrate out of the gland and onto the surface epithelium. Single-cell analysis with spatial validation depicted marker genes enriched in certain compartments. However, apart from some basal specific genes, these marker genes could be modulated to be expressed outside their primary compartment. Consistent with compartmentalization of the extramural glands, lineage tracing based on the proliferative marker Mki67 resulted in enhanced pit labeling and gave rise to progressive labeling of the surface epithelium over time whereas base directed labeling with Olfm4 or Muc6 led to labeling within the gland, but not the surface epithelium. Our results show that the peribiliary pits and bases function as privileged zones that together coordinate biliary renewal in the extrahepatic bile ducts.
RESULTS
Proliferation primarily localizes to the pits of extramural peribiliary glands.
The large branches of the biliary tree are distinct from the smaller branches and can be identified by the presence of columnar epithelia and glands, a feature only seen in the extrahepatic tree of mice (Figures 1A and 1B). We utilized the murine common bile duct (CBD) to study the interplay between the peribiliary glands and surface epithelium at homeostasis. These glands are heterogenous structures that are mural (simple tubular glands extending superficially into the mesenchyme) or extramural (complex tubulo-alveolar glands extending deep into the mesenchyme), with mural glands comprising a minority of the gland population16. Since glands have a heterogenous structure, we developed a conceptual framework to organize a gland. In this framework, a given gland was subdivided into three parts, a basal aspect, the body, and the pit. The body makes up the majority of cells within a gland (~60%), with the base and pit encompassing a smaller fraction (~20% each, Figure 1C). Lgr5 marks intestinal and ampullary stem cells within the base of their glands17,18; however, we found little to no expression within the peribiliary glands (Figure S1A–S1C). Axin2, another Wnt target gene that is highly expressed in intestinal and ampullary stem cells18,19, could be seen within the peribiliary glands with greater expression toward the gland base; however, at a far lower expression level than seen in the small intestine (Figure S1D, S1E, S1G, S1H). Axin2 could also be seen heterogeneously expressed within patches of the CBD’s surface epithelium (Figure S1F).
Figure 1. Localizing Proliferation in the Extrahepatic Bile Duct.
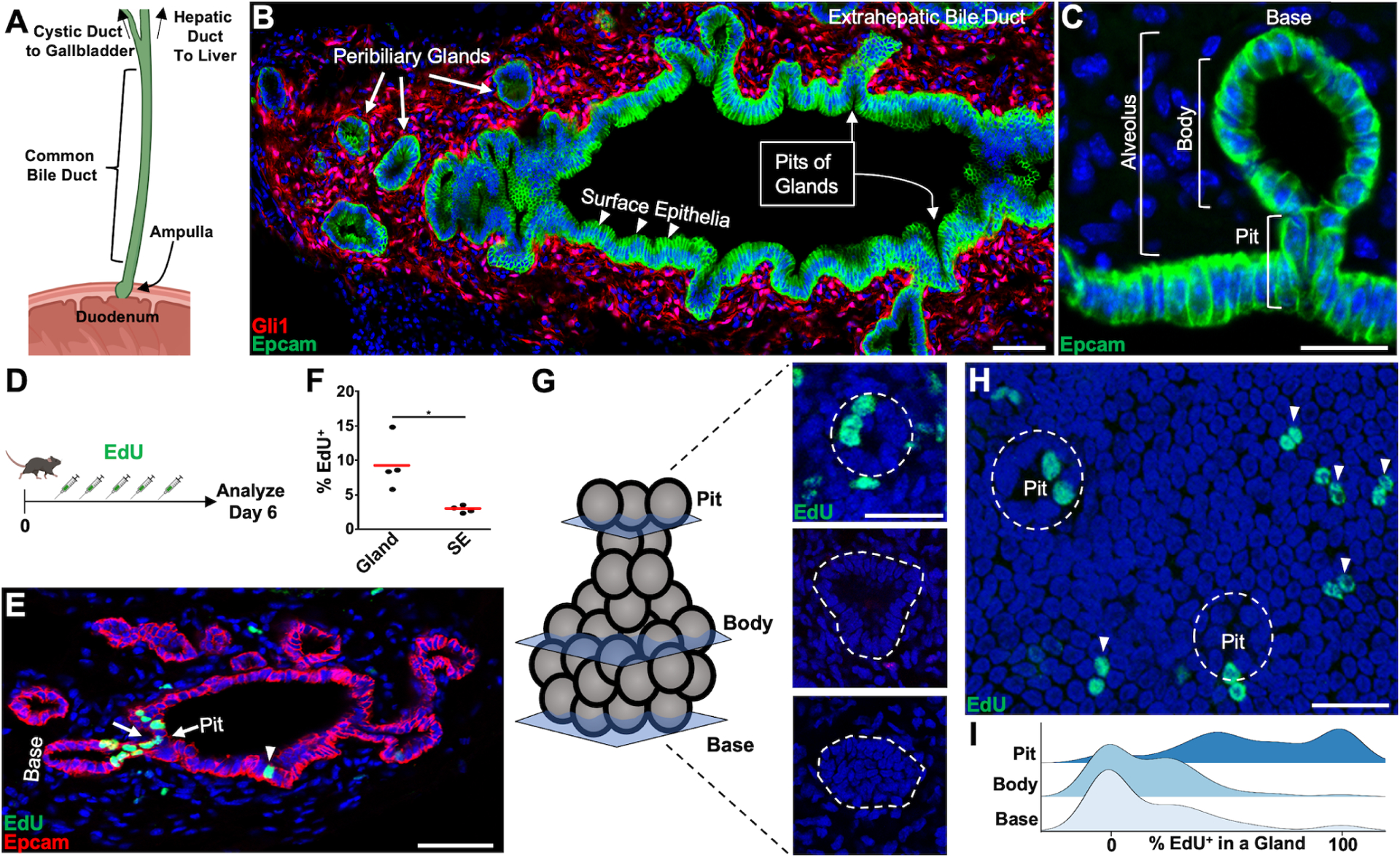
(A) Schematic of the extrahepatic biliary tree. (B) The CBD from a Gli1:CreERT2; Rosa26:lox-STOP-lox-tdTomato animal where mesenchyme is labeled. Peribiliary glands (thick arrows) connect with the surface epithelia (arrowheads) through pits (thin arrows). (C) A single alveolar gland with gland pit, body, and base diagramed. (D) Animals were pulsed with EdU daily for 5 total doses and analyzed 1 day after. (E) A cluster of EdU+ cells is seen around the pit of a gland (arrows), with lower labeling on the surface epithelium (arrowhead). (F) Quantification of EdU+ cholangiocytes seen in surface and glandular epithelium from sections (n = 4 animals). (G) Optical sections through the gland base, body, and pit. (H) EdU+ surface epithelia (arrowheads) and pit cholangiocytes (dashed line encompasses gland pit). (I) Ridge plot showing percentage of EdU+ cholangiocytes found in a gland by region (102 glands from n = 4 animals). Scale bars, 50 μm (B, E); 25 μm (C, G, H). * p<0.05, by Students t-test.
Peribiliary glands had higher rates of EdU incorporation compared to surface epithelia after 5 daily doses of EdU (9.2 ± 1.9% vs 2.8 ± 0.2%, Average ± SEM, p <0.05, n = 4, Figures 1D, 1F). Notably, instances of labeling were found that could extend around the pit of a gland (Figure 1E). The 3-dimensional complexity of peribiliary glands prevented a reliable assessment of regionalized proliferation in tissue sections. Therefore, we performed whole mount imaging of optically cleared CBDs to assess for regionalized proliferative preferences after 5 daily doses of EdU. Within a given extramural gland, the majority of EdU+ epithelial cells were localized to the gland pit, whose openings could be visualized in 2-dimensional sheets in continuity with the surface epithelium (Figure 1G–1I).
Survival and expansion of pit and basal clones
To determine whether the pits of extramural peribiliary glands harbor epithelia with renewal potential, we performed unbiased lineage tracing using Rosa26:CreERT2; Rosa26:Confetti mice and performed whole mount imaging of optically cleared CBDs (Figure 2A). We did not detect leaky recombination in corn oil treated animals over 1 year (Figure S2A). The surface epithelium was analyzed as a 2-dimensional sheet, where multiple small clones were visualized 14 days after tamoxifen (Figure 2B). Over the course of 1.5 years, the number of these small clones on the surface epithelium steadily decreased (Figure 2C–2G). Within the glands, there was also a steady decrease in the number of clones over time (Figure S2B). When quantifying the proportion of “anchored” clones (where one cell in a clone was located in either the pit or base compartment of a gland), we found that pit or base anchored clones increased over time (Figure 2H and Figure S2C). At 6 months post-labeling, clones could be found that wrapped circumferentially around a pit with clonally related cells on the surface epithelium (Figure 2I). At 1- and 1.5-year timepoints, large clones on the surface epithelium were anchored to the gland pit and spread across the surface epithelium with evidence for clone fragmentation (Figure 2F, 2J–2L, and S2D). There was also evidence of clones being once attached to the pit and having then completely detached from the pit and spread out onto the surface epithelium (Figure 2F and S2E). Clones anchored at the base of extramural glands were never seen in continuity with the surface epithelium through the pit. Instead, there was evidence of clonality within alveoli and contributions to the gland body (Figure S2F–S2H). The exception to this trend were the minority of mural glands, where clones could be seen extending from the base to the surface epithelium (Figure S2I and S2J). These data suggested that the peribiliary pits were the primary source of surface epithelium renewal over the long term, whereas epithelia within the gland base were responsible for turnover of the base and body regions.
Figure 2. Non-biased Lineage Tracing of the Common Bile Duct.
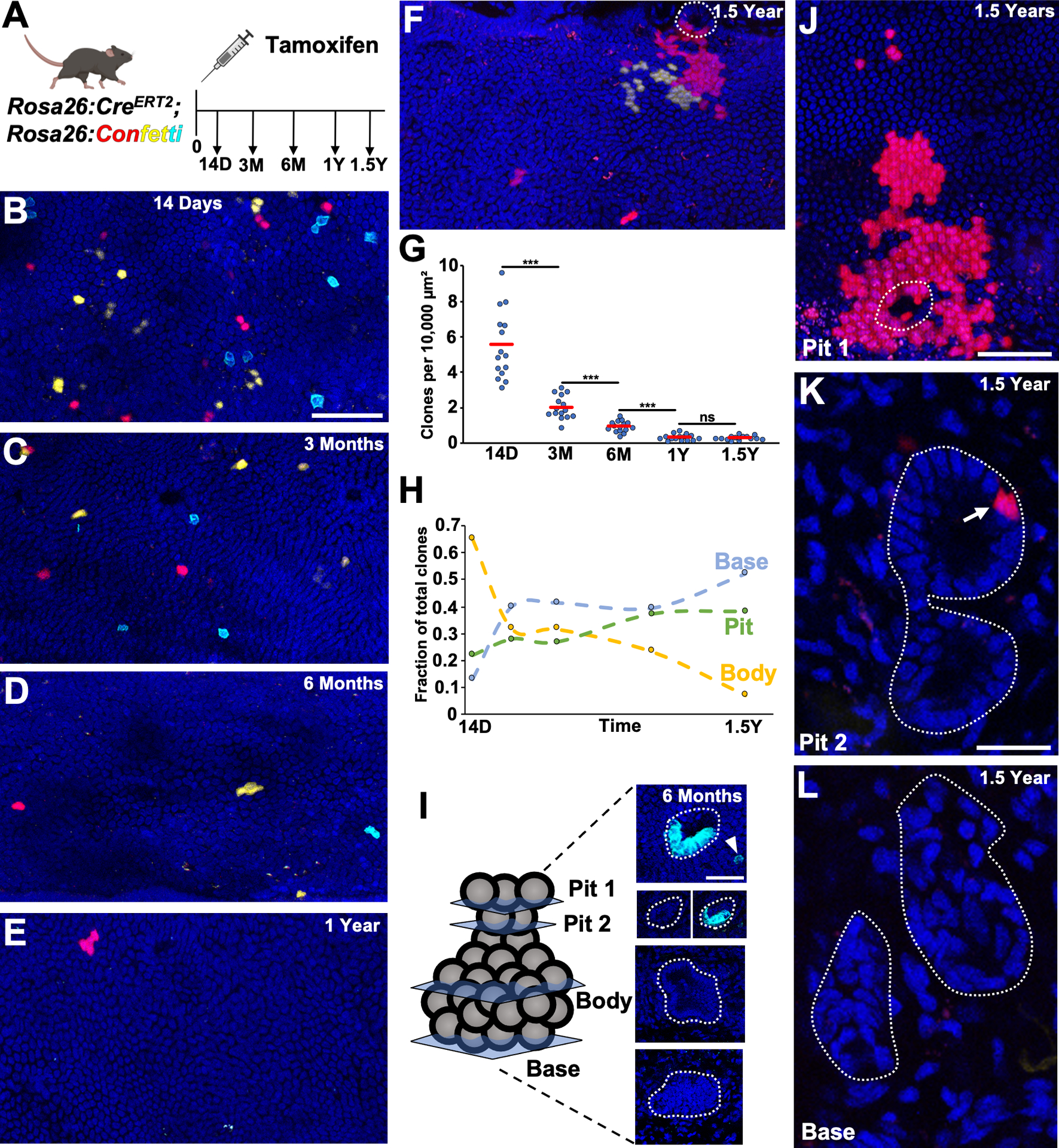
(A) Schematic of experimental design. (B-F) Surface epithelial panels at indicated timepoints with (F) showing a red clone coming from the pit of a gland (in dashed outline) adjacent to a yellow clone. (G) Number of clones per area quantified from 5 luminal panels per animal (n = 3 animals for each timepoint). (H) Fraction of body encompassed, pit and base anchored clones (97 to 157 glands, n = 3 animals per timepoint). (I) Example of a 6 month gland where a cyan clone has expanded around the gland pit (Pit 2 panels with and without cyan channel, just above surface epithelium) and a corresponding cyan surface epithelial cell is in close proximity (Pit 1, arrowhead). (J-L) A two alveoli extramural gland (L, dashed outline around gland) with a red clone seen in the proximal gland pit (K, arrow), which has spread over the surface epithelium (J, dashed outlines indicates pit). Scale bars, 50 μm (B, C-F same scale as B, J); 25 μm (I, K, L same scale as K). *** p<0.001, ns = not significant, by Students t-test.
Single-cell transcriptomics and trajectory analysis identifies graded gene expression across compartments
To characterize the cellular heterogeneity within the epithelium, whole CBD was digested and profiled with the 10X Genomics single-cell sequencing platform. Epithelial cells were identified by the presence of Onecut2, Prom1, and Hnf1β (Figure 3A and 3B). In all epithelia, there was low level expression of Pdx1 (Figure 3B), which is normally expressed in pancreatobiliary progenitors and the duodenum20–22. We found that epithelia could be generally divided into 4 groups (Figure 3A, 3B, S3A). Tuft cells, marked by high expression of Dclk1 and Rgs13, were transcriptomically distinct and could be spatially identified by staining for Dclk1 (Figure 3B and S3B). Interestingly, tuft cells within the extrahepatic bile duct showed expression of Sct, typically seen in enteroendocrine (S) cells of the duodenum (Figure 3B). Proliferating epithelia were identified by high expression of Mki67 and Stmn1; however, this population did not contain any specific marker genes (Figure 3B and S3A).
Figure 3. Single-Cell and Spatial Characterization of Biliary Epithelia.
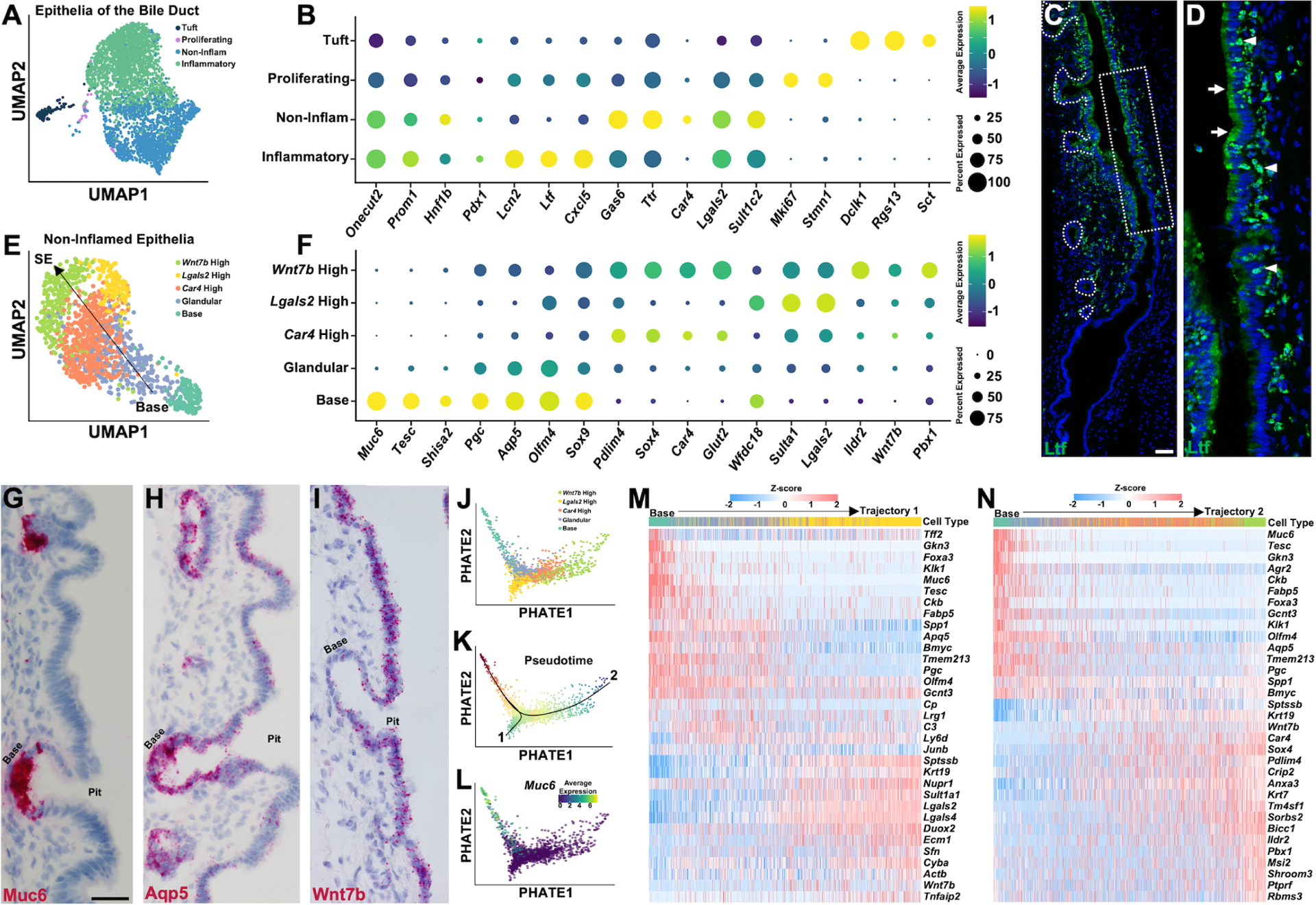
(A) UMAP reduction showing epithelia of the CBD. (B) Dot plot showing expression level of common and marker genes for each cluster from (A). (C) Ltf staining showing an area of high expression that stops abruptly. (D) Zoom of dashed box from (C), where cholangiocytes show Ltf expression in apical aspect (arrows) and small Ltf+ cells (arrowheads) are infiltrating in-between epithelial cells. (E) UMAP reduction showing non-inflamed epithelial cells. (F) Dot plot showing marker gene expresion for each cluster from (E). (G-I) Muc6 (G), Aqp5 (H), and Wnt7b (I) in situs on CBD. (J) Two-dimensional PHATE embeddings of non-inflamed epithelial cells colored by identified cell types in (E). (K) Two trajectories inferred by Slingshot along pseudotime. (L) Expression level of Muc6 along trajectory. (M and N) Heatmap showing dynamically expressed genes over pseudotime along each trajectory. Scale bars, 50 μm (H and I same scale as G).
One large cluster of epithelial cells had high expression of inflammatory markers; particularly Lactoferrin (Ltf) (Figure 3B), a secreted protein with antimicrobial activity found in secretory fluids23. Ltf staining showed areas with strong epithelial expression that could start and stop abruptly (Figure 3C). These areas also had Ltf+ cells that could be seen within the mesenchymal space and infiltrating through the epithelium (Figure 3D). Analysis of the immune populations from the entire single-cell dataset showed these Ltf+ cells as high in S100a9 and Csf3r, identifying them as neutrophils (Figure S3C and S3D).
Inflammatory epithelia had a lack of compartmental specificity by Ltf staining. We therefore examined the non-inflamed population of epithelia to determine if compartmental identities could be uncovered. The non-inflamed epithelia could be clustered into 5 groups (Figure 3E and 3F). One cluster had unique expression of Muc6 and Tesc, with Muc6 being a known marker of basally located stem cells within the gastric pylorus24. Muc6 in situ showed this population localizing to the base of peribiliary glands (Figure 3G). This cluster also had the highest expression of Olfm4 and Aqp5 (Figure 3F), which are expressed within basally located stem cells within the pylorus24 (Aqp5) and intestinal tract/ampulla18,25,26 (Olfm4). Unlike Muc6, spatial localization of Olfm4 and Aqp5 demonstrated predominance in the gland base; however, their expression decreased toward the surface epithelia in a graded manner (Figure 3H and S3G). Basal epithelia also had high expression of Pepsinogen C (Pgc), a marker of gastric chief cells, as well as the stem cell marker Sox9 (Figure 3F). Staining for Sox9 confirmed areas of higher expression toward the gland base, with lower-level expression seen within the surface epithelium as has been reported in the murine extrahepatic duct by Matusi et al15 (Figure S3H). Our analysis demonstrated that the Muc6+ cluster marked an exclusive base phenotype, whereas glandular epithelia located in-between the putative surface epithelia and the base had intermediate expression levels of Olfm4, Aqp5, and other genes (Figure 3E and 3F).
The other three epithelial clusters did not contain unique marker genes, but had certain genes enriched in each. One had enrichment of transport proteins such as Glut2 and Car4 (Figure 3F). Another cluster showed high expression of Lgals2, a protein expressed within the stomach surface mucosa27. Dual staining for Lgals2 and Car4 showed epithelia that could have high expression of either protein or have intermediate expression of both (Figure S3I–S3K). The last cluster showed high expression of Pbx1, Ildr2, and Wnt7b, but also contained lower levels of Car4 and Lgals2 expression. Wnt7b showed little to no expression within the base cluster and in situ examination of Wnt7b confirmed a gradual decrease in expression from the surface epithelium toward the gland base (Figure 3I).
With the exception of Muc6+ base cells, epithelial cells were not segregated by unique marker genes. Rather, gene expression gradually changed from the base phenotype to the surface epithelial phenotype. To further analyze this, we used PHATE28 combined with Slingshot29 to perform a trajectory analysis (Figure 3J–3L). This in silico analysis identified two separate trajectories, one toward a Lgals2 high fate and another toward a Car4/Wnt7b high fate (Figure 3J and 3K). Across these trajectories, we could find opposing gradients of gene expression defining basal and surface epithelial cells (Figure 3M and 3N). We regressed out cell cycle genes to determine where proliferating cells were enriched upon this trajectory, where the majority clustered around the interface of the glandular and surface epithelial identities (Figure S3E).
Our results demonstrate a distinct basally oriented epithelial cell type, whose enriched genes decrease in expression toward the surface epithelium. The surface epithelium is comprised of cells depicting transcriptional enrichment in certain genes, but also with genes that show opposing gradients to those enriched at the gland base.
Fasting and feeding states control epithelial identity and glandular proliferation
The low-level expression of Pdx1 and the lack of compartmentally specific expression of the stem cell markers Sox9, Axin2, Aqp5, and Olfm4 suggested a relatively plastic cell state. Additionally, proliferating cells did not express genes that localized them to a particular compartment. Flow of bile, a caustic substance, through the CBD is controlled by gastrointestinal hormones regulated by food intake. We hypothesized that diurnal regulation of bile flow through the biliary tree could modulate these gradients and alter biliary proliferation and cellular identity. To test this, we fasted animals for two days and then allowed them to eat again (Figure 4A). Fasting led to an increase in gallbladder size, which collapsed upon refeeding (Figure S4A and S4B). During the fasting period, animals lost an average of 23% of their starting body weight that recovered during refeeding (Figure S4C).
Figure 4. Fasting and Refeeding Modulate Cellular Identity and Glandular Proliferation.
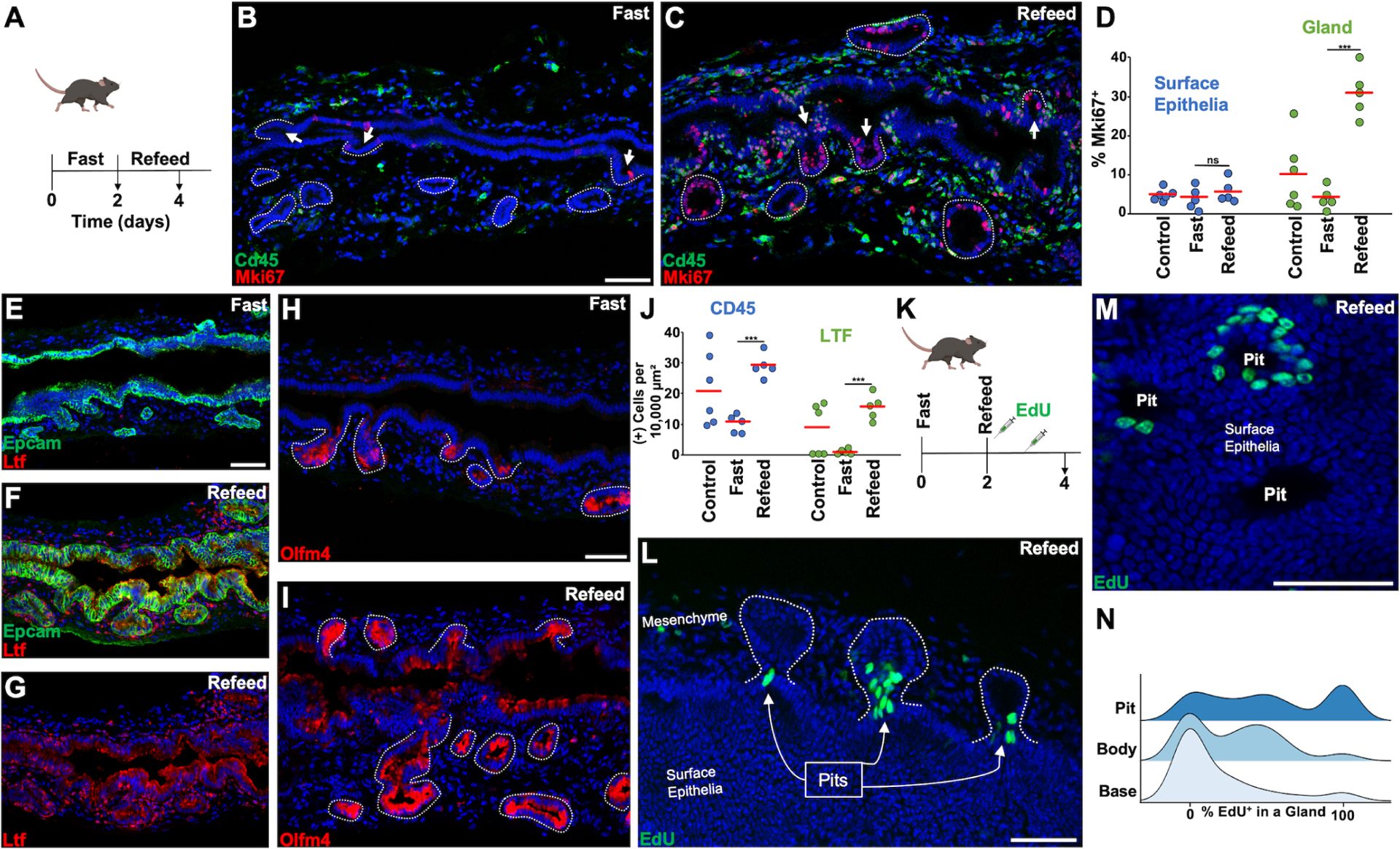
(A) Fasting animals were fasted for two days before harvesting and refeed animals were allowed to eat ad libitum for two days before harvesting. (B and C) Cd45 and Mki67 staining with arrows indicating pits and dashed lines encircling glands. (D) Quantification of Mki67+ surface and glandular epithelia in randomly selected animals versus animals after fasting, and fast/refeeding (n = 5–6 animals per group). (E and F) Ltf staining in fasting and refeed states. (G) Ltf channel from (F). (H and I) Olfm4 staining in fasting and refeed states. (J) Quantification of Cd45+ and Ltf+ cells located within the mesenchymal space in the indicated states (n = 5–6 animals per group). (K) Animals were pulsed with EdU 6hrs after refeeding and 24hrs after refeeding. (L) 3 glands (in dashed outlines) with EdU+ pit cells 2 days after refeeding. (M) The pits of 3 glands seen from the aspect of the surface epithelium. (N) Ridge plot showing percentage of EdU+ cholangiocytes found in a gland by region in the refeed state (102 glands from n = 4 animals). Scale bars, 50 μm (C same scale as B, F and G same scale as E, I same scale as H). *** p<0.001, ns = not significant, by Students t-test.
We found that 2 days of fasting led to a reduction in Mki67+ glandular epithelial cells that increased upon refeeding without significant effect on surface epithelia (Figure 4B–4D). These prandial states modulated epithelial identity as fasting caused a reduction in Ltf expression and Olfm4 was expressed more specifically within the glands (Figure 4E and 4H). Refeeding lead to areas of Ltf and Olfm4 expression in the surface cholangiocytes (Figure 4F–4J). In addition, refeeding caused an increase in Cd45+ and Cd45+Ltf+ cells (Figure 4J and S4D–S4F). Lgals2, which normally has higher expression in the surface epithelium, showed elevated expression within the glandular compartment in the refed state (Figure S4G and S4H). Refeeding led to the complete elimination of Car4 expression (Figure S4I and S4J). Aqp5 and Axin2 expression increased in the glandular compartment, but not the surface epithelium, after refeeding (Figure S4K–S4Q). The expression of Lgr5 and Muc6 had no significant change after refeeding (Figure S4O–S4Q).
EdU was dosed after refeeding with whole mount imaging to localize proliferation (Figure 4K). EdU+ cells localized to the gland pit, which could be seen coming out onto the surface epithelium (Figure 4L–4N). The amount of labeled cells within a gland was heterogenous with some glands being almost entirely labeled (Video S1). Multialveolar glands could have EdU+ cells within only one alveolus (Video S2). Additionally, we found glands that had labeled cells at the base and labeled cells at the pit with a clear break in the body of a gland, suggesting compartmentalization (Video S3).
Pit targeted lineage tracing enhances surface epithelial labeling
Given the developmental origin of the extrahepatic bile duct and the presence of gastric chief cell markers, which are located in the gastric corpus, we hypothesized that renewing epithelial cells in the peribiliary pits may be analogous to gastric corpus isthmus stem cells. In common with peribiliary pit cells, gastric isthmus stem cells are not characterized by unique marker genes. Their identity has been only inferred through lineage tracing with proliferative markers such as Mki67, Stmn1, and Iqgap330,31.
We used Mki67:CreERT2; Rosa26:lox-STOP-lox-tdTomato mice to target proliferative cells in the peribiliary pits and analyzed the kinetics over 180 days (Figure 5A). We quantified labeling in the glands and surface epithelium as follows (see Methods for details): To visualize the surface epithelium in 2-dimensions, max intensity projections were created from 1–5 optical slices. The fractional labeling of the surface epithelium was determined by calculating the ratio of the total area covered by tdTomato+ nuclei. For gland quantifications, two separate optical sections from a z-stack were taken from the pit, body, and base regions leading to the quantification of 6 optical sections per gland. For each of these optical sections, the total number of epithelia and tdTomato+ epithelia were counted to determine the average labeling fraction within that optical section. In the absence of tamoxifen, no labeling was detected over 180 days (Figure S5A). A single dose of tamoxifen labeled peribiliary pits and bases at a rate of ~9%, almost double that of epithelia within the gland body at 14 days post tamoxifen (Figure 5I). In contrast, only ~2.9% of surface epithelial cells were labeled at 14 days post-induction (Figure 5G). Notably, labeled pits could be observed at the gland orifice in continuity with the surface epithelium (Figure 5B). At 30 days post-tamoxifen, localized expansion circumferentially around the pits and onto the surface epithelium could be seen (Figure 5C and 5D). By 180 days, there were areas of confluent labeling with average surface epithelial labeling reaching ~30%, higher than the average gland labeling of ~24% (Figure 5E–5H), and similar to the average labeling fraction in the pit (~29%). These findings were consistent with the notion that pit cells slowly replenish surface epithelial cells. We found that fasting and refeeding Mki67:CreERT2; Rosa26:lox-STOP-lox-tdTomato animals, in a cyclical fashion, could accelerate surface epithelial labeling (Figure S5B and S5C).
Figure 5. Peribiliary Pit Directed Lineage Tracing.
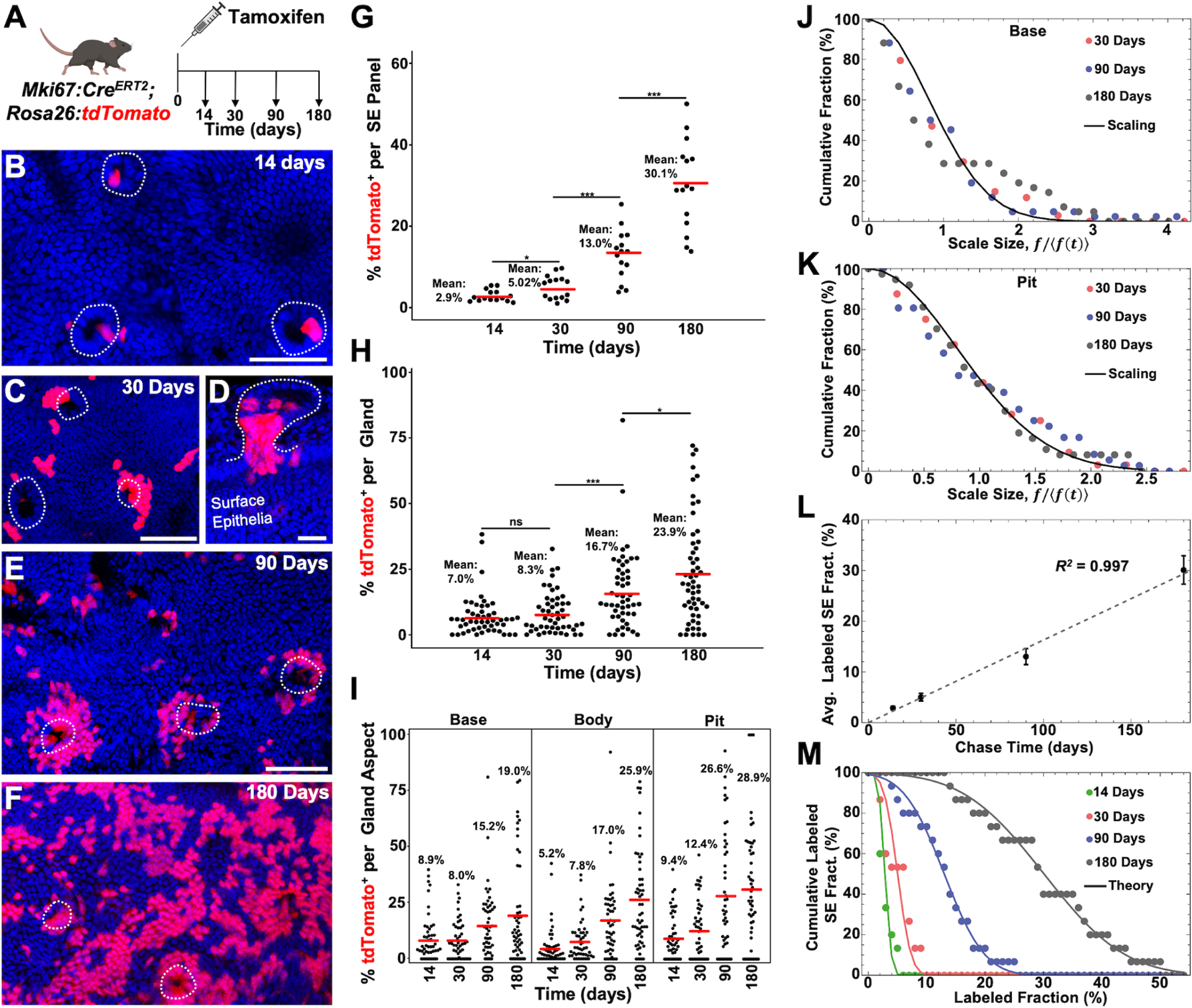
(A) Schematic showing experimental design. (B and C) Surface epithelial panels from 14 (B) and 30 (C) days post tamoxifen in which gland pits (dashed lines) can be seen with labeled cells. (D) Gland (outlined) from 30 days post tamoxifen in continuity with the surface epithelium where high pit labeling is seen. (E and F) Surface epithelial panels from 90 (E) and 180 (F) days post tamoxifen with peribiliary pits outlined. (G) Percent labeling of the surface epithelium from 5 low power panels (n = 3 animals per timepoint). (H) Percent labeling of peribiliary glands (49 to 58 total glands from n = 3 animals per timepoint). (I) Percent labeling of the gland base, body, and pit. (J and K) Cumulative distribution of fractional labeling in the gland base (J) and pit (K) as a function of the rescaled fraction, f/〈f (t)〉, where 〈f(t)〉 denotes the average fraction of cells in glands with non-zero base labeling at chase time t. The line shows the parameter-free statistical scaling curve exp[−πx2/4] predicted for neutral clone dynamics around the circumference of the base and pits regions (for details, see Supplementary Theory). (L) Average labeled cell fraction on the surface epithelium as a function of chase time. Note that the average size grows approximately linearly with time, as predicted by a model based on the neutral competition of surface epithelial cells, fed by migrating cells from the pit. Points show data (error bars denotes standard error. Line shows a linear fit with a slope of 0.16 per day). (M) Cumulative distribution of labeled cell fraction in the surface epithelium. The points show data and lines shows the Gaussian-like prediction of the model at the four chase times (14 days, 30 days, 90 days and 180 days from left to right). For a Gaussian distribution, the cumulative distribution translates to the Error function, , where the factor 0.063 is inferred from the linear fit to the standard deviation of the fraction distribution and t denotes the chase time. Scale bars, 50 μm (B,C,E, F same scale as E); 25 μm (D). * p<0.05, *** p<0.001, ns = not significant, by Students t-test.
To gain further insight into the organization and fate behavior of renewing cells, we questioned whether clonal information could be inferred from this lineage tracing data (Supplemental Theory). From the frequency of unlabeled glands, we estimated that the majority of labeled glands involved just 1 or 2 clonal induction events in the base and pit regions with, respectively, ~20% and ~16% of labeled glands having more than 2 clones in either region at the time of induction (Figure S5D). Notably, a persistent lack of correlation between the fraction of labeled cells in the base/body and pit regions of individual glands over time suggested that these two compartments are maintained independently under homeostatic conditions (Figure S5E–S5G). Based on this reasoning, we questioned whether the lineage tracing data could be used to infer the pattern of renewal in the two compartments.
Previously, lineage tracing studies of the intestinal epithelium have shown that tissue maintenance involves neutral stem cell competition for niche access at the intestinal crypts32,33. As stem cells divide, others become displaced from the crypt base, leading to a “neutral drift” of labeled cells around the circumference of the crypt until the clone is altogether lost or the crypt becomes monoclonal and fixed. A signature of this dynamic is found in the convergence of clone sizes to a statistical scaling behavior in which the chance of finding a clone larger than some multiple of the average becomes independent of the chase time and given by a defined Gaussian size dependence32. Importantly, since such dynamics lead to the rapid elimination of the majority of clones through stochastic loss, we reasoned that, at the level of induction used here, the vast majority of labeled glands at intermediate (30 days) and longer chase times would involve only clonal labeling events (Supplementary Theory). Consistently, quantitative analysis of the labeled cell fractions in both the base and pit regions showed evidence for statistical scaling behavior (Figure 5J and 5K), suggesting that, in common with the intestinal epithelium, both base/body and pit regions are supported independently through a process of cell competition for niche access, leading to neutral drift of clones around the circumference of the gland (Figure S5L).
Based on the inferred dynamics of the pit, we then questioned whether the growth dynamics and distribution of the labeled surface epithelial population could be predicted. From the observed Gaussian-like dependence of the distribution of labeled cell fractions on the surface epithelium, as well as the linear-like time-dependence of both the average fraction and its standard deviation, we could use a statistical modeling-based argument to infer that the maintenance of this compartment involves the slow and continuous migration of epithelial cells from the pit regions, which thereafter undergo a process of stochastic loss and replacement on the surface epithelium (Figure 5L, 5M, S5H, and Supplementary Theory).
Our single-cell sequencing and fasting/refeeding experiments suggested that lineage fate and proliferation are dynamic processes. Lineage tracing with Mki67:CreERT2 had preferential targeting of the gland pit and base, but did not capture the entirety of these regions. To examine whether there was further induction of proliferation in non-labeled cells or whether proliferation was confined to lineage labeled cells, we examined tissue sections of Mki67:CreERT2; Rosa26:lox-STOP-lox-tdTomato mice 30 days post tamoxifen and looked at Mki67 expression by antibody staining. When looking at Mki67 lineage positive only, Mki67 antibody positive only, and Mki67 lineage/Mki67 antibody double positive epithelia within glands, the majority of cells were Mki67 lineage positive only (~65%) (Figure S5I–S5K). ~12% were lineage and antibody double positive (Figure S5I and S5K). Another 22% were only antibody positive for Mki67 (Figure S5J and S5K). This suggests that while renewal has a predilection for certain areas, it is a dynamic process, being induced with need.
To further test the idea that cholangiocyte proliferation was inducible, we sought to understand if this hierarchal preference held true during states of increased biliary proliferation. Bile duct ligation causes a marked increase in proliferation of upstream epithelial cells (Figure S6N). Therefore, 14 days after labeling Mki67:CreERT2; Rosa26:lox-STOP-lox-tdTomato mice, we performed bile duct ligations and looked at surface epithelial labeling 4 days after ligation (Figure S6M). While evidence of positive cells streaming out of a gland could be seen (Figure S6O), the vast majority of the surface epithelium remained unlabeled, suggesting that during obstructive injury, proliferation was not regionally localized (Figure S6P and S6Q).
Base targeted lineage tracing results in contribution to the gland base and body
To target cells at the gland base, we utilized Olfm4:CreERT2; Rosa26:lox-STOP-lox-tdTomato mice and analyzed them at 14, 90 and 180 days post tamoxifen (Figure 6A). In the absence of tamoxifen, we did not detect leaky recombination out to 180 days (Figure S6A). After a single low dose of tamoxifen, there was a low rate of surface epithelial labeling at 14 days post tamoxifen (Figure 6B,E, 3.5% vs 2.9% for Mki67:CreERT2). Unlike Mki67:CreERT2, Olfm4:CreERT2 labeled the base at 17.3% (8.9%, Mki67:CreERT2), body at 12.9% (5.2%, Mki67:CreERT2) and pit at 2.4% (9.4%, Mki67:CreERT2) with an average glandular compartment labeling that was almost double that seen with Mki67:CreERT2 (Figure 6F and 6G). Over the course of the chase, surface epithelial labeling reached an average of 12.1% at 180 days (Figure 6C–6E), far lower than the 30.1% seen with Mki67:CreERT2. Average labeling of the glands increased overtime; however, the fraction of unlabeled pits remained relatively constant, indicating a lack of significant immigration of cells from the base/body into the pit region of extramural glands (Figure 6G and S6B). Because of the low contribution from the pits of extramural glands, there was a lack of confluence of labeled cells on the surface epithelium at 180 days (Figure 6D). As such, the contribution of the minority of mural glands was more readily detectable, where the entirety of the gland was often labeled and could be seen spreading circumferentially onto the surface epithelium (Figure S6C).
Figure 6. Peribiliary Base Directed Lineage Tracing.
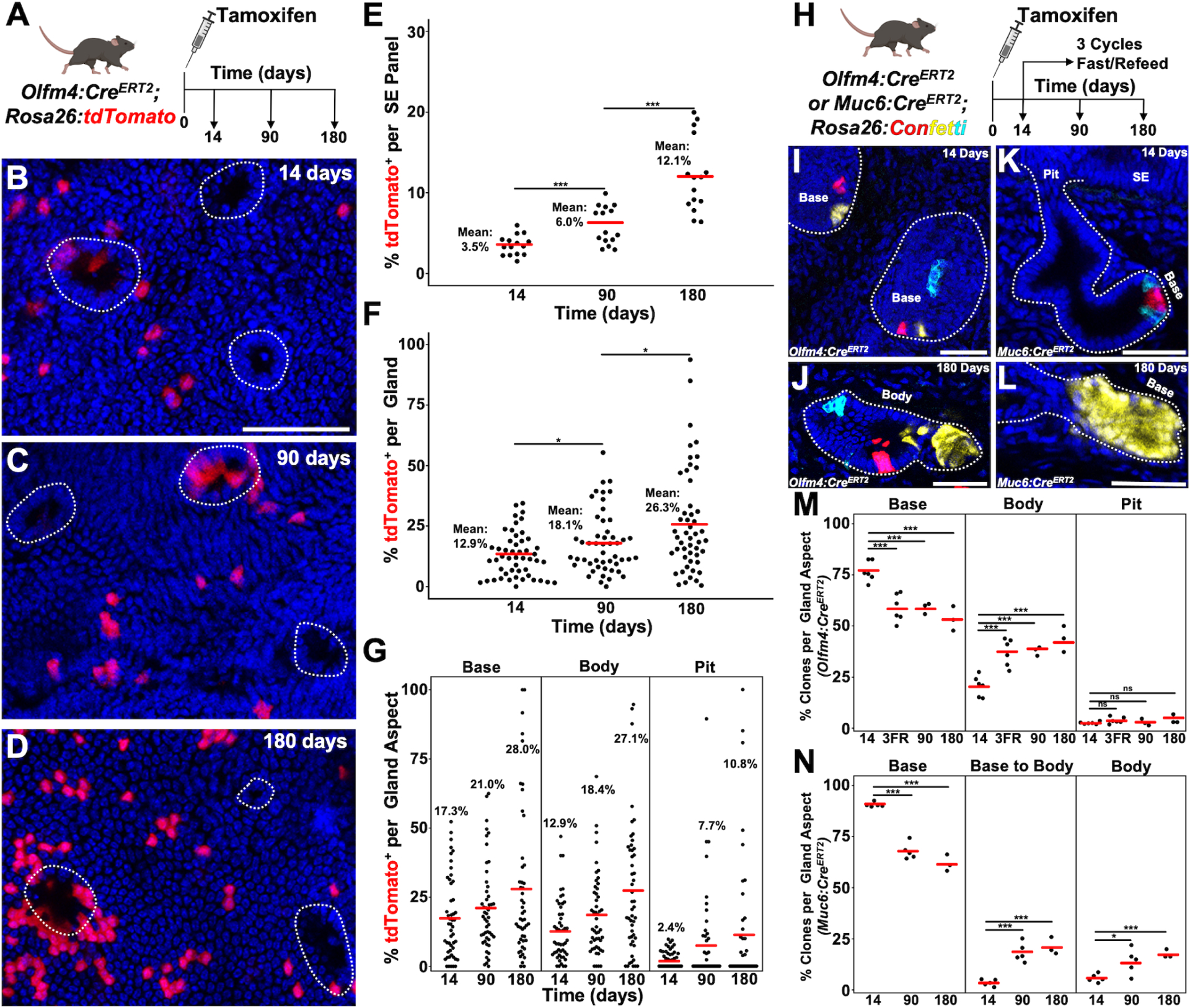
(A) Schematic of experimental design. (B-D) Surface epithelial panels from 14 (B), 90 (C) and 180 (D) days with peribiliary pits marked with dashed outlines. (E) Percent labeling of the surface epithelium from 5 low power panels (n = 3 animals per timepoint). (F) Percent labeling of peribiliary glands (50 total glands from n = 3 animals per timepoint). (G) Percent labeling of the gland base, body, and pit. (H) Schematic showing experimental design. (I) Image of the bases of outlined peribiliary glands 14 days after tamoxifen in a Olfm4:CreERT2; Rosa26:Confetti animal. (J) Example of a gland in which a small alveolus is clonally labeled. (K) Sectional view of a Muc6:CreERT2; Rosa26:Confetti gland 14 days post tamoxifen where recombined cells are only located at the base. (L) Example of a gland at 180 days after tamoxifen in which an alveolus is clonally labeled. (M and N) Quantification of the percent of clones found within a glandular compartment from Olfm4:CreERT2 (M) or Muc6:CreERT2 (N) (at least 50 clones per animal, n = 3–6 animals per timepoint). Scale bars, 50 μm. * p<0.05, *** p<0.001, ns = not significant, by Students t-test.
To further confirm this, we turned to a clonal approach utilizing Olfm4:CreERT2; Rosa26:Confetti animals. In the absence of tamoxifen, there was no recombination seen (Figure S6D). We confirmed that these clones were induced in Olfm4 expressing areas of the gland by Olfm4 antibody staining (Figure S6E–S6G). Tamoxifen induction resulted in ~1.5 clones per gland that were primarily located within the gland base 14 days post tamoxifen (Base = 76% +/− 2%, Body = 20% +/− 2%, Pit = 2.6% +/−0.2%, Average ± SEM, n = 6, Figure 6I and 6M). Animals were chased over 180 days or subjected to 3 cycles of fasting and refeeding (Figure 6H). Similar to lineage tracing with Rosa26:CreERT2; Rosa26:Confetti, we found evidence for clonality in alveoli (Figure 6J). In rare instances, when pit and base labeled clones were within one gland, pit clones expanded onto the surface epithelium; however, the vast majority of the surface epithelium remained unlabeled (Video S4 and Figure S6H–S6J). An analysis of base clones (defined as those clones entirely within the base or extending from the base into the body) and body encompassed clones showed that there was a progressive reduction in the fraction of base clones and a reciprocal increase in the fraction of body-encompassed clones consistent with the body gaining clones through base loss (Figure 6M). The fraction of pitanchored clones did not change significantly (Figure 6M). We also found that mural glands had base anchored clones that migrated out onto the surface epithelium (Figure S6N).
As Olfm4:CreERT2; Rosa26:Confetti animals had some induction of clones within the gland body and pit, we generated a Muc6:CreERT2 line by inserting an IRES-CreERT2 sequence within the 3’ UTR of the endogenous Muc6 locus. In the absence of tamoxifen, no recombination was seen in Muc6:CreERT2; Rosa26:Confetti animals out to 180 days post vehicle (Figure S6K). 14 days after a single dose of tamoxifen, induced clones were seen primarily at the gland base (90% +/− 1%, Average ± SEM, n = 5, Figure 6K and 6N). A minority of clones could be seen extending from the base into the body or located entirely within the body (Base to Body = 4% +/− 1%, Body = 6% +/−1%, Average ± SEM, n = 5, Figure 6N). Unlike Olfm4:CreERT2, there was no labeling within the pit or surface epithelium (Figure 6K and S6L). When chased over a period of 180 days, there was a decrease in the number of base clones with a reciprocal increase in base to body and body clones (Figure 6N and Video S5). As in our previous lineage tracing experiments, we saw clonality within alveoli (Figure 6L). There was no labeling of the surface epithelium over 180 days, with the exception of contributions from mural glands, and consistent with the base/body region of extramural glands functioning as a primarily closed anatomical unit (Figure S6M and S6O).
Increased canonical Wnt signaling causes acquisition of gastric chief cell markers and loss of surface epithelial markers
While sparse, the base of peribiliary glands showed the highest level of Axin2 expression, matching architecturally the epithelial linings of the stomach, intestine, and colon. However, by in situ staining, the expression level was found to be lower than that seen in the intestine (Figure S1D–S1F). We therefore questioned whether increased canonical Wnt signaling would affect cellular identity and proliferation. To target epithelia in both surface and glandular compartments, we used a Prom1:CreERT2, which causes recombination in all epithelial cells of the bile duct after tamoxifen mediated recombination as seen in Prom1:CreERT2; Rosa26:lox-STOP-lox-tdTomato mice (Figure S7A and S7B). To increase canonical Wnt signaling within epithelia, we employed Prom1:CreERT2; Ctnnb1loxEx3/+ mice (Figure 7A). We found that recombination was mosaic with patchy epithelial areas showing confluent in situ expression of Lgr5 and Axin2 (Figure 7B–7F, S7C). In situs of Muc6 showed increased expression within glands, but without expression in the surface epithelium (Figure S7D, S7E, and S7H). The in situ expression level of Aqp5 did not change in either the glands or surface epithelium (Figure S7F, S7G, and S7I). Staining with Mki67 showed areas of high expression within the surface epithelium and glands (Figure 7G and 7H).
Figure 7. Epithelial β-catenin Gain of Function.
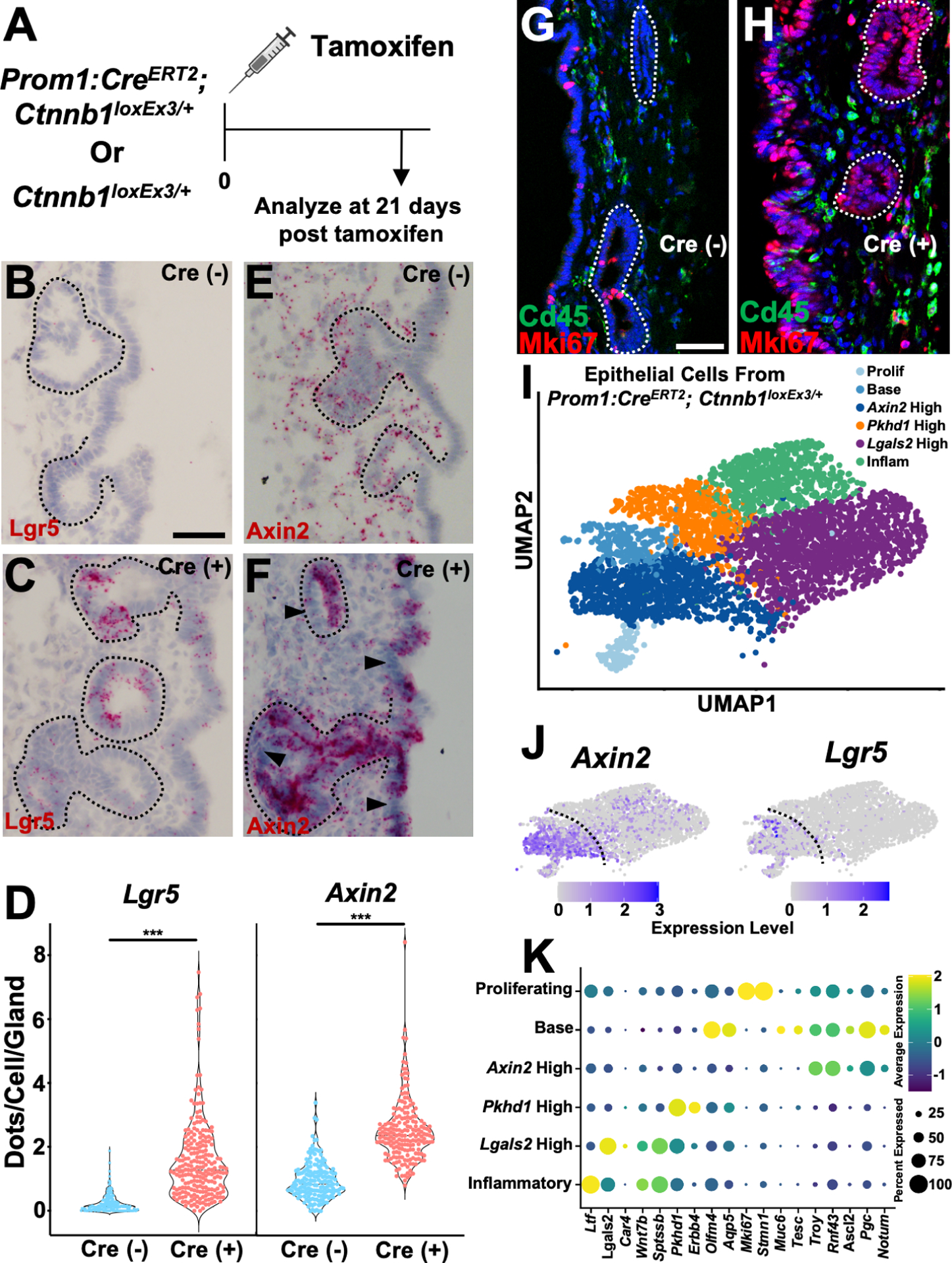
(A) Schematic showing experimental design. (B and C) Lgr5 in situ in Cre (−) and Cre (+) animals with glands outlined. (D) Quantification of the amount of Lgr5 and Axin2 in situ signal in glands (37–67 glands per animal, n = 4 animals). (E and F) Axin2 in situ in Cre (−) and Cre (+) animals with glands outlined. Arrowheads (F) point to Axin2 negative epithelia to contrast more confluent staining in epithelia with β-catenin gain of function. (G and H) Mki67 and Cd45 staining in Cre (+) and Cre (−) animals. (I) UMAP of epithelial cells from Prom1:CreERT2; Ctnnb1loxEx3/+ animals 21 days after tamoxifen. (J) Feature plots showing expression of Axin2 and Lgr5. (K) Dot plot of indicated genes from Prom1:CreERT2; Ctnnb1loxEx3/+ animals. Scale bars, 50 μm (A-D); 25 μm (G; H same scale as G). *** p<0.001, by Student’s t-test.
To disaggregate the mosaic recombination and assess transcriptional changes in recombined cells, we performed single-cell profiling of Prom1:CreERT2; Ctnnb1loxEx3/+ animals 21 days after tamoxifen induction. Examination of epithelia, without tuft cells, showed 6 broad classes. Like our initial dataset, there was still significant transcriptional overlap (Figure 7I). When the expression of Axin2 and Lgr5 was assessed, it was highest in 3 clusters (Figure 7J). These clusters contained proliferating epithelia, a population with base markers, and a group that was high in Axin2. Interestingly, these three clusters had high expression of Pgc, Tnfrsf19 (Troy), Rnf43, and Ascl2, which are gastric chief cell marker genes (Figure 7K and S7L). When the Axin2 high population was compared to the non-recombined epithelial populations (Pkhd1 high, Lgals2 high, Inflammatory), there was a significant decrease in surface epithelial enriched genes such as Lgals2, Wnt7b, and Sptssb (Figure 7K). Additionally, there was evidence of Wnt antagonism seen with the increased expression of Notum (Figure 7K).
To confirm these findings, we compared the Prom1:CreERT2; Ctnnb1loxEx3/+ base and Axin2 high populations to base, glandular, Lgals2 high, Car4 high and Wnt7b high populations from our wildtype dataset. Markers of gastric chief cells were still elevated and there was still a decrease in surface epithelial markers (Figure S7M–S7O). The expression of Sox9 still maintained a higher level of expression within the glandular compartment (Figure S7P and S7Q).
DISCUSSION
Here, we used non-targeted clonal and targeted higher density lineage tracing combined with single-cell analysis to define the homeostatic dynamics of the extrahepatic bile duct. In common with the stomach corpus gland and the epidermis30,34, the extramural glands show hallmarks of compartmentalization. In this model, pits of extramural glands progressively renew the surface epithelium, whereas those within the gland base and body undergo localized turnover. The pit can therefore be thought of as a privileged niche, similar to the isthmus region of the stomach corpus30,31. Quantitative analysis of the clonal dynamics in the base and pit regions provided evidence that both regions undergo a process of stochastic renewal based on neutral competition for niche access, reminiscent of that reported in the intestinal epithelium.
While often concentrated within the pits of glands, there was heterogeneity in proliferative activity with glands adjacent to each other showing varying levels of proliferation. The environment of the extrahepatic bile duct is unique in that it receives diurnal pulses of caustic/proinflammatory bile, which can also cause stretch upon the duct itself. It is possible that the proliferative heterogeneity between glands is a byproduct of differences in the gland microenvironment. Those able to respond first to the stimuli are able to meet the demands and could alleviate the need for additional proliferative responses. Notably, surface epithelial cells proliferate during homeostasis, which raises the question of why do surface epithelial clones not survive over the long term? We suspect that this is due to greater loss of epithelia from the surface epithelium compared to the glands. Indeed, in humans there is evidence that surface epithelial cells are more sensitive to injury than epithelia in the peribiliary glands35. One can imagine that oscillating biliary flow causes greater stochastic clone loss on the surface epithelium with reconstitution driven by the pits of extramural glands allowing pit-derived clones to win out over long time frames. In contrast to the extramural glands, mural glands showed evidence of base-derived clones contributing to the entire gland and surface epithelium, in a manner reminiscent of intestinal crypts and stomach pyloric glands24,33. Our results show that as the complexity of glands increases, there is a change in how they contribute to homeostasis.
How does stem cell theory play into these findings? On one hand, the turnover of the extrahepatic biliary epithelium appears to conform to a hierarchical organization in which gland pits and bases show a survival advantage. However, unlike a classical stem cell driven paradigm, where differentiated lineages derive predominantly from a stem cell compartment marked by a unique expression signature, cholangiocytes are capable of modifying their identity and behavior in response to demand, potentially allowing for greater adaptation to a changing microenvironment.
The biliary epithelium showed a striking response to β-catenin gain of function. This pushed biliary epithelia toward a gastric chief cell fate, and increased Muc6 expression, while decreasing the expression of surface epithelial markers. This shows that canonical Wnt signaling is a key component in regulating positional identity within the extrahepatic duct. This is important translational information, as peribiliary glands are hyperproliferative in diseases such as primary sclerosing cholangitis with increased Muc6 expression seen during biliary epithelial neoplasia14,36. Further dissecting out the role of Wnt signaling on the maintenance and dysregulation of the peribiliary glands will be important for therapeutic approaches.
Our results provide a new framework with which to understand proliferation, homeostasis, and cellular identity in the extrahepatic bile ducts of the liver. Future studies examining proliferative regulation will be crucial to gain insights into diseases of the large ducts such as primary sclerosing cholangitis and cholangiocarcinoma.
Limitations of the Study
A limitation of our analysis is the inability to identity a pit-specific marker that would enable cells to be traced from this region. We attribute this to the dynamic nature of cellular identity within the extrahepatic bile duct which, like proliferation, appears to be modulated by changes in the microenvironment. Our study also relies on density labeling of cells within a region and examining a different duct at a later timepoint. It is possible that migration and proliferation from a lower density labeled area could dominate. Such a limitation could be overcome with live imaging of the duct to trace individual cells.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the Lead Contact, Dr. Vikas Gupta (vikas.gupta@yale.edu).
Materials availability
Transgenic mice generated in this study are available for research purposes upon request.
Data and code availability
Sequencing data generated can be downloaded from the Gene Expression Omnibus under accession number GSE223099. All other data are available from the corresponding author upon reasonable request. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
Gli1:CreERT2 mice43 were generously donated by Dr. Alexandra L. Joyner (Memorial Sloan Kettering Center). Ctnnb1loxEx3 mice44 were generously donated by Drs. David T. Breault (Boston Children’s Hospital) and Makoto Mark Taketo (Kyoto University). Olfm4:CreERT2 mice26 were generously donated by Drs. Linda C. Sameulson (University of Michigan) and Hans Clevers (Hubrecht Institute). Rosa26:Confetti33, Mki67:CreERT245, Prom1:CreERT246, Rosa26:lox-STOP-lox-tdTomato47, and Rosa26:CreERT248 mice were obtain from The Jackson Laboratory. The Muc6-IRES-CreERT2 mouse model was generated via CRISPR/Cas9-mediated genome editing. Templates for sgRNA synthesis were generated by PCR and tested for activity by zygote electroporation. A recombination template plasmid was constructed of 1 kb Muc6 5’ and 3’ homology arms into a vector containing an IRES-CreERT2 cassette. Genotype screening of tissue biopsies from founder pups was performed by PCR amplification and Sanger sequencing to verify the knockin allele. Male and female mice were used for all experiments at the ages of 8–10 weeks. Up to 5 mice were housed in ventilated cages in a pathogen free facility under a 12-h light and dark cycle at an ambient temperature of 22°C with food and water ad libitum. Mice were kept in a C57BL/6 or C57BL/6 X 129S1 mixed background.
METHODS DETAILS
Mice treatments
Tamoxifen (Sigma) was diluted in corn oil at a concentration of 20 mg/ml and the dose was titrated for recombination efficiency for each experiment as follows (X mg of tamoxifen for 25 g of mouse body weight). Rosa26:CreERT2; Rosa26:Confetti animals were given 2 mg once; Muc6:CreERT2; Rosa26:Confetti and Mki67:CreERT2; Rosa26:lox-STOP-lox-tdTomato animals were given 1.5 mg once; Olfm4:CreERT2; Rosa26:lox-STOP-lox-tdTomato animals were given 0.5 mg once; Olfm4:CreERT2; Rosa26:Confetti animals were given 2 mg daily dose for two days; Gli1:CreERT2; Rosa26:lox-STOP-lox-tdTomato and Prom1:CreERT2; Ctnnb1loxEx3/+ animals were given 2.5 mg daily for three days. Bile duct ligation was performed as previously described by Gupta et al49. For fasting and refeeding experiments, animals were placed in metabolic caging without solid food and grates to prevent coprophagia. After 48 hours, solid food was reintroduced. Animals had continuous access to water. Animal weights were measured daily during this experiment. All animal experiments were performed on at least two separate occasions. All animal experiments procedures, breeding, and ethical use were performed in accordance with the guidelines set by the Institutional Animal Care and Use Committee at the Yale School of Medicine.
Histology and Immunofluorescence
For tissue sections, bile duct was freshly isolated and fixed in 4% paraformaldehyde, cryoprotected in 30% sucrose, embedded in OCT (Tissue-Tek; Sakura Finetek), and sectioned at 8 μm. Sections were blocked for 40 minutes at room temperature in blocking solution (5% goat serum or 5% donkey serum, 2% fish gelatin, 0.2% BSA, 0.3% Triton-X, 1X PBS) and incubated with the following primary antibodies: rat monoclonal anti-Epcam (1:500, BD Biosciences), rabbit polyclonal anti-Mki67 (1:500, Abcam), rat monoclonal anti-Cd45 (1:500, BD Biosciences), rabbit monoclonal anti-Ltf (1:300, Sinobiological), rabbit monoclonal anti-Olfm4 (1:500, Cell Signaling Technology), rabbit polyclonal anti-Lgals2 (1:200, Sigma), goat polyclonal anti-Car4 (1:200, R&D Systems), rabbit polyclonal anti-Sox9 (1:500, EMD milipore), rabbit polyclonal anti-Dclk1 (1:500, Abcam), and rat monoclonal anti-RFP (1:200, Chromotek). Secondary Alexa Fluor antibodies (Thermo) were used for detection. For Mki67 and Cd45, sections underwent antigen retrieval at 85°C for 15 minutes with Citric Acid pH6 buffer (70°C for Car4, Olfm4, Lgals2). For Sox9, sections underwent antigen retrieval at 95°C for 15 minutes with Tris-EDTA pH8 buffer.
Confocal Imaging
Fluorescent images were taken with a Leica SP8 or Stellaris confocal microscope using a 20 to 25X objective with LAS AF software. Images were acquired at a resolution of 1024 X 1024, 8 bit. Scanning frequency was set to 600 Hz. Sections for counting were processed and analyzed with ImageJ.
Whole Mount Staining and Imaging
Whole bile duct was carefully dissected out and the lumen flushed with ice cold PBS. The duct was fixed for 3 hours at 4°C on a rocker and then placed into a solution containing 0.3% Triton-X, 10 μg/ml Hoechst 33342 or Draq7 (Confetti), and 1X PBS and rocked overnight at 4°C. The duct was washed 2 × 15 minutes with PBS. Ducts were then placed into Ce3d50 and rocked overnight at 4°C to clear before mounting between two coverslips. Z stacks of a duct were acquired on a Leica SP8 or Stellaris microscope with a 20 to 25X lens at a resolution of 1024 × 1024, 8 bit. Scanning frequency was set to 600 Hz with line averaging to 1. Z step size was set at 1.5 μm.
RNAscope in situ hybridization
7 μm fixed cryosections were prepared according to manufacturer’s instructions (Advanced Cell Diagnostics TN 320534/Rev B and 322360-USM). The 2.5 HD Reagent Red Kit was used for detection. The following probes were used: Mm-Muc6, Mm-Lgr5, Mm-Aqp5, Mm-Axin2.
Proliferation Assays
To measure cell proliferation, EdU (Lumiprobe, 10540) was dissolved in PBS at a concentration of 4 mg/ml and 1 mg was injected intraperitoneally for a dose. Detection was performed with “click” chemistry51 using a Sulfo-Cyanine5 azide (Lumiprobe, A3330). Briefly, sections or whole mount tissue was incubated for 15 minutes in a solution containing 100 mM Tris buffer, 100 mM ascorbic acid, 2 mM CuSO4, 50 μm azide dye to detect EdU.
Single-Cell Library Preparation
Whole bile ducts were isolated and micro dissected in petri dishes to remove all adjoining tissue. Ducts were placed in 1 ml of HBSS with 1.8 mM Ca, 0.8 mM Mg, 1 mg dispase (Roche), 2 mg protease from Bacillus licheniformis (Sigma) and cut into small pieces with microdissecting scissors. These were incubated for 15 minutes at 37°C. This solution was removed and the remaining bile duct chunks were placed in a solution with 1X PBS, 5mM EDTA, and 0.5% BSA and incubated at 37°C for another 10 minutes. This was filtered through a 100 μm filter. This single cell suspension was washed twice with a PBS, 0.04% BSA solution prior to GEM and library generation according to manufacturer’s protocol (10X Genomics, Chromium Single Cell 3’ Reagent Kits User Guide, v3 Chemistry). Libraries were sequenced on an Illumina HiSeq 4000 and an Illumina Novaseq.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of Single-Cell Data
The single cell RNA sequencing fastq data was processed according to 10X Genomics instructions in CellRanger to generate a gene expression matrix for the sample. The filtered expression matrix was then imported into R package Seurat for analysis. Filtering was then performed on the expression matrix with cutoffs of greater than 1800 and less than 9000 genes and mitochondrial RNA content threshold set at 10 to 20% per cell (Figure S3F and S7K) . This left 4273 and 4002 cholangiocytes in the wild type and Prom1:CreERT2; Ctnnb1loxEx3/+ datasets, respectively. The resulting UMI counts were normalized using the R package scran (v1.26.2) and batch-corrected with FastMNN wrapped in R package SeuratWrappers (v0.3.1), with default parameters. The top 30 principal components of reconstructed assays derived from FastMNN were used to perform unsupervised clustering analysis with Louvain (resolution set as 0.6) to give biologically relevant cell type content to each cluster. UMAP was used to generate two-dimensional visualizations with default parameter settings. Python package PHATE (v1.0.10) was adopted to capture and visualize biological structure in high-dimensional scRNA-seq data. Based on PHATE-embeddings, trajectory inference was then performed with R package slingshot (v2.6.0) and temporally dynamic genes were detected with R package tradeSeq (v1.12.0).
Statistical Analysis of Sectional Cell Counting
For animal studies involving cell counting from sections, at least 3 or more separate animals were used, and measurements were made on at least five 20X images of the bile duct for a given animal. For in situ counting, each dot was counted and the total number was normalized by area or cell number. For parametric data involving cell counting experiments, significance was analyzed using a one-tailed unpaired Student’s t-test performed in Microsoft Excel.
Statistical and Compartmental Analysis of Whole Mount Imaging
For labeling experiments from Mki67:CreERT2; Rosa26:lox-STOP-lox-tdTomato and Olfm4:CreERT2; Rosa26:lox-STOP-lox-tdTomato animals, we quantified the labeling fraction in each compartment by the following method. To visualize the surface epithelium in 2-dimensions, max intensity projections were created from 1–5 slices, subtracting any mesenchyme. The fractional labeling of the surface epithelium was determined by calculating the ratio of the total area covered by tdTomato+ nuclei and Hoechst+ nuclei. For quantifications of labeling in glands, two separate sections from a z-stack were taken from the pit, body, and base regions leading to the quantification of a total of 6 sections per gland. To define these compartments, a trajectory starting from the lumen of the pit and into the lumen of each alveolus was used to identify the most distant portions of the gland away from the surface epithelium. To demarcate the pit region, a section was chosen in continuity with the surface epithelium to define the upper boundary while the lower boundary was taken as a section ~5 to 10 μm into the gland depending upon the gland size to represent approximately 20% of the gland. To define the base region, the lower boundary was defined as the most distal section capturing the aspect of the gland furthest away from the surface epithelium (as defined by the initial trajectory) with the upper boundary positioned at ~5 to 10 μm into the gland depending upon the gland size to represent approximately 20% of the gland. For glands with multiple alveoli, the base of each alveolus was counted separately and averaged. Body images were obtained as two equally spaced slices between the pit and base regions. For each of these optical sections, the total number of epithelia and tdTomato+ epithelia were counted to determine the average labeling fraction within that optical section. The same procedure was followed when counting EdU+ epithelial cells from whole mount stained ducts.
For labeling experiments involving multicolor Rosa26:CreERT2; Rosa26:Confetti and Olfm4:CreERT2; Rosa26:Confetti animals, a clone was considered to be surface bound if it was entirely contained within the surface epithelium. A pit clone was one that wrapped around the pit of the gland and could have portions on the surface epithelium. A body clone was one that was contained within the body of the gland and did not extend to the pit or base region. A base clone was one that contained cells at the most basal aspect of the gland and could extend into the body region of the gland. For Muc6:CreERT2; Rosa26:Confetti animals a base clone was one that contained cells only within the base of the gland; a base to body clone was one that had a clone extending from the base and into the body; and a body clone was one that was contained entirely within the body of the gland.
Supplementary Material
Video S1. Z-stack of gland (dashed outline) after refeeding in which the entire gland is positive for EdU (Red), related to Figure 4.
Video S2. Z-stack of a gland (dashed outline) after refeeding in which 1 alveolus of a trialveolar gland is positive for EdU (Red), related to Figure 4.
Video S3. Z-stack of a gland (dashed outline) after refeeding in which there are EdU+ cells at the base and EdU+ cells at the pit with a clear break in a gland (dashed outline), related to Figure 4.
Video S4. Z-stack of a gland 90 days post tamoxifen from a Olfm4:CreERT2; Rosa26:Confetti animal in which a yellow fragmented clone is seen at the base of 3 different alveoli and a red clone anchored at the pit is seen expanding onto the surface epithelium, related to Figure 6.
Video S5. Z-stack of an gland 180 days post tamoxifen from a Muc6:CreERT2; Rosa26:Confetti animal in which a red clone can be seen extending from the base of one alveoli and into the body of the gland, related to Figure 6.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-Epcam | BD Biosciences | Cat# 552370 |
| Rat monoclonal anti-Cd45 | BD Biosciences | Cat# 553076 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat# ab15580 |
| Rabbit monoclonal anti-Ltf | Sinobiological | Cat# 11096-R006 |
| Rat monoclonal anti-Olfm4 | CST | Cat# 39141 |
| Rabbit polyclonal anti-Lgals2 | Sigma | Cat# HPA003536 |
| Goat polyclonal anti-Car4 | R&D Systems | Cat# AF2414 |
| Rabbit polyclonal anti-Sox9 | EMD Milipore | Cat# AB5535 |
| Rat monoclonal anti-RFP | Chromotek | Cat# 5f8 |
| Rabbit polyclonal anti-Dclk1 | Abcam | Cat# ab31704 |
| Goat anti-Rabbit IgG (H+L) Secondary, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21244 |
| Goat anti-Rabbit IgG (H+L) Secondary, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21429 |
| Goat anti-Rat IgG (H+L) Secondary, Alexa Fluor 555 | Thermo Fisher Scientific | Cat # A-21434 |
| Goat anti-Rat IgG (H+L) Secondary, Alexa Fluor 647 | Thermo Fisher Scientific | Cat # A-21247 |
| Donkey anti-Goat IgG (H+L) Secondary, Alexa Fluor 647 | Thermo Fisher Scientific | Cat # A-21447 |
| Chemicals, peptides, and recombinant proteins | ||
| Normal Goat Serum | Abcam | Cat# ab7481 |
| Tamoxifen | Sigma | Cat# T5648 |
| Tween-20 | Sigma | Cat# P9416 |
| Triton-X | Sigma | Cat# T8787 |
| Hoechst 33258 | Thermo Fisher Scientific | Cat# H3569 |
| Normal Donkey Serum | Abcam | Cat# ab7475 |
| EdU | Lumiprobe | Cat# 10540 |
| Sulfo-Cyanine5 azide | Lumiprobe | Cat# A3330 |
| BSA | Sigma | Cat# A9647 |
| Tri-Sodium Citrate Dihydrate | Sigma | Cat# 6132-04-3 |
| Draq 7 | Abcam | Cat# ab109202 |
| Sucrose | Sigma | Cat# S8501 |
| Fluoromount-G | SouthernBiotech | Cat# 0100–01 |
| DNAase | Roche | Cat# 4716728001 |
| Protease from Bacillus licheniformis | Sigma | Cat# 9014-01-1 |
| Dispase | Roche | Cat# 4942078001 |
| Collagenase IV | Thermo Fisher Scientific | Cat# 17104019 |
| DMEM, high glucose, HEPES, no phenol red | Thermo Fisher Scientific | Cat# 21063029 |
| N-methylacetamide | Sigma | Cat# M26305 |
| Histodenz | Sigma | Cat# D2158 |
| 1-Thioglyceral | Sigma | Cat# M1753 |
| Fish Gelatin | Sigma | Cat# G7765 |
| Critical commercial assays | ||
| RNAscope 2.5 HD Red | Advanced Cell Diagnostics | Cat# 322350 |
| Chromium Single Cell 3’ Kit | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/library-prep/doc/user-guide-chromium-single-cell-3-reagent-kits-user-guide-v2-chemistry |
| Deposited data | ||
| Single Cell RNA Seq | This paper | GEO: GSE223099 |
| Experimental models: Organisms/strains | ||
| Olfm4:Cre ERT2 | Donated by Drs. Linda C. Sameulson (University of Michigan) and Hans Clevers (Hubrecht Institute) | MGI:5805245 |
| Rosa26:Confetti | The Jackson Laboratory | JAX: 013731 |
| Mki67:Cre ERT2 | The Jackson Laboratory | JAX: 029803 |
| Rosa26:lox-STOP-lox-tdTomato | The Jackson Laboratory | JAX: 007909 |
| Gli1:Cre ERT2 | Donated by Dr. Alexandra L. Joyner (Memorial Sloan Kettering Center) | JAX: 007913 |
| Prom1:Cre ERT2 | The Jackson Laboratory | JAX: 017743 |
| Rosa26:Cre ERT2 | The Jackson Laboratory | JAX: 008875 |
| Ctnnb1 loxEx3 | Donated by Drs. David T. Breault (Boston Children’s Hospital) and Makoto M. Taketo (Kyoto University) | JAX: 032770 |
| Muc6-IRES-Cre ERT2 | This paper | N/A |
| Oligonucleotides | ||
| Mm-Axin2-C1 | Advanced Cell Diagnostics | Cat# 400331 |
| Mm-Lgr5-C1 | Advanced Cell Diagnostics | Cat# 312171 |
| Mm-Aqp5-C1 | Advanced Cell Diagnostics | Cat# 430021 |
| Mm-Muc6-C1 | Advanced Cell Diagnostics | Cat# 448481 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 201237 | https://imagej.nih.gov/ij/ |
| Cell Ranger v5.0 | Zheng et al., 201738 | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat (v4.0.1) | Butler et al., 201839 | https://github.com/satijalab/seurat |
| Scran (v1.26.2) | Lun et al., 201640 | https://github.com/MarioniLab/scran |
| PHATE (v1.0.10) | Moon et al., 201928 | https://github.com/KrishnaswamyLab/PHATE |
| Slingshot (v2.6.0) | Street et al., 201829 | https://github.com/kstreet13/slingshot |
| tradeSeq (v1.12.0) | Van den Berge et al., 202041 | https://github.com/statOmics/tradeSeq |
| FastMNN (SeuratWrappers v0.3.1) | Haghverdi et al., 201842 | http://bioconductor.org/packages/devel/bioc/vignettes/batchelor/inst/doc/correction.html |
Highlights.
Pits of extramural peribiliary glands homeostatically renew the surface epithelium
Bases of mural peribiliary glands homeostatically renew the surface epithelium
Graded gene expression defines compartments, which could be modulated diurnally
Canonical Wnt signaling controls biliary epithelial identity
Acknowledgements:
We would like to thank the staff of the Yale Liver Center Imaging and Morphology Core as well as the Yale Genome Editing Center for generating the Muc6-IRES-CreERT2 mouse. Some illustrations were created with BioRender. Grant Support: V.G. is supported by 1K08DK128585. This work was supported by the Yale Liver Center award NIH P30 DK034989. B.D.S. is supported by the Wellcome Trust (219478/Z/19/Z) and a Royal Society EP Abraham Research Professorship (RP/R1/180165 and RP\R\231004). This research was funded in whole, or in part, by the Wellcome Trust (203151/Z/16/Z, 203151/A/16/Z) and the UKRI Medical Research Council (MC_PC_17230).
For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
REFERENCES
- 1.Clevers H, Loh KM, and Nusse R (2014). Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012. 10.1126/science.1248012 [DOI] [PubMed] [Google Scholar]
- 2.Rompolas P, and Greco V (2014). Stem cell dynamics in the hair follicle niche. Seminars in cell & developmental biology 25, 34–42. 10.1016/j.semcdb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzales KAU, and Fuchs E (2017). Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev Cell 43, 387–401. 10.1016/j.devcel.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison SJ, and Scadden DT (2014). The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334. 10.1038/nature12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanpain C, and Simons BD (2013). Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol 14, 489–502. 10.1038/nrm3625 [DOI] [PubMed] [Google Scholar]
- 6.Lemaigre FP (2020). Development of the Intrahepatic and Extrahepatic Biliary Tract: A Framework for Understanding Congenital Diseases. Annu Rev Pathol 15, 1–22. 10.1146/annurev-pathmechdis-012418-013013 [DOI] [PubMed] [Google Scholar]
- 7.Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, Karns RA, Chen F, Rezvani M, Luu HY, Mattis AN, Rougemont AL, Rosenthal P, Huppert SS, and Willenbring H (2018). De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature 557, 247–251. 10.1038/s41586-018-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, Ridgway RA, Kendall T, Williams MJ, Jamieson T, Raven A, Hay DC, Iredale JP, Clarke AR, Sansom OJ, and Forbes SJ (2015). Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 17, 971–983. 10.1038/ncb3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O’Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, Kotelevtsev Y, Ffrench Constant C, Boulter L, and Forbes SJ (2017). Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354. 10.1038/nature23015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, and Clevers H (2015). Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312. 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aloia L, McKie MA, Vernaz G, Cordero-Espinoza L, Aleksieva N, van den Ameele J, Antonica F, Font-Cunill B, Raven A, Aiese Cigliano R, Belenguer G, Mort RL, Brand AH, Zernicka-Goetz M, Forbes SJ, Miska EA, and Huch M (2019). Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat Cell Biol 21, 1321–1333. 10.1038/s41556-019-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardinale V, Wang Y, Carpino G, Mendel G, Alpini G, Gaudio E, Reid LM, and Alvaro D (2012). The biliary tree--a reservoir of multipotent stem cells. Nat Rev Gastroenterol Hepatol 9, 231–240. 10.1038/nrgastro.2012.23 [DOI] [PubMed] [Google Scholar]
- 13.Cardinale V, Wang Y, Carpino G, Cui CB, Gatto M, Rossi M, Berloco PB, Cantafora A, Wauthier E, Furth ME, Inverardi L, Dominguez-Bendala J, Ricordi C, Gerber D, Gaudio E, Alvaro D, and Reid L (2011). Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology 54, 2159–2172. 10.1002/hep.24590 [DOI] [PubMed] [Google Scholar]
- 14.Carpino G, Nevi L, Overi D, Cardinale V, Lu WY, Di Matteo S, Safarikia S, Berloco PB, Venere R, Onori P, Franchitto A, Forbes SJ, Alvaro D, and Gaudio E (2020). Peribiliary Gland Niche Participates in Biliary Tree Regeneration in Mouse and in Human Primary Sclerosing Cholangitis. Hepatology 71, 972–989. 10.1002/hep.30871 [DOI] [PubMed] [Google Scholar]
- 15.Matsui S, Harada K, Miyata N, Okochi H, Miyajima A, and Tanaka M (2018). Characterization of Peribiliary Gland-Constituting Cells Based on Differential Expression of Trophoblast Cell Surface Protein 2 in Biliary Tract. Am J Pathol 188, 2059–2073. 10.1016/j.ajpath.2018.05.016 [DOI] [PubMed] [Google Scholar]
- 16.de Jong IEM, van Leeuwen OB, Lisman T, Gouw ASH, and Porte RJ (2018). Repopulating the biliary tree from the peribiliary glands. Biochim Biophys Acta Mol Basis Dis 1864, 1524–1531. 10.1016/j.bbadis.2017.07.037 [DOI] [PubMed] [Google Scholar]
- 17.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- 18.Hayata Y, Nakagawa H, Kurosaki S, Kawamura S, Matsushita Y, Hayakawa Y, Suzuki N, Hata M, Tsuboi M, Kinoshita H, Miyabayashi K, Mizutani H, Nakagomi R, Ikenoue T, Hirata Y, Arita J, Hasegawa K, Tateishi K, and Koike K (2021). Axin2+ Peribiliary Glands in the Periampullary Region Generate Biliary Epithelial Stem Cells That Give Rise to Ampullary Carcinoma. Gastroenterology 160, 2133–2148.e6. 10.1053/j.gastro.2021.01.028 [DOI] [PubMed] [Google Scholar]
- 19.van Amerongen R, Bowman AN, and Nusse R (2012). Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400. 10.1016/j.stem.2012.05.023 [DOI] [PubMed] [Google Scholar]
- 20.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, and Wright CV (1996). PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983–995. 10.1242/dev.122.3.983 [DOI] [PubMed] [Google Scholar]
- 21.Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, and Wells JM (2009). Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell 17, 62–74. 10.1016/j.devcel.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiPaola F, Shivakumar P, Pfister J, Walters S, Sabla G, and Bezerra JA (2013). Identification of intramural epithelial networks linked to peribiliary glands that express progenitor cell markers and proliferate after injury in mice. Hepatology 58, 1486–1496. 10.1002/hep.26485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kell DB, Heyden EL, and Pretorius E (2020). The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria. Front Immunol 11, 1221. 10.3389/fimmu.2020.01221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan SH, Swathi Y, Tan S, Goh J, Seishima R, Murakami K, Oshima M, Tsuji T, Phuah P, Tan LT, Wong E, Fatehullah A, Sheng T, Ho SWT, Grabsch HI, Srivastava S, Teh M, Denil SLIJ, Mustafah S, Tan P, Shabbir A, So J, Yeoh KG, and Barker N (2020). AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature 578, 437–443. 10.1038/s41586-020-1973-x [DOI] [PubMed] [Google Scholar]
- 25.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, and van Oudenaarden A (2011). Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol 14, 106–114. 10.1038/ncb2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuijers J, van der Flier LG, van Es J, and Clevers H (2014). Robust cremediated recombination in small intestinal stem cells utilizing the olfm4 locus. Stem Cell Reports 3, 234–241. 10.1016/j.stemcr.2014.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nio-Kobayashi J, Takahashi-Iwanaga H, and Iwanaga T (2009). Immunohistochemical localization of six galectin subtypes in the mouse digestive tract. J Histochem Cytochem 57, 41–50. 10.1369/jhc.2008.952317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, Yim K, Elzen AVD, Hirn MJ, Coifman RR, Ivanova NB, Wolf G, and Krishnaswamy S (2019). Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol 37, 1482–1492. 10.1038/s41587-019-0336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S, Fink J, Jörg DJ, Lee E, Yum MK, Chatzeli L, Merker SR, Josserand M, Trendafilova T, Andersson-Rolf A, Dabrowska C, Kim H, Naumann R, Lee JH, Sasaki N, Mort RL, Basak O, Clevers H, Stange DE, Philpott A, Kim JK, Simons BD, and Koo BK (2019). Defining the Identity and Dynamics of Adult Gastric Isthmus Stem Cells. Cell Stem Cell 25, 342–356.e7. 10.1016/j.stem.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuo J, Douchi D, Myint K, Mon NN, Yamamura A, Kohu K, Heng DL, Chen S, Mawan NA, Nuttonmanit N, Li Y, Srivastava S, Ho SWT, Lee NYS, Lee HK, Adachi M, Tamura A, Chen J, Yang H, Teh M, So JB, Yong WP, Tan P, Yeoh KG, Chuang LSH, Tsukita S, and Ito Y (2021). Iqgap3-Ras axis drives stem cell proliferation in the stomach corpus during homoeostasis and repair. Gut 70, 1833–1846. 10.1136/gutjnl-2020-322779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Garcia C, Klein AM, Simons BD, and Winton DJ (2010). Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825. 10.1126/science.1196236 [DOI] [PubMed] [Google Scholar]
- 33.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, and Clevers H (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 34.Page ME, Lombard P, Ng F, Göttgens B, and Jensen KB (2013). The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13, 471–482. 10.1016/j.stem.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jong IEM, Matton APM, van Praagh JB, van Haaften WT, Wiersema Buist J, van Wijk LA, Oosterhuis D, Iswandana R, Suriguga S, and Overi D (2019). Peribiliary glands are key in regeneration of the human biliary epithelium after severe bile duct injury. Hepatology 69, 1719–1734. 10.1002/hep.30365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okumura M, Yamanoi K, Uehara T, and Nakayama J (2020). Decreased alpha 1,4-linked N-acetylglucosamine glycosylation in biliary tract cancer progression from biliary intraepithelial neoplasia to invasive adenocarcinoma. Cancer Sci 111, 4629–4635. 10.1111/cas.14677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, and Bielas JH (2017). Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lun AT, Bach K, and Marioni JC (2016). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 17, 75. 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, and Clement L (2020). Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 11, 1201. 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn S, and Joyner AL (2004). Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516. 10.1016/j.cell.2004.07.023 [DOI] [PubMed] [Google Scholar]
- 44.Harada N, Tamai Y, Ishikawa T. o., Sauer B, Takaku K, Oshima M, and Taketo MM (1999). Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. The EMBO journal 18, 5931–5942. 10.1093/emboj/18.21.5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basak O, Krieger TG, Muraro MJ, Wiebrands K, Stange DE, Frias-Aldeguer J, Rivron NC, van de Wetering M, van Es JH, and van Oudenaarden A (2018). Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proceedings of the National Academy of Sciences 115, E610–E619. 10.1073/pnas.1715911114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, and Jacks T (2007). Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665. 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- 47.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, and Zeng H (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badea TC, Wang Y, and Nathans J (2003). A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. Journal of Neuroscience 23, 2314–2322. 10.1523/jneurosci.23-06-02314.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta V, Gupta I, Park J, Bram Y, and Schwartz RE (2020). Hedgehog Signaling Demarcates a Niche of Fibrogenic Peribiliary Mesenchymal Cells. Gastroenterology 159, 624–638.e9. 10.1053/j.gastro.2020.03.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Germain RN, and Gerner MY (2019). High-dimensional cell-level analysis of tissues with Ce3D multiplex volume imaging. Nat Protoc 14, 1708–1733. 10.1038/s41596-019-0156-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salic A, and Mitchison TJ (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105, 2415–2420. 10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Z-stack of gland (dashed outline) after refeeding in which the entire gland is positive for EdU (Red), related to Figure 4.
Video S2. Z-stack of a gland (dashed outline) after refeeding in which 1 alveolus of a trialveolar gland is positive for EdU (Red), related to Figure 4.
Video S3. Z-stack of a gland (dashed outline) after refeeding in which there are EdU+ cells at the base and EdU+ cells at the pit with a clear break in a gland (dashed outline), related to Figure 4.
Video S4. Z-stack of a gland 90 days post tamoxifen from a Olfm4:CreERT2; Rosa26:Confetti animal in which a yellow fragmented clone is seen at the base of 3 different alveoli and a red clone anchored at the pit is seen expanding onto the surface epithelium, related to Figure 6.
Video S5. Z-stack of an gland 180 days post tamoxifen from a Muc6:CreERT2; Rosa26:Confetti animal in which a red clone can be seen extending from the base of one alveoli and into the body of the gland, related to Figure 6.
Data Availability Statement
Sequencing data generated can be downloaded from the Gene Expression Omnibus under accession number GSE223099. All other data are available from the corresponding author upon reasonable request. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


