Small RNA molecules play major roles in a number of cellular functions. Depending on their predominant cellular location, small RNAs have been classified as nuclear (sn), nucleolar (sno) or cytoplasmic (sc). The nucleoplasmic species include the spliceosomal snRNAs (U1, U2, U4, U5 and U6) and the U7 snRNA required for histone pre-mRNA 3′-end formation. The snoRNAs are key players in the processing of ribosomal RNA (rRNA), and scRNAs function in protein synthesis. Most small RNAs have a complex life cycle, being synthesized as precursors that must undergo a series of processing events in order to become functionally mature. Many of these post-transcriptional modifications are ‘guided’ by other small RNAs and occur in specific cellular compartments such as the nucleolus (Filipowicz and Pogacic, 2002). Two related papers in The EMBO Journal from the groups of Tamás Kiss and Edouard Bertrand now highlight a role for Cajal bodies in the post-transcriptional modification of spliceosomal snRNAs and snoRNAs. Kiss and co-workers report a new family of small RNAs that localize specifically to Cajal bodies and are involved in the maturation of spliceosomal snRNAs (Darzacq et al., 2002), and Bertrand and co-workers show that the final processing steps involved in the biogenesis of U3 snoRNA take place in Cajal bodies (Verheggen et al., 2002).
Despite being identified around 100 years ago by the neurocytologist Ramon y Cajal, the function of Cajal bodies has so far proved elusive. When viewed with the electron microscope, these subnuclear ‘organelles’ appear to consist of a tangle of coiled threads and were hence also referred to as coiled bodies. Since they contain U7 snRNA and components of the three RNA polymerase transcription complexes, they have been proposed to be involved in the assembly of the transcriptional machinery and U7-dependent histone mRNA 3′-end processing (Gall, 2000). Furthermore, Cajal bodies are highly enriched in spliceosomal snRNAs, which contain many 2′-O-ribose-methylated nucleotides and pseudouridines (Reddy and Busch, 1988). Kiss and co-workers have now discovered a novel family of Cajal-body-specific small RNAs (termed scaRNAs) that have the potential to base-pair with the U snRNAs and are likely to function as guides for the site-specific 2′-O-ribose methylation and pseudouridine formation in U1, U2, U4 and U5 snRNAs (Darzacq et al., 2002; Figure 1A). Two of these authors have recently described an unusual small RNA, the U85 RNA (Jády and Kiss, 2001); now, in their latest paper, the group report three novel U85-related RNAs, called U87, U88 and U89. What makes these small RNAs unusual is the simultaneous presence of short sequence elements termed box C/D and box H/ACA domains in the same molecule. Boxes C and D were initially described in snoRNAs that guide ribose methylation in rRNA, whereas conserved H and ACA boxes were identified in a separate group of snoRNAs that act as guides for pseudouridylation in rRNA (Filipowicz and Pogacic, 2002). These latest novel RNAs are therefore predicted to function both in 2′-O methylation and pseudouridylation of the U5 and U4 snRNAs. Darzacq et al. (2002) also identify U90, U91 and U92 as three further novel RNAs that localize to Cajal bodies. The U90 and U91 RNAs contain the conserved C and D boxes (but not box H/ACA) and may guide methylation of the A70 and C8 residues in the U1 and U4 snRNAs, respectively. The U92 RNA lacks C and D boxes but contains box H/ACA and is, in theory, capable of directing pseudouridylation of residue U44 in the U2 snRNA (Table I).
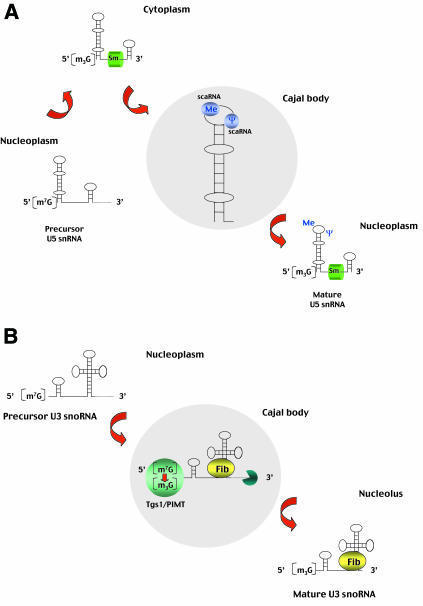
Fig. 1. (A) Processing of spliceosomal U snRNAs. The U1, U2, U4 and U5 spliceosomal snRNAs are synthesized in the nucleoplasm by RNA polymerase II as immature precursors containing a 5′-terminal 7-mono-methylguanosine cap and extra nucleotides at the 3′ end (for simplicity, only the U5 snRNA is depicted). These RNAs are rapidly exported to the cytoplasm, where they bind to Sm proteins. The association of Sm proteins allows subsequent hyper-methylation of the 5′-cap and 3′-end maturation of the snRNA. The newly assembled snRNPs (small nuclear ribonucleoprotein particles) are then transported back into the nucleus, where the snRNAs further undergo 2′-O-ribose methylation (Me) and pseudouridine formation (Ψ). The new data reported by Darzacq et al. (2002) suggest that these modifications are guided by the U85, U87, U88, U89, U90, U91 and U92 small RNAs located in the Cajal body (scaRNAs). Modified nucleotides are confined to the functionally important regions of snRNAs. (B) Processing of the U3 snoRNA. The U3 snoRNA is synthesized in the nucleoplasm by RNA polymerase II as a 3′-extended precursor with a mono-methylated 5′ cap. Cap tri-methylation is catalysed by the methyl-transferase Tgs1/PIMT, which localizes to the Cajal body as shown by Verheggen et al. (2002). Following 3′-end processing, which also takes place within the Cajal body, the U3 RNA binds fibrillarin (Fib) and other core snoRNP proteins. The mature U3 snoRNPs leave the Cajal body to accumulate in the nucleolus.
Table I. Classes of small Cajal body (sca) RNAs.
| scaRNAs | Function | Conserved motifs | Common proteins |
|---|---|---|---|
| U85, U87, U88, U89 | Specify site of both ribose methylation and pseudouridine formation in U5 and U4 snRNAs | Box C/D, Box H/ACA | Fibrillarin, Gar1p |
| U90, U91 | Specify site of ribose methylation in U1 and U4 snRNAs | Box C/D | Fibrillarin |
| U92 | Specify site of pseudouridine formation in U2 snRNA | Box H/ACA | Gar1p |
Among other small RNAs containing box C/D motifs, U3 is one of the most abundant species. It is required for pre-rRNA cleavage during ribosome biogenesis and, at steady state, is predominantly detected in the nucleolus. However, U3 and other box C/D snoRNAs transiently associate with Cajal bodies before they localize to nucleoli (Narayanan et al., 1999), and the most recent data from Bertrand and co-workers suggest that U3 matures while passing through Cajal bodies (Verheggen et al., 2002). This detailed dissection of the U3 maturation pathway shows that immature precursor species are present at transcription sites and in Cajal bodies, but not in nucleoli. In contrast, mature U3 RNAs are present both in Cajal bodies and in nucleoli. Furthermore, the nuclear methyl-transferase required for converting the 5′-terminal 7-mono-methylguanosine cap of precursor U3 snoRNA into the tri-methyl cap of the mature form (Mouaikel et al., 2002) localizes specifically to the vertebrate Cajal body and to a corresponding structure of the yeast nucleus, the nucleolar body. As cap hyper-methylation is one of the final steps in the processing of U3, the data suggest that U3 snoRNA completes its maturation in the Cajal body (Figure 1B).
Cajal bodies were first proposed to represent a maturation station for small RNAs following the observation that spliceosomal snRNAs and U3 snoRNA accumulate in these subnuclear structures before moving on to their final destination in the nucleoplasm or in the nucleolus, respectively (Narayanan et al., 1999; Sleeman and Lamond, 1999). The groups of Kiss and Bertrand have now provided evidence that spliceosomal snRNAs transcribed by RNA polymerase II are indeed methylated and pseudouridylated in the Cajal body (Darzacq et al., 2002) and that this is also the site for 3′-end processing and cap hyper-methylation of the U3 snoRNA (Verheggen et al., 2002). What remains unknown is how scaRNAs and U3 snoRNA processing enzymes are targeted to the Cajal body. Are they recruited by the accumulation of cognate substrates, or are they themselves responsible for the recruitment of small RNA substrates? In other words, what is the origin of Cajal bodies? Continued efforts to identify the activities of new molecular components of the Cajal body and to determine their kinetic behaviour in living cells are likely to contribute further clues to this persistent enigma of nuclear organization.

Maria Carmo-Fonseca
References
- Darzacq X., Jády, B.E., Verheggen, C., Kiss, A.M., Bertrand, E. and Kiss, T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. and Pogacic, V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Jády B.E. and Kiss, T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen, C., Bertrand, E., Tazi, J. and Bordonne, R. (2002) Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell, 9, 891–901. [DOI] [PubMed] [Google Scholar]
- Narayanan A., Speckmann, W., Terns, R. and Terns, M. (1999) Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R. and Busch, H. (1988) Small nuclear RNAs: RNA sequences, structure, and modifications. In Birnstiel, M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer, Berlin, Germany, pp. 1–37.
- Sleeman J. and Lamond, A.I. (1999) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine, D.L.J., Samarsky, D., Mouaikel, J., Blanchard, J.-M., Bordonné, R. and Bertrand, E. (2002) Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J., 21, 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]