Abstract
Stimulation of p21-activated kinase-1 (Pak1) signaling promotes motility, invasiveness, anchorage-independent growth and abnormal mitotic assembly in human breast cancer cells. Here, we provide new evidence that, before the onset of mitosis, activated Pak1 is specifically localized with the chromosomes during prophase and on the centrosomes in metaphase and moves to the contraction ring during cytokinesis. To identify mitosis-specific substrates of Pak1, we screened a synchronized G2–M expression library by using a glutathione transferase Pak1 solid-phase-based kinase reaction. This analysis identified histone H3 as a substrate of Pak1 both in vitro and in vivo, and it specifically interacted with Pak1 but not Pak2 or Pak3. Site-directed mutagenesis indicated that Pak1 phosphorylates histone H3 on Ser10. Expressions of the wild-type, or catalytically active, Pak1 caused it to appear at the poles corresponding to mitotic centrosomes in a variety of mammalian cells. Together, these results suggest for the first time that Pak1 interacts with and phosphorylates histone H3 and may thus influence the Pak1–histone H3 pathway, which in turn may influence mitotic events in breast cancer cells.
INTRODUCTION
The formation and functions of motile structures in mammalian cells are regulated by the small GTPases, including Cdc42 and Rac1 (Manser et al., 1997; Daniels and Bokoch, 1999). In mammalian cells, p21-activated kinases (Paks) are one of the targets of Cdc42 and Rac1, and binding of GTP-bound GTPases to Pak stimulates its kinase activity via autophosphorylation (Manser et al., 1997). Activation of Pak1 by diverse signals leads to autophosphorylation at several sites, including Thr423 within the activation loop of the kinase (Sells et al., 1997). Pak1 phosphorylation at Thr423 has been linked with its activation, since substitution of the acidic residue glutamic acid at this site produces a constitutively active T423E Pak1 mutant enzyme (Sells et al., 2000). We recently generated a human breast cancer cell line expressing T423E Pak1 and showed a close linkage between the expression of T423E Pak1 and an abnormal formation of mitotic spindles (Vadlamudi et al., 2000), which suggests that Pak1 plays a role in mitosis and the formation of chromosomal abnormalities.
Chromosomal and cytoskeleton-linked functions during mitosis are regulated by a class of proteins termed chromosomal passengers (Earnshaw and Bernat, 1991). These proteins, such as the inner centromere protein (INCENP), the Aurora-B, the polo-like kinases and survivin, associate with chromosomes along their length during prophase and then concentrate at the inner centromere and later by metaphase move to the centrosome. At anaphase, the passengers abruptly move to the center of the mitotic spindle and the cell cortex, where the contractile ring originates (Adams et al., 2001).
The eukaryotic genome is compacted with histone and other proteins to form chromatin, which consists of repeating units of nucleosomes (Wolffe and Kurumizaka, 1998). The chromatin structure plays an important role in gene regulation and is a point where signaling pathways converge (Cheung et al., 2000). The eukaryotic chromosomes contain five major classes of histones: H1, H2A, H2B, H3 and H4. H3 histones are divided into three subtypes: H3.1, H3.2 and H3.3. H3.3 is encoded by two distinct genes: H3.3A and H3.3B, which code identical proteins and are structurally similar to histones H3.1 and H3.2 except for the addition of the first initiation codon, methionine, and three amino acid substitutions.
Cellular signaling pathways modify histones by a number of methods, including phosphorylation of the histone tails, thereby controlling gene expression (Cheung et al., 2000; Jenuwein and Allis, 2001). In addition, histone H3 Ser10 phosphorylation, which has been linked with the functions of chromosomal passengers, regulates mitotic chromosome assembly, congregation at the metaphase plate and segregation (Earnshaw and Bernat, 1991; de la Barre et al., 2000; Adams et al., 2001).
Despite the well-known role of histones in chromatin structure and function, very little is known about their role in the Pak1 pathway of mammalian cells. In the experiments described below, we provide new evidence that Pak1, besides playing an important role in the mitosis of eukaryotic cells, phosphorylates histone H3.
RESULTS AND DISCUSSION
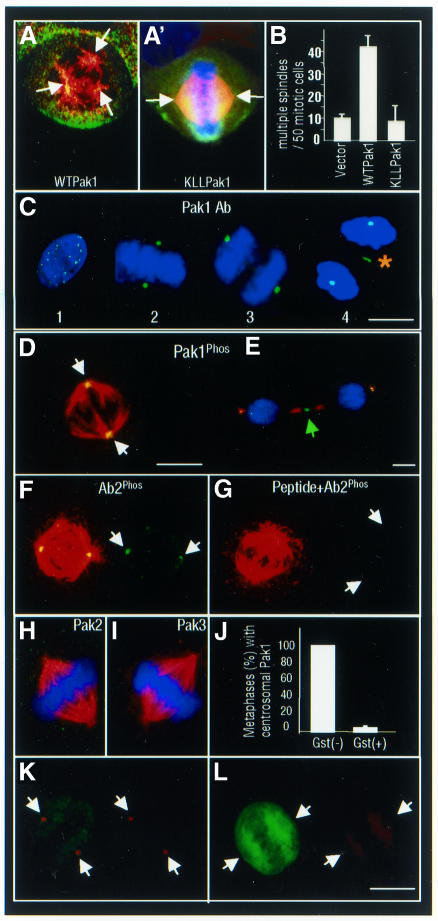
Deregulation of Pak1 activation promotes the formation of abnormal mitotic spindles in human breast cancer cells (Vadlamudi et al., 2000). To understand the potential role of activated Pak1 in mitotic spindle formation, we first determined the location of activated Pak1 in mitotic MCF-7 breast cancer cells transiently transfected with myc-tagged Pak1. About 40–50% of Pak1-transfected mitotic cells but <10% of the mock-transfected cells had three or more spindle poles (Figure 1A and B). myc-Pak1 was localized on and around the spindles pole in the centrosomal region (Figure 1A’).
Fig. 1. Subcellular localization of Pak1 during mitosis. (A and A¢) Merged confocal image of α-tubulin (red) and myc-tagged wild-type (WT) Pak1 or myc-tagged kinase-dead K299LL (KLL) Pak1 (green) overexpression in MCF-7 cells. (B) Quantification of the multiple spindle poles induced by myc-tagged WT or KLL Pak1 expression in MCF-7 cells. (C) Interphase (1), metaphase (2), anaphase (3) and telophase (4) of NIH 3T3 cells showing intranuclear, centrosomal and midbody (asterisk) localization of Pak1 (green). (D) Centrosome (white arrows) localization of phosphorylated Pak1 in mitotic MDA-MB231 cells. (E) Midbody (green arrow) localization of phosphorylated Pak during cytokinesis. (F and G) Pak1Phos at the spindle poles (white arrows) of metaphase MDA-MB231 cells by another anti-Pak1Phos antibody (Ab2) in the absence or presence of a competing peptide used to generate this antibody. (H and I) Absence of Pak2 or Pak3 on centrosomes in mitotic MDA-MB231 cells. (J) Centrosomal Pak1 localization requires Pak1 activity. MCF-7 cells were transfected with GST–Pak1 inhibitor (amino acids 83–149). Quantification of centrosomal Pak1 expression in the GST-positive (GST+) and GST-negative (GST–) metaphase cells. (K) Example of mitotic (GST–) cell with centrosomal Pak1. (L) Example of mitotic (GST+) cell with no centrosomal Pak1. Scale bars = 4 µm.
Endogenous Pak1 was localized inside the nucleus in 18–24% of interphase cells (Figure 1C, panel 1), on the centrosomes and/or metaphase plate in 100% of the metaphase-to-anaphase cells (Figure 1C, panels 2 and 3) and on the midbody of 85% of the telophase cells (Figure 1C, panel 4). This cell-cycle-related Pak1 translocation was observed in several eukaryotic cell lines, as well as in NIH 3T3 murine fibroblasts (Figure 1C), in MCF-7 (Figure 1F and G), in MDA-MB231 (Figure 1D and E) and in ZR-75R breast cancer cells (data not shown).
To examine the activation status of Pak1 in mitotic cells, we next determined the distribution of endogenous activated Pak1 in mitotic cells using an anti-Pak1Phos antibody that recognizes phosphorylated Thr423 Pak1 (Sells et al., 2000). Pak1Phos staining had a strong, punctated pattern at the mitotic spindle poles (Figure 1D, white arrows) and at the midbody (Figure 1E, green arrow). Centrosomal Pak1 reactivity was reproduced by another Pak1 antibody, Ab2 (from Cell Signaling), which was raised against the phosphorylated Thr423 site (Figure 1F); it specifically competed with the immunizing peptide (Figure 1G). Centrosomal Pak1Phos was specific for Pak1, as neither Pak2 (Figure 1H) nor Pak3 (Figure 1I) localized on the centrosome, as determined by specific antibodies. Furthermore, Pak1 translocation to the centrosomes depended on its kinase activity, as this localization (Figure 1K) was blocked by the expression of a glutathione S-transferase (GST)-tagged Pak1 inhibitor (amino acids 83–149) (Figure 1L and J). Similar experiments using the dominant-negative Cdc42 construct failed to block Pak1 centrosomal localization (data not shown).
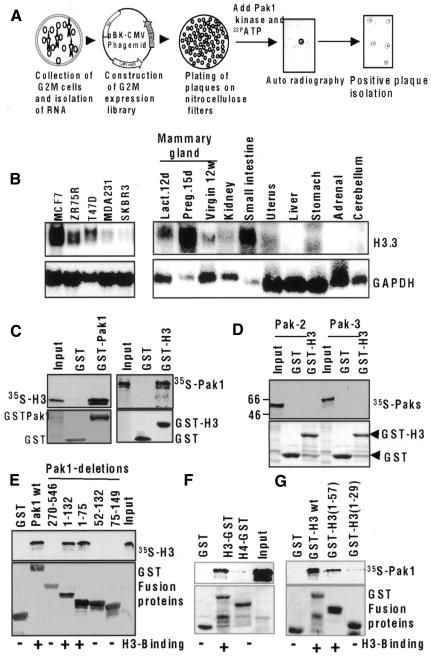
The newly observed localization of activated Pak1 raised the possibility that Pak1 phosphorylates mitosis-specific substrates. To identify such substrates, we screened a synchronized G2–M expression library from HeLa cells using a GST–Pak1 solid-phase-based kinase reaction (Figure 2A). After the third screening, several clones were identified, and isolates from one of the clones were 100% identical to histone H3.3A (DDBJ/EMBL/GenBank accession No. BC001124), which is structurally similar to histone H3.1 and H3.2 except for the addition of the first initiation codon, methionine, and three amino acids substitutions. Histone H3.3 was strongly expressed in the human breast cancer cells and in a number of murine tissues and was at its highest level in mammary gland during pregnancy day 15 (Figure 2B). Since the N-terminal regions of histone H3 isoforms are highly conserved and are similar to those of the histone H3.1 and histone H3.2 isoforms, H3.3 may not retain its first methionine; therefore, we will refer to the H3.3 isoform of histone as H3 in this report.
Fig. 2. Identification of histone H3.3A as a Pak1 substrate. (A) G2–M expression library screening by Pak1. (B) mRNA expression of H3.3A in breast cancer cell lines and murine tissues. (C) Direct interaction of Pak1 with histone H3 in a GST pull-down assay. In vitro-translated H3 bound to GST–Pak1 (left panel), and in vitro-translated Pak1 bound to GST–H3 (right panel). (D) Non-interaction of H3 with Pak 2 and Pak3. cDNAs were translated in vitro, incubated with GST–H3 and analyzed by a GST pull-down assay. (E) Interaction of Pak1 domains with histone H3. Pak1 GST-fusion proteins containing full-length Pak1 amino acids (aa 1–546), the kinase domain (aa 270–546), the N-terminal domain (aa 1–132), the Nck-binding domain (aa 1–75), the Cdc42/Rac-interacting domain (aa 52–132) and the autoinhibitory domain (aa 75–149) were all incubated with in vitro-translated H3, and binding was analyzed by a GST pull-down assay. (F) Pak1 specifically interacts with H3 but not H4 in TNT pull-down assays. GST pull-down assay using 35S-labeled Pak1 and GST fusion of H3 or H4. (G) Pak1 interacts with H3 tail. GST pull-down assay using 35S-labeled Pak1 and GST fusions of H3 1–57 or H3 1–29.
To determine whether histone H3 interacts with Pak1, we next examined the ability of an in vitro-translated Pak1 protein to bind with the GST-tagged histone H3. As shown in Figure 2C, Pak1 efficiently interacted with GST–H3 but not with GST alone in GST pull-down assays. Conversely, an in vitro-translated histone H3 protein also specifically interacted with GST–Pak1. To evaluate the specificity of the histone H3–Pak1 interaction, we examined whether histone H3 could interact with the Pak2 and Pak3 proteins. It did not interact with Pak2 or Pak3 (Figure 2D). To examine the H3 binding domain of Pak1, we used a series of GST-fusion proteins expressing Pak1 domains (Figure 2E). Histone H3 did not interact with the kinase domain, the Cdc42/Rac1-interacting domain or the Pak1 autoinhibitory domain. However, N-terminal region fusion proteins containing amino acids 1–132 and 1–75 bound strongly to histone H3 (Figure 2E), indicating that a histone H3 binding site was located in the N-terminal 75 amino acids, a region involved in Nck binding to Pak1 (Lu et al., 1997). The noticed histone H3 interaction with Pak1 was specific, as Pak1 failed to interact with histone H4 (Figure 2F). The Pak1 binding to histone H3 was localized within the histone H3 tail region amino acids 1–57 (Figure 2G).
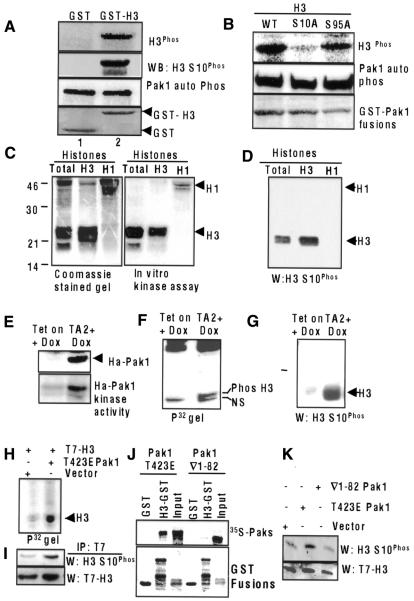
To determine whether histone H3 was a substrate of Pak1, we performed in vitro kinase assays using a purified Pak1 enzyme and GST–H3. As shown in Figure 3A (upper panel), recombinant Pak1 efficiently phosphorylated histone H3. We next explored whether a Pak1-phosphorylated histone H3 could be recognized by a well-characterized anti-H3 Ser10Phos antibody (Zeitlin et al., 2001). Interestingly, Pak1-mediated phosphorylation of histone H3 was readily recognized by the anti-H3 Ser10Phos antibody (Figure 3A, second panel), raising the possibility of a close relationship between Pak1 and histone H3 phosphorylation. Lack of phosphorylation in lane 1 was not due to lack of Pak1 activity, since Pak1 autophosphorylation status was similar in both lanes (Figure 3A, third panel). The mutation of histone H3 Ser10 to alanine (S10A) completely blocked the ability of Pak1 to phosphorylate H3, whereas the mutation of histone H3 Ser95 to alanine (S95A) had no effect (Figure 3B), confirming that Ser10 of H3 is the Pak1 phosphorylation site. Accordingly, the recombinant Pak1 enzyme (Figure 3C) efficiently phosphorylated both native histone and highly purified histone H3 but not histone H1. The anti-H3 Ser10Phos antibody also readily recognized the Pak1-mediated phosphorylation of purified histone H3 on Ser10 (Figure 3D).
Fig. 3. Histone H3 is a substrate of Pak1. (A) Pak1 phosphorylation of histone H3. Phosphorylation of GST–H3 by purified Pak1 (upper panel). Phosphorylated GST–H3 was immunoblotted with an anti-H3 Ser10Phos antibody. Autophosphorylated Pak1 and GST–H3 are shown as controls (lower panels). (B) Ser10 of H3 is the Pak1 phosphorylation site. In vitro kinase assay of mutated histone H3 (H3.3 S10A and S95A) GST-fusion proteins by Pak1 enzyme. (C) Pak1 phosphorylation of native and purified histones in vitro. (D) Purified native histones H3 and H1 were phosphorylated in vitro using a Pak1 enzyme, blotted with an anti-H3 Ser10Phos antibody. (E) Tet vector or Ha-T423E Pak1 vector stable MCF-7 cells were treated with doxycyclin (1 µg/ml) for 24 h to induce expression of Pak1. Expression of Ha-T423EPak1 was analyzed by western blotting (upper panel) and its activity was analyzed by in vitro kinase assay (lower panel). (F) Ser10 phosphorylation of the endogenous histone H3 in T423E Pak1 (TA2) expressing cells as determined by in vivo labeling with [32P]orthophosphoric acid. Histones were isolated and phosphorylation of H3 was analyzed by autoradiography. Histone H3 (upper band). NS, non-specific band present in both lanes. (G) Ser10 phosphorylation of H3 in T423E expressing TA2 cells as shown by blotting with an anti-H3 Ser10Phos antibody. (H) T423E Pak1 stimulates phosphorylation of T7-H3. MCF-7 cells cotransfected with T7-H3 and T423E Pak1 or vectors were labeled with 32P-labeled orthophosphoric acid, and cell lysates were immunoprecipitated with a T7 antibody. Phosphorylated histone H3 is shown. (I) The same blot was reprobed with anti-H3 Ser10Phos and T7 antibodies. (J) H3 fails to bind ∇1–82 Pak1 which lacks N-terminal 82 aa. T423E Pak1 or ∇1–82 Pak1 was translated in vitro and GST pull-down assays were performed with GST–H3. (K) Pak1 mutant lacking H3 binding region fails to phosphorylate H3 on H3 Ser10Phos. MCF-7 cells were cotransfected with T423E Pak1 or ∇1–82 Pak1 along with T7-H3. The status of H3 Ser10Phos on T7-H3 was analyzed by western blotting.
To demonstrate that Pak1 phosphorylates histone H3 in vivo, we examined the regulation of endogenous histone H3 phosphorylation in MCF-7 cells, which express the catalytically active mutant Ha-tagged T423E Pak1 in the presence of doxycyclin (TA2 clones; Vadlamudi et al., 2000) (Figure 3E). The results of both in vivo labeling of cells with 32P-labeled orthophosphoric acid (Figure 3F, upper band; NS, non-specific band present in both lanes) and western blotting with the anti-H3 Ser10Phos antibody (Figure 3G) demonstrated that overexpression of catalytically active Pak1 was accompanied by increased H3 phosphorylation on Ser10. To further confirm that H3 was indeed phosphorylated in vivo by Pak1, we transfected MCF-7 cells with the T7-tagged histone H3 and myc-tagged T423E Pak1. Immunoprecipitation of histone H3 with an anti-T7 monoclonal antibody showed that histone H3 was indeed phosphorylated in cells expressing T423E Pak1 but not in vector-transfected cells (Figure 3H). The anti-H3 Ser10Phos antibody also recognized this phosphorylation of T7-H3 (Figure 3I). Expression of a Pak1 construct lacking the histone H3 binding region (∇1–82 Pak1) into MCF-7 cells failed to bind histone H3 (Figure 3J), as well as failing to induce histone H3 phosphorylation (Figure 3K), suggesting a close relationship between Pak1 interaction with histone H3 and its phosphorylation by Pak1. Together, these results suggest that Pak1 phosphorylates histone H3 and that histone H3 is a substrate of Pak1 that interacts with Pak1 and may influence its functioning in mitotic cells.
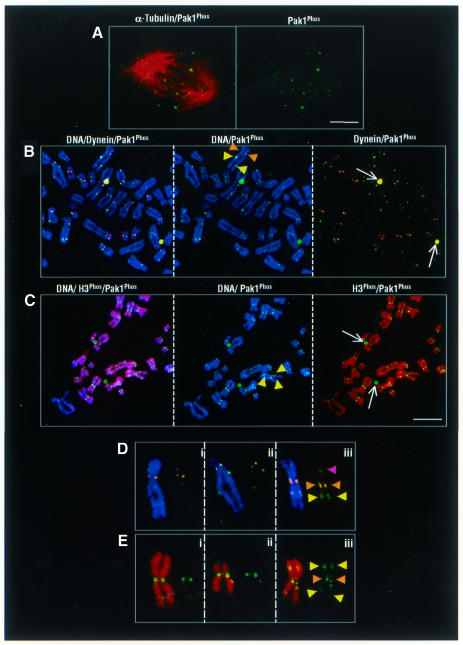
We observed a punctated pattern of Pak1 localization in the nucleus (Figure 1C, panel 1), which suggested that activated Pak1 might be associated with the kinetochore complex/centromeric region of mitotic chromosomes during prophase. To support such a possibility, we show the presence of the Pak1 protein at the metaphase plates (20–30%), as shown in a representative tangentially ‘cut’ confocal section of a metaphase plate with a high Pak1 immunoreactivity (Figure 4A). A similar pattern has been reported for another component of the kinetochore component dynein (Echeverri et al., 1996). To explore whether the microtubules and Pak1 compete for the kinetochore in MDA-MB231 cells, we next eliminated the possible contribution of microtubules using a tubulin-depolymerizing agent (colcemid) and immunostaining the resulting chromosomal spread preparations for either histone H3 Ser10Phos or dynein (Steuer et al., 1990) and activated Pak1. Representative examples of these studies, shown in Figure 4B (brown arrowheads), indicated the presence of centromeric Pak1 colocalized with the dynein intermediary chain. These findings suggested the possibility that Pak1 leaves the kinetochores after the microtubules attach. It is possible that under normal circumstances an abundance of Pak1/dynein complex at the kinetochores may help capture the microtubules and drive the chromosome movement, after which Pak1 leaves the kinetochore as part of a putative complex. Examples of individual chromosomes with different patterns are shown in Figure 4D. Some displayed either an occasionally extracentromeric parallel localization of the activated Pak1 (Figure 4D, yellow arrowheads) or non-parallel pattern (Figure 4D, purple arrowhead). The extracentromeric Pak1 was colocalized with the abundantly phosphorylated histone H3 Ser10Phos on the chromosome arms (Figure 4C, yellow arrowheads). No colocalization of Pak1Phos and H3 Ser10Phos was detected on the two centrosomes (Figure 4C, white arrows) or the centromeres (Figure 4E, brown arrowheads). These results suggest that Pak1 may be a transitory component of kinetochores in mammalian cells and that its association with the kinetochore was sensitive to the presence of the microtubules. Pak1 extracentromeric localization, as well as H3 binding and specific ability to phosphorylate histone H3 on Ser10, may identify novel functions of Pak1 during mitosis.
Fig. 4. Chromosomal localization of activated Pak1. (A) Metaphase plate localization of Pak1. Confocal micrographs of a single tangentially cut plane through a metaphase cell. Pak1Phos alone (right panel) and merged image with α-tubulin (left panel). (B) Centromeric and extracentromeric patterns of chromosome-bound activated Pak1, as revealed by three-color confocal microscopy of chromosome spreads obtained from colcemid-treated MDA-MB231 cells. Centromeric colocalization of dynein (red) and activated Pak1 (green) (brown arrowheads); extracentromeric localization of Pak1Phos (yellow arrowsheads). Dynein and Pak1 colocalization on centrosomes (white arrows) and in the centromeric region (brown arrowheads). (C) Centromeric and extracentromeric localization of activated Pak1 on chromosome spreads from MDA-MB231 cells. Activated Pak1 associated with condensed chromosomes containing histone H3 Ser10Phos. Histone H3 Ser10Phos (red), activated Pak1 (green); Topro-3 DNA staining (blue). Representative spreads are shown in merged colors in the left panel and in two colors in the middle and right panels. Note the activated Pak1 on both the centrosomes and chromosomes (white arrows and yellow arrowheads, respectively). Scale bar = 4 µm. (D) Extracentromeric localization of activated Pak1 immunoreactivity without dynein labeling (yellow arrowheads); Pak1 and dynein colocalization in the centromeric region (brown arrowheads). Unilateral Pak1 localization on the chromosome arm (purple arrowhead). (E) Representative examples of individual chromosomes in which activated Pak1 colocalized with histone H3 Ser10Phos in the extracentromeric (yellow arrowheads) regions.
In summary, our results show a putative chromosomal function of Pak1. We have shown that, before the onset of mitosis, Pak1 becomes activated and translocates to the nucleus. From this point on, Pak1 behaves like a chromosomal passenger protein. Results presented here raise new questions about the functions of Pak1 during mitosis, including its potential involvement in the condensation, capture and/or movement of chromosomes during mitosis, all of which are required for proper chromosomal segregation. Furthermore, in addition to Pak1’s potential role in chromatin rearrangement at the chromosomal level, this process may also be influenced by the potential substrates that participate in microtubule dynamics. Because histone H3 phosphorylation has been shown to be required for the initiation of chromosome condensation (Cheung et al., 2000) and cell-cycle progression, our present findings suggest that Pak1, through histone H3 phosphorylation, may play a role in regulating mitotic events.
METHODS
Cell cultures and reagents. MCF-7 breast cancer cells were maintained in DMEM-F12 (1:1, v/v) supplemented with 10% fetal calf serum. Antibodies against Pak1, Pak2 and Pak3 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Zymed Laboratories (San Francisco, CA). Antibodies against T7 tag and histone H3 Ser10Phos were purchased from Novagen (Madison, WI) and United Biochemical Inc. (Hauppage, NY). Antibodies against phospho-T423E Pak1 (Sells et al., 2000) were purchased from Cell Signaling (Beverly, MA).
Screening of G2–M expression library. HeLa cells were synchronized with a double thymidine block and release protocol (Kumar et al., 1994). The cDNA expression library was established using the ZAP Express cDNA Synthesis Kit and the ZAP Express cDNA Gigapack III Gold Cloning Kit (Stratagene, La Jolla, CA), as described previously (Fukunaga and Hunter, 1997).
Plasmid construction. H3 expression vectors were constructed by amplifying the coding region of H3.3 of the positive clone obtained from expression library screening, by PCR amplification and by subcloning into the EcoRI and XhoI sites of pcDNA3.1 (T7-H3) and the PGEX5 vector (GST–H3). Mutations of H3.3, S10A and S95A were created using the Stratagene Quik Change kit and pcDNA 3.1-H3 in the vector as a template. ∇1–82 Pak1 was constructed by PCR amplification of Pak1 coding region containing amino acids 83–546 and subcloning into pcDNA3.1 HIS vector (Invitrogen).
In vitro kinase and GST pull-down assays. In vitro kinase assays using either purified histones or (GST)–H3 protein were performed as described previously (Mazumdar et al., 2001).
Confocal studies and chromosome spreads. Confocal and chromosome spreading studies were performed as described previously (Saffery et al., 2000; Vadlamudi et al., 2000).
Acknowledgments
ACKNOWLEDGEMENTS
This work is supported by NIH grant CA80066, Cancer Center core grant CA16672, M.D. Anderson Cancer Therapeutic Discovery Program (to R.K.) and M.D. Anderson Intramural Research Grant (to L.A.).
REFERENCES
- Adams R.R., Carmena, M. and Earnshaw, W.C. (2001) Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol., 11, 49–54. [DOI] [PubMed] [Google Scholar]
- Cheung P., Allis, C.D. and Sassone, C. (2000) Signaling to chromatin through histone modifications. Cell, 103, 263–271. [DOI] [PubMed] [Google Scholar]
- Daniels R.H. and Bokoch, G.M. (1999) p21-activated protein kinase: a crucial component of morphological signaling? Trends Biochem. Sci., 24, 350–355. [DOI] [PubMed] [Google Scholar]
- de la Barre A.E., Gerson, V., Gout, S., Creaven, M., Allis, C.D. and Dimitrov, S. (2000) Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J., 19, 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C. and Bernat, R.L. (1991) Chromosomal passengers: toward an integrated view of mitosis. Chromosoma, 100, 139–146. [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal, B.M., Vaughan, K.T. and Vallee, R.B. (1996) Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol., 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R. and Hunter, T. (1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J., 16, 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis, C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kumar R., Korutla, L. and Zhang, K. (1994) Cell cycle-dependent modulation of α-interferon-inducible gene expression and activation of signaling components in Daudi cells. J. Biol. Chem., 269, 25437–25441. [PubMed] [Google Scholar]
- Lu W., Katz, S., Gupta, R. and Mayer, B.J. (1997) Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr. Biol., 7, 85–94. [DOI] [PubMed] [Google Scholar]
- Manser E., Huang, H.Y., Loo, T.H., Chen, X.Q., Dong, J.M., Leung, T. and Lim, L. (1997) Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol., 17, 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar A., Wang, R.A., Mishra, S.K, Adam, L., Bagheri-Yarmand, R., Mandal, M., Vadlamudi, R.K. and Kumar, R. (2001) Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat. Cell Biol., 3, 30–37. [DOI] [PubMed] [Google Scholar]
- Saffery R., Irvine, D.V., Griffiths, B., Kalitsis, P., Wordeman, L. and Choo, K.H. (2000) Components of the human spindle checkpoint control mechanism localize specifically to the active centromere on dicentric chromosomes. Hum. Mol. Genet., 9, 175–185. [DOI] [PubMed] [Google Scholar]
- Sells M.A., Knaus, U.G., Bagrodia, S., Ambrose, D.M., Bokoch, G.M. and Chernof, J. (1997) Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol., 7, 202–210. [DOI] [PubMed] [Google Scholar]
- Sells M.A., Pfaff, A. and Chernoff, J. (2000) Temporal and spatial distribution of activated Pak1 in fibroblasts. J. Cell Biol., 151, 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer E.R., Wordeman, L., Schroer, T.A. and Sheetz, M.P. (1990) Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature, 345, 266–268. [DOI] [PubMed] [Google Scholar]
- Vadlamudi R.K., Adam, L., Wang, R.A., Mandal, M., Nguyen, D., Sahin, A., Chernoff, J., Hung, M.C. and Kumar, R. (2000) Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J. Biol. Chem., 275, 36238–36240. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Kurumizaka, H. (1998) The nucleosome: a powerful regulator of transcription. Prog. Nucleic Acid Res. Mol. Biol., 61, 379–422. [DOI] [PubMed] [Google Scholar]
- Zeitlin S.G., Barber, C.M., Allis, C.D., Sullivan, K.F. and Sullivan, K. (2001) Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci., 114, 653–661. [DOI] [PubMed] [Google Scholar]