Abstract
Mini-chromosome maintenance (MCM) proteins form a conserved family found in all eukaryotes and are essential for DNA replication. They exist as heteromultimeric complexes containing as many as six different proteins. These complexes are believed to be the replicative helicases, functioning as hexameric rings at replication forks. In most archaea a single MCM protein exists. The protein from Methanobacterium thermoautotrophicum (mtMCM) has been reported to assemble into a large complex consistent with a dodecamer. We show that mtMCM can assemble into a heptameric ring. This ring contains a C-terminal helicase domain that can be fit with crystal structures of ring helicases and an N-terminal domain of unknown function. While the structure of the ring is very similar to that of hexameric replicative helicases such as bacteriophage T7 gp4, our results show that such ring structures may not be constrained to have only six subunits.
INTRODUCTION
DNA replication is fundamental to all organisms, and studies using prokaryotic and viral models have contributed greatly to our understanding of the mechanistic details. An important element involves processive replicative helicases that are initially assembled at replication origins (Dean et al., 1992). The replicative helicase is typically a complex of six identical subunits assembled into a ring-shaped structure. Strong evidence exists for hexameric helicases, such as bacteriophage T7 gp4 (Yu et al., 1996) and Escherichia coli DnaB (Kaplan, 2000), that unwinding involves the ring encircling and moving along one of the DNA strands. The prime candidate for the replicative helicase in eukaryotes is a complex of six non-identical but highly conserved subunits known as MCM 2-7 (Tye, 1999). Hexamers containing stoichiometric amounts of each of the six mini-chromosome maintenance (MCM) proteins can be readily purified as a globular complex from a number of eukaryotes (Adachi et al., 1997). Although genetic evidence strongly implicates the MCM hexamer as the replicative helicase that melts origin DNA and unwinds growing forks (Aparicio et al., 1997; Labib et al., 2000), the purified MCM hexamer failed to demonstrate such an activity in vitro (Ishimi, 1997). Interestingly, weak helicase activity can be reconstituted from a metastable hexamer containing only three of the six subunits (Ishimi, 1997; Lee and Hurwitz, 2001). To understand how the MCM helicase works, it is important to study the structure of the assembled helicase. However, in order to study the anatomy of a functional helicase, successful assembly of an active and stable helicase is necessary.
The best model for the eukaryotic MCM helicase may be the archaeal homolog, a 3′–5′ ATP-dependent helicase presumed to contain six identical subunits (Kelman et al., 1999; Chong et al., 2000; Shechter et al., 2000). The eukaryotic MCM helicase has no known bacterial homolog, but there is a single highly conserved progenitor homolog in all archaea studied (Tye, 2000). The Methanobacterium thermoautotrophicum MCM (mtMCM) protein is 34% identical and 51% similar in sequence to the MCM2-7 protein family with the strongest homology concentrated in three conserved motifs, two of which are the Walker A and B nucleotide binding sequences (Koonin, 1993). mtMCM, a single polypeptide of 75 kDa, self-assembles on purification into a large-molecular-weight complex that contains robust helicase activity.
RESULTS
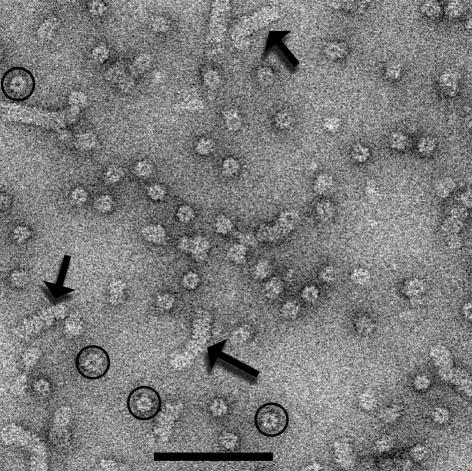
Electron micrographs of mtMCM display fields of abundant ring-like structures (Figure 1, circles). While most of the objects are consistent with projections of randomly-oriented rings, many larger aggregates are also seen (Figure 1, arrows). These larger structures are consistent with side views of ‘stacks’ of rings. An extensive analysis (see Supplementary data available at EMBO reports Online) involving 81 749 images of the single rings has shown that these single rings are heptameric. This analysis involved images of 55 925 rings from one preparation (with an N-terminal His tag), and 25 824 rings from a different construct (with a C-terminal His tag), prepared and purified in a different laboratory.
Fig. 1. An electron micrograph showing both single rings (circles) and larger aggregates (arrows) of the mtMCM protein. The scale bar is 1000 Å.
A reconstruction generated from 14 785 images (Figure 2) showed that the mtMCM ring was ∼100 Å high (along the symmetry axis) and ∼150 Å wide (perpendicular to the symmetry axis). Different reconstructions were generated by forcing the number of images to be comparable in each of the projection directions, or by first averaging the projections and then using these averages for back projection. The size of the central channel was found to be variable among these different reconstructions, while all other features remained relatively constant. Thus, no reliable conclusions can be drawn about the diameter of this central channel.
Fig. 2. Surfaces (A) and cross-sections (B) from the 3D reconstruction of mtMCM. The surface at the top is oriented so that the 7-fold axis is vertical, and the axial position of the sections (B) at –40, 0 and 40 Å are indicated. A very small detached density was present in the central channel, but was removed since simulations showed that it was consistent with noise. The heptamer is ∼100 Å in length along the 7-fold axis. The surface in the center corresponds to a ‘top’ view (showing the helicase domain, Figure 4).
Molecular modeling
Two crystal structures now exist for ring helicases: bacteriophage T7 gp4 (Singleton et al., 2000) and the plasmid-encoded RepA helicase (Niedenzu et al., 2001). The gp4 protein actually contains two domains, a helicase and a primase (Bernstein and Richardson, 1988), but the crystal structure is of the helicase domain alone. The gp4 helicase domain (Singleton et al., 2000), the full-length gp4 (Egelman et al., 1995) and RepA (Scherzinger et al., 1997) assemble into hexameric rings. A sequence alignment of MCM, gp4 and RepA (Figure 3) shows that the nucleotide binding site (involving the Walker A and B motifs) in both MCM and gp4 is in the C-terminal half of the protein.
Fig. 3. A comparison between the location of the Walker A and B motifs in mtMCM and in two helicase proteins that form hexameric rings, RepA and T7 gp4 (A). The T7 gp4 protein has a primase domain located on the N-terminal end of the protein and a small linker region located between these two domains (Singleton et al., 2000). The mtMCM protein has an N-terminal domain of unknown function, with no homology to any protein other than MCMs. (B) The location of the Walker A (red) and B (green) motifs are shown for the RepA (Niedenzu et al., 2001) and T7 gp4 (Singleton et al., 2000) crystal hexamers.
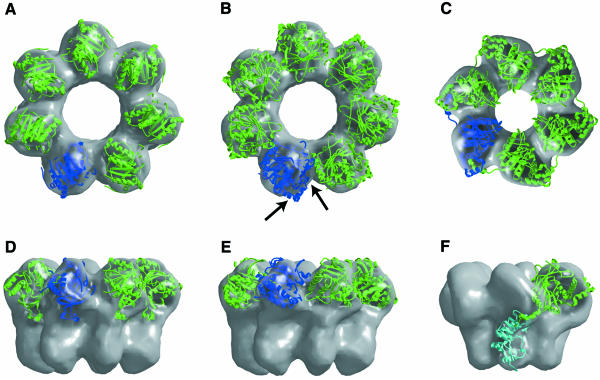
We have identified the helicase domain in the MCM heptameric ring by positioning X-ray crystal structures of both RepA (Figure 4A and D) and the T7 gp4 helicase domain (Figure 4B and E) into the three-dimensional (3D) reconstruction of MCM. In the best fit of the gp4 helicase domain into the reconstruction, the crystal structure protruded significantly out of the electron microscopic (EM) surface, and steric clashes were present with neighboring subunits in the heptamer. Unlike T7 gp4, the RepA helicase crystallized as a hexameric ring with perfect 6-fold symmetry. We extracted one subunit from the RepA hexamer and docked this structure into the MCM reconstruction. The RepA structure provided a significantly better fit to the MCM surface (Figure 4A and D) than did the gp4 structure (Figure 4B and E). In a model of the MCM heptamer generated using the best fit orientation of the RepA structure, the subunit–subunit interface was very similar to that in the RepA hexamer structure. This suggests that, as seen in RepA (Figure 3B), the nucleotide binding site in MCM lies near the subunit–subunit interface. Interestingly, neither RepA (Scherzinger et al., 1997) nor MCM requires a nucleotide cofactor for ring assembly, in contrast to ring helicases such as SV40 large T antigen (Bullock, 1997), where the nucleotide cofactor is essential for ring assembly, or helicases such as T7 gp4 (Patel and Hingorani, 1993) and E. coli DnaB (Jezewska and Bujalowski, 1996), where the nucleotide cofactor is important for the stabilization of the ring.
Fig. 4. Locating the helicase domain in the mtMCM heptamer. Crystal structures of both the RepA (A and D) and T7 gp4 (B and E) helicase domains have been docked into the mtMCM reconstruction (gray surface). To help in visualization, one subunit is shown in blue and the remainder are in green. For comparison, a model for the location of the helicase (green) and primase (cyan) domains in the EM reconstruction of the T7 gp4 hexamer (VanLoock et al., 2001) is shown in (C) and (F). The arrows in (B) indicate those regions of the gp4 helicase structure that penetrate the MCM EM reconstruction surface.
DISCUSSION
Contrary to the predictions of previous studies and the expectations from the hexameric eukaryotic enzyme, electron microscopy and image analysis indicates that the archaeal MCM helicase can exist as a heptameric ring. An obvious question arises as to whether the heptameric rings could be due to a contaminant, such as GroEL, which does form heptameric rings (Chen et al., 1998). We can exclude this possibility for four reasons. (i) The purified mtMCM protein runs as a single band on Coomassie Blue-stained SDS gels at the same time that EM fields (Figure 1) show large numbers of rings. If the heptameric rings were due to a contaminant, this contaminant could only be present as a very small fraction of the total protein and could not account for the highly abundant rings. We can eliminate the possibility that a GroEL contaminant is comigrating with MCM and therefore not resolved on SDS gels, since the molecular weight of GroEL is 57 kDa, while the molecular weight of the mtMCM is 76.5 kDa. (ii) GroEL forms double heptameric rings, while we observe single heptameric rings. Due to the dihedral symmetry of the double GroEL rings (head-to-head packing), end views of the GroEL rings show no chirality (handedness) (Falke et al., 2001). In contrast, we observe a chirality in such end views (see Supplementary figure D) that likely corresponds to projections of single rings. (iii) The use of an antibody raised against the mtMCM protein shows binding to these rings in electron micrographs (data now shown). Since the binding was largely disordered, these complexes were not amenable to further image analysis. (iv) The same heptameric structure is seen after purification of the protein in two different laboratories (Cornell and CARB) using three different clones (with an N-terminal His tag, a C-terminal His tag and a Zn2+-domain mutation). However, preliminary observations of mtMCM protein from yet another clone show that both heptameric and hexameric rings can form (X. Yu and E.H. Egelman, unpublished data), suggesting that there are unknown factors that might affect this polymorphism.
The finding that the archaeal MCM rings can be heptameric is unexpected, given that the eukaryotic MCM proteins have been observed to assemble into hexameric rings. In Xenopus egg extracts, three different heterohexamers have been observed, containing either MCM(2 + 3 + 4 + 5+ 6 + 7), MCM(2 + 3 + 4 + 6 + 7 + 7) or MCM(4 + 6 + 7)2 subunits (Prokhorova and Blow, 2000). A hexameric complex of the human MCM(4 + 6 + 7)2 subunits has been described (Sato et al., 2000). The observation that different MCM complexes formed heterohexamers was interpreted as being consistent with the role of MCM proteins in being replicative helicases. However, other proteins containing the RecA-like nucleotide binding core present in all helicases have been observed to form rings with different symmetries. The meiotic eukaryotic RecA homolog, Dmc1, forms rings with eight subunits, and it has been suggested that this is the active form of the protein (Passy et al., 1999). While the Thermus aquaticus RecA protein forms rings with six subunits (in addition to helical filaments) (Yu et al., 1995), the E. coli RecA protein and the human Rad51 protein form rings with eight subunits (X. Yu and E.H. Egelman, unpublished data). Both the E. coli and Thermus thermophilus RuvB protein form hexameric rings when bound to DNA (Stasiak et al., 1994; Miyata et al., 2000). Remarkably, heptameric rings of the T. thermophilus RuvB protein have been observed when the rings are not bound to DNA (Miyata et al., 2000). This raises the possibility that the mtMCM protein might form a hexameric ring when bound to DNA. Since we have been unable to obtain conditions for electron microscopy where the MCM rings are unambiguously bound to DNA, this will need to be tested in future work.
Although we cannot exclude the presence of double heptamers in our preparations, the majority of the rings appear to be single heptamers. Previous studies of mtMCM protein by gel filtration or glycerol gradient centrifugation (Kelman et al., 1999; Chong et al., 2000; Shechter et al., 2000), as well as by scanning transmission electron microscopy (Chong et al., 2000), have reported a single high-molecular-weight species, interpreted as being a double hexamer. The resolution of these techniques may not be great enough to distinguish between a double hexamer and a double heptamer. For example, a single mutation in the zinc-finger domain of the mtMCM protein causes a shift of the putative double hexamer to a significantly lower apparent molecular mass, as judged by gel filtration (Poplawski et al., 2001). In contrast to what has been observed for the mtMCM, a study of the Sulfolobus solfataricus MCM protein using gel filtration and glycerol gradient centrifugation found two species, and these were interpreted as a monomer and a hexamer (Carpentieri et al., 2002).
At this time, we are unable to verify the physiological relevance of the heptameric structure of the archaeal MCM helicase or the subunit composition of the active eukaryotic MCM helicase. Assuming that the heptamer is the physiological form of the archaeal MCM helicase, an important lesson to be learned from this study may be that the essence of a processive helicase lies not in the 3- or 6-fold symmetry of the ring but in the topological constraints imposed by the ring itself when it encircles DNA. If the MCM rings can exist in two states, with either six or seven subunits (consistent with our unpublished observations), an interesting possibility arises for the loading of these rings on DNA. A topological problem exists for binding any preformed ring to other than the ends of DNA, and the inability of preformed SV40 large T rings to bind origin DNA was used as an argument that these rings encircle the DNA (Dean et al., 1992). The possibility that preformed MCM rings with seven subunits lose one subunit, encircle DNA and then close to form a ring with six subunits is highly speculative, but interesting. It has previously been shown that the hexameric E. coli ρηο transcription-termination rings can actually be found with a missing subunit, as well as with an opened cleft induced by binding RNA to an external high-affinity RNA binding site (Yu et al., 2000). A ring-opening model for the hexameric T7 helicase primase has also been proposed, based upon kinetic data (Ahnert et al., 2000). Taken together, this provides a testable hypothesis that needs to be considered in future studies of MCM rings.
METHODS
Sample preparation.
The mtMCM protein was overexpressed and purified from E. coli as described previously (Poplawski et al., 2001). Samples were applied to carbon-coated grids and negatively stained with 2% uranyl acetate. Specimens were examined in an FEI Tecnai 12 electron microscope at an accelerating voltage of 80 keV and a nominal magnification of 30 000×.
Image analysis and modeling.
Electron micrographs were scanned on a Leaf 45 densitometer at a raster corresponding to 3.9 Å/pixel. The SPIDER software package (Frank et al., 1996) was used for most of the image processing and O (Jones et al., 1991) was used for the molecular modeling. Models were built by first generating 3D densities from atomic coordinates of individual subunits, and filtering these volumes to a resolution comparable to that of the MCM reconstruction. We then docked the low-resolution surfaces into the MCM reconstruction using shape as our primary guide. Transformations used in docking the surfaces were applied to the structure coordinates. The enantiomorphic ambiguity present in the 3D reconstruction was resolved by choosing the structure that gave the best fit with the crystal models. BOBSCRIPT (Esnouf, 1999) was used for displaying molecular models within EM surfaces.
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Xiaojiang Chen for helpful discussions. This work was supported by NIH grants GM35269 (to E.H.E.) and GM34190 (to B.K.T.). Z.K. is an Invitrogen Professor.
REFERENCES
- Adachi Y., Usukura, J. and Yanagida, M. (1997) A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells, 2, 467–479. [DOI] [PubMed] [Google Scholar]
- Ahnert P., Picha, K.M. and Patel, S.S. (2000) A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J., 19, 3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein, D.M. and Bell, S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Bernstein J.A. and Richardson, C.C. (1988) A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc. Natl Acad. Sci. USA, 85, 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P.A. (1997) The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol., 32, 503–568. [DOI] [PubMed] [Google Scholar]
- Carpentieri F., De Felice, M., De Falco, M., Rossi, M. and Pisani, F.M. (2002) Physical and functional interaction between the MCM-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem., 277, 12118–12127. [DOI] [PubMed] [Google Scholar]
- Chen S., Roseman, A.M. and Saibil, H.R. (1998) Electron microscopy of chaperonins. Methods Enzymol., 290, 242–253. [DOI] [PubMed] [Google Scholar]
- Chong J.P., Hayashi, M.K., Simon, M.N., Xu, R.M. and Stillman, B. (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 97, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F.B., Borowiec, J.A., Eki, T. and Hurwitz, J. (1992) The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem., 267, 14129–14137. [PubMed] [Google Scholar]
- Egelman E.H., Yu, X., Wild, R., Hingorani, M.M. and Patel, S.S. (1995) Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA, 92, 3869–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Falke S., Fisher, M.T. and Gogol, E.P. (2001) Structural changes in GroEL effected by binding a denatured protein substrate. J. Mol. Biol., 308, 569–577. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. and Leith, A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Jezewska M.J. and Bujalowski, W. (1996) Global conformational transitions in Escherichia coli primary replicative helicase DnaB protein induced by ATP, ADP and single-stranded DNA binding. Multiple conformational states of the helicase hexamer. J. Biol. Chem., 271, 4261–4265. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kaplan D.L. (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol., 301, 285–299. [DOI] [PubMed] [Google Scholar]
- Kelman Z., Lee, J.K. and Hurwitz, J. (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DH contains DNA helicase activity. Proc. Natl Acad. Sci. USA, 96, 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. (1993) A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res., 21, 2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., Tercero, J.A. and Diffley, J.F. (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Lee J.K. and Hurwitz, J. (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA, 98, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yamada, K., Iwasaki, H., Shinagawa, H., Morikawa, K. and Mayanagi, K. (2000) Two different oligomeric states of the RuvB branch migration motor protein as revealed by electron microscopy. J. Struct. Biol., 131, 83–89. [DOI] [PubMed] [Google Scholar]
- Niedenzu T., Roleke, D., Bains, G., Scherzinger, E. and Saenger, W. (2001) Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol., 306, 479–487. [DOI] [PubMed] [Google Scholar]
- Passy S.I., Yu, X., Li, Z., Radding, C.M., Masson, J.Y., West, S.C. and Egelman, E.H. (1999) Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl Acad. Sci. USA, 96, 10684–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.S. and Hingorani, M.M. (1993) Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J. Biol. Chem., 268, 10668–10675. [PubMed] [Google Scholar]
- Poplawski A., Grabowski, B., Long, S.E. and Kelman, Z. (2001) The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J. Biol. Chem., 276, 49371–49377. [DOI] [PubMed] [Google Scholar]
- Prokhorova T.A. and Blow, J.J. (2000) Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem., 275, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Gotow, T., You, Z., Komamura-Kohno, Y., Uchiyama, Y., Yabuta, N., Nojima, H. and Ishimi, Y. (2000) Electron microscopic observation and single-stranded DNA binding activity of the Mcm4, 6, 7 complex. J. Mol. Biol., 300, 421–431. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Ziegelin, G., Barcena, M., Carazo, J.M., Lurz, R. and Lanka, E. (1997) The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J. Biol. Chem., 272, 30228–30236. [DOI] [PubMed] [Google Scholar]
- Shechter D.F., Ying, C.Y. and Gautier, J. (2000) The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicumδ H minichromosome maintenance protein. J. Biol. Chem., 275, 15049–15059. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Sawaya, M.R., Ellenberger, T. and Wigley, D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Tsaneva, I.R., West, S.C., Benson, C.J.B., Yu, X. and Egelman, E.H. (1994) The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc. Natl Acad. Sci. USA, 91, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- Tye B.K. (2000) Insights into DNA replication from the third domain of life. Proc. Natl Acad. Sci. USA, 97, 2399–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanLoock M.S., Chen, Y.J., Yu, X., Patel, S.S. and Egelman, E.H. (2001) The primase active site is on the outside of the hexameric bacteriophage T7 gene 4 helicase-primase ring. J. Mol. Biol., 311, 951–956. [DOI] [PubMed] [Google Scholar]
- Yu X., Angov, E., Camerini-Otero, R.D. and Egelman, E.H. (1995) Structural polymorphism of the RecA protein from the thermophilic bacterium Thermus aquaticus. Biophys. J., 69, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Hingorani, M.M., Patel, S.S. and Egelman, E.H. (1996) DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol., 3, 740–743. [DOI] [PubMed] [Google Scholar]
- Yu X., Horiguchi, T., Shigesada, K. and Egelman, E.H. (2000) Three-dimensional reconstruction of transcription termination factor ρ: orientation of the N-terminal domain and visualization of an RNA-binding site. J. Mol. Biol., 299, 1279–1287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.