Abstract
CSL [CBF-1, Su(H), Lag-1]-type transcription factors are the primary effectors of the Notch pathway, a signal transduction cascade that is essential for the development of all metazoan organisms. Interestingly, CSL proteins were originally classified as transcriptional repressors in vertebrates, but as transcriptional activators in model invertebrate organisms. Resolution of this paradox came with the realization that repression and activation by CSL proteins occurs in both systems and that the switch involves recruitment of distinct co-repressor and co-activator complexes. Although CSL proteins appear to utilize a common co-activator complex of largely similar constitution, recent studies have demonstrated that vertebrate and Drosophila CSL interact with a variety of distinct co-repressor complexes. This review highlights differences in composition and similarities in function of different CSL co-repressor complexes, which actively repress Notch pathway target genes in the absence of Notch pathway activity.
Introduction—CSL proteins and the N pathway
CSL [CBF-1, Su(H), Lag-1] proteins are transcription factors containing a unique DNA-binding domain encoded by only one or a few genes in any given genome. In metazoan organisms, CSL proteins are the primary nuclear effectors of the Notch signaling pathway. This pathway mediates cell–cell interactions via the transmembrane proteins DSL (Delta, Serrate, Lag-2) and Notch (N), which act as a ligand and receptor, respectively. The N pathway has been evolutionarily conserved and is essential for a variety of developmental processes, including asymmetric cell-fate decisions, boundary formation and cell proliferation (reviewed in Artavanis-Tsakonas et al., 1999).
Involvement of a CSL protein in the N pathway was first demonstrated in Drosophila. The key observations that allowed its ortholog Suppressor of Hairless [Su(H)] to be placed in this pathway are that (i) its function is obligate in developmental settings that require N (Schweisguth and Posakony, 1992), (ii) it displays genetic and physical interactions with N (Fortini and Artavanis-Tsakonas, 1994) and (iii) it directly activates gene expression upon activation of N by binding to the regulatory DNA of target genes (Bailey and Posakony, 1995; Furukawa et al., 1995; Lecourtois and Schweisguth, 1995). Although the functional significance of the physical association between the intracellular domain of N (NIC) and Su(H) was controversial for some years, it is now generally accepted that activation of N by its ligand results in the cleavage and release of NIC, which then functions as a nuclear transcriptional co-activator for Su(H) (reviewed in Mumm and Kopan, 2000).
A vertebrate CSL ortholog was independently identified by multiple groups prior to the cloning of Su(H). These researchers did not then recognize its connection to the N pathway and purified CSL solely on the basis of its high-affinity DNA-binding activity towards YRTGDGAD motifs (Tun et al., 1994; Barolo et al., 2000). This resulted in the repeated identification of the same protein as RBP-Jκ (recombination signal binding protein of the Jκ immunoglobulin gene; Matsunami et al., 1989), KBF2 (MHC enhancer κB binding factor; Brou et al., 1994), LMP-2 [Epstein–Barr virus (EBV) latent membrane promoter binding protein] (Grossman et al., 1994) and CBF-1 (EBV latency C promoter binding factor; Henkel et al., 1994). Vertebrate CSL will henceforth be referred to as CBF-1. Although the purification of CBF-1 in the first two studies eventually proved fortuitous or indeed artifactual (in the first study, the relevant binding site was unknowingly created by the introduction of a flanking restriction enzyme site), the collected work is a testament to the specificity and strength of DNA recognition by CSL proteins.
CSL proteins and repression
The initial characterization of CBF-1 in tissue culture cells suggested that it normally functions as a transcriptional repressor (Dou et al., 1994; Waltzer et al., 1995). However, during EBV-mediated cell immortalization, the viral protein Epstein–Barr nuclear antigen-2 (EBNA-2) subverts CBF-1 function by binding to it and converting it into a transcriptional activator, in part by masking the CBF-1 repression domain (Hsieh and Hayward, 1995). Interestingly, it was found that NIC has an analogous activity and similarly converts CBF-1 from a repressor into an activator (Hsieh et al., 1996). Studies such as these led to the proposal that activation of the N receptor and translocation of cleaved NIC to the nucleus causes a switch in CBF-1 transcriptional activity from negative to positive.
For many years, this model was difficult to reconcile with the in vivo activity of Su(H), as this protein seemed to activate all of its initially identified targets. However, it was later observed that ectopic Su(H) represses the expression of some target genes (Furriols and Bray, 2000; Klein et al., 2000), and detailed studies now definitively demonstrate a role for Su(H) in transcriptional repression of several targets previously known to be activated by it. For example, mutation of Su(H) binding sites in cis or of Su(H) in trans results not only in a quantitative reduction in single-minded (sim) gene or reporter expression [consistent with an activating function for Su(H)], but also in a broader domain of sim expression [consistent with a general repressive role for Su(H) in the spatial control of sim activity] (Morel and Schweisguth, 2000). Studies of Su(H) itself during asymmetric cell divisions in the sensory organ lineage also revealed dual functions in transcriptional auto-regulation: it is auto-activating in the socket cell but auto-repressive in the sibling shaft cell (Barolo et al., 2000). Finally, studies with artificial enhancers revealed that Su(H) binding sites and Su(H) gene activity can confer repression in tissues not actively involved in N signaling and, conversely, activation in settings of endogenous or forced N activity (Furriols and Bray, 2001).
The accumulated data now strongly support a general ‘switch’ model for CSL proteins, in which they actively repress target gene expression in the absence of signaling, but then activate them upon stimulation of the N receptor. Curiously, although the composition of CSL co-activator complexes appears to be conserved and includes NIC and the nuclear proteins Mastermind/LAG-3 (Petcherski and Kimble, 2000; Wu et al., 2000; Kitagawa et al., 2001; Figure 1D), the composition of CSL co-repressor complexes characterized to date differs significantly between vertebrates and Drosophila (Figure 1A–C). The following sections will review proteins involved in repression by CSL proteins.
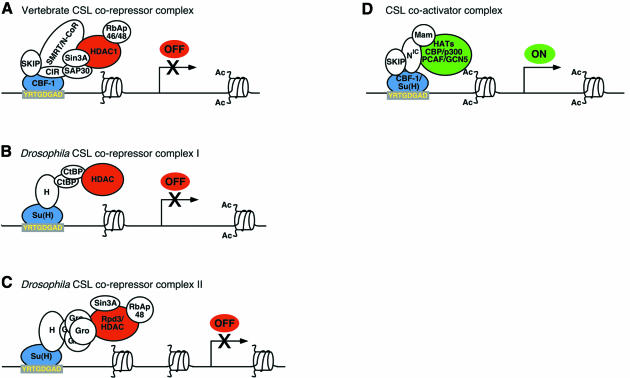
Fig. 1. CSL proteins associate with co-repressor complexes containing HDAC activity and a co-activator complex containing HAT activity. CSL proteins (blue) bind with high affinity to the consensus site YRTGDGAD (certain other sites included by the broader consensus RTGRGAR are bound with lower affinity). The vertebrate ortholog CBF-1 interacts with the CIR+SMRT/N-CoR co-repressor complex (A), while Drosophila Su(H) uses Hairless (H) to recruit both dCtBP (B) and Gro (C) co-repressor complexes. Drosophila SMRTER associates with Su(H), but has not yet been shown to mediate repression by Su(H) in vivo. All of these co-repressors have been functionally linked to HDACs (red), whose activity leads to a transcriptionally repressed chromatin state. Note that the composition of these repression complexes has been extrapolated from multiple studies and is tentative. Other components of these co-repressor complexes may exist, particularly for the Gro and CtBP complexes, which have not been as extensively characterized as the SMRT/Sin3A complex. In Drosophila, Gro represses transcription over greater distances than does dCtBP (as depicted by a greater extent of histone deacetylation), but it remains to be determined if these co-repressors mediate separable modes of ‘short-range’ and ‘long-range’ repression by Su(H). HDAC-independent modes of repression might also exist for these co-repressors or for CBF-1 (data not shown, see text for details). (D) Vertebrate and Drosophila CSL proteins interact with a co-activator complex that includes NIC, Mam and HATs (green).
Vertebrate CSL co-repressors
Multiple transcription factors often recruit the same co-repressor. Such is the case for the related co-repressors SMRT (silencing mediator for retinoid and thryoid receptor) and N-CoR (nuclear receptor co-repressor). These were originally identified as proteins that bound to unliganded nuclear hormone receptors and repressed their ability to activate transcription (Chen and Evans, 1995; Horlein et al., 1995). Both proteins contain a pair of SANT domains, a motif found in a variety of other chromatin-associated transcriptional co-regulators. SMRT and N-CoR have since been found to be co-repressors for several other unrelated classes of transcription factors, including CBF-1 (Figure 1A). In support of this, both proteins can bind directly to CBF-1 and antagonize the ability of NIC to stimulate gene expression via CBF-1 (Kao et al., 1998). A second class of CBF-1 co-repressor is represented by CIR (CBF-interacting repressor; Figure 1A), a novel protein identified in a yeast two-hybrid screen (Hsieh et al., 1999). As is the case with SMRT/N-CoR, CIR mediates transcriptional repression when targeted to DNA either through its interaction with CBF-1 or as a fusion protein with the Gal4 DNA-binding domain. SMRT, N-CoR and CIR all fail to bind repression-defective mutants of CBF-1, further demonstrating their inclusion in the CBF-1 repression complex.
The Ski-interacting protein (SKIP) is an adaptor protein that exhibits mutually exclusive associations with co-repressor (via direct contacts with CBF-1 and SMRT) and co-activator (via direct contacts with CBF-1 and NIC) complexes (Zhou et al., 2000). This protein may facilitate switching of transcriptional activity of CBF-1. In addition, interactions of CBF-1 with the SKIP/CIR/SMRT co-repressor complex appear to be important for nuclear localization of CBF-1 (Zhou and Hayward, 2001).
Drosophila CSL co-repressors
Su(H) was originally identified on the basis of its dominant suppression of the Hairless (H) phenotype, and a variety of genetic evidence indicates that H negatively regulates the activity of Su(H) and the N pathway (Schweisguth and Posakony, 1994; Bang et al., 1995). At least three Su(H)-regulated enhancers [vgBE, sim and Su(H)ASE] are activated ectopically under H loss-of-function conditions (Barolo et al., 2000; Furriols and Bray, 2000; Klein et al., 2000; Morel et al., 2001), indicating that H normally antagonizes Su(H) function as a transcriptional activator.
These nuclear proteins bind directly to each other in vitro, and initial studies suggested that H might prevent Su(H) from binding DNA (Brou et al., 1994). However, other data were at odds with this model. For example, H misexpression never de-represses N target gene expression, as might be predicted from a model in which H blocks the DNA-binding capacity of Su(H). In addition, co-misexpression of Su(H) and H does not result in a mutually suppressed phenotype, as one might expect for two proteins with opposing activities. Instead, its effect closely resembles a strong loss in N pathway function (Furriols and Bray, 2000; Morel et al., 2001).
Re-evaluation of H function demonstrated that it does not in fact occlude Su(H) DNA-binding activity, since H can bind the Su(H)–DNA complex under appropriate conditions (Morel et al., 2001; Barolo et al., 2002). Importantly, it was shown that the H C-terminal sequence PLNLS interacts with the well-characterized co-repressor dCtBP (Drosophila C-terminal binding protein) and that deletion of this sequence interferes with the ability of H to antagonize N signaling when misexpressed, either by itself or in combination with Su(H) (Morel et al., 2001; Barolo et al., 2002). Genetic interaction data further indicate that dCtBP contributes positively to H function (Barolo et al., 2002). Taken together, these data suggest that H is an adaptor protein that recruits the co-repressor dCtBP to DNA-bound Su(H) (Figure 1B).
Interestingly, dCtBP accounts for only a subset of H’s repressive function. This appears to be due to the independent utilization of the co-repressor Groucho (Gro) by H (Barolo et al., 2000; Figure 1C). This finding is unexpected, since previous genetic analysis defined Gro, a protein containing seven WD40 repeats, as a positive component of the N pathway. Notably, gro mutant embryos and adult clones display, among other phenotypes, an excess of neural differentiation, which is indicative of a failure of N-mediated lateral inhibition. Gro functions as a co-repressor for the seven basic helix–loop–helix (bHLH) repressor proteins encoded by the Enhancer of split Complex [E(spl)-C], which are among the most well characterized transcriptional targets of the N pathway; Gro binds directly to the WRPW motif found at the extreme C-termini of these bHLH repressors. However, it also binds directly to the H sequence YSIHSLLG, an ‘eh1’-type Gro-binding motif. Genetic interactions strongly support the in vivo significance of this in vitro observation. As is the case for dCtBP, gro mutations dominantly enhance H and suppress N phenotypes during sensory organ development (Barolo et al., 2002). These observations now suggest a dual role for Gro, whereby it functions as a positive component of N-regulated processes when signaling is active [as a co-repressor for E(spl)bHLH proteins], but as an antagonist of the same processes when signaling is absent [as a co-repressor for the Su(H)–H complex, which would in turn repress E(spl)bHLH expression].
Like SMRT/N-CoR, both dCtBP and Gro directly interact with a wide variety of other unrelated transcription factors in Drosophila to mediate transcriptional repression. Interestingly, two of these, Hairy and Brinker, also recruit both Gro and dCtBP (Paroush et al., 1994; Poortinga et al., 1998; Hasson et al., 2001). Since Gro mediates ‘long-range’ silencing while dCtBP mediates ‘short-range’ repression (reviewed in Courey and Jia, 2001), utilization of multiple co-repressors might allow for mechanistic flexibility in the regulation of different promoters. A final twist is that in some cases, dCtBP actually antagonizes Gro-mediated repression when both co-repressors are simultaneously recruited to the same promoter (Zhang and Levine, 1999; Phippen et al., 2000). It remains to be determined whether interactions of Gro and dCtBP with the Su(H)–H complex are mutually exclusive and whether these co-repressors mediate qualitatively different modes of repression by Su(H).
Different means to the same end? CSL recruits HDAC
A unifying property of these different CSL co-repressor complexes appears to be their recruitment of histone deacetylases (HDACs). The acetylation state of chromatin is correlated with its transcriptional competency: reduction of histone acetylation by HDACs is linked to repressed, transcriptionally inactive chromatin, while elevation of histone acetylation by histone acetyltransferases promotes a transcriptionally active chromatin state. Although it is undoubtedly a gross oversimplification to reduce the transcriptional capacity of a given gene to its local acetylation status, it is nonetheless intriguing to find that all of the CSL co-repressor complexes described above have been functionally linked to HDAC activity.
SMRT/N-CoR and CIR are components of a large co-repressor complex that has been the subject of intense biochemical scrutiny (Figure 1A); this complex includes Sin3A, SAP18, SAP30, RbAp46/48 and HDAC (reviewed in Glass and Rosenfeld, 2000). Both CIR and SMRT/N-CoR are key bridging components between CBF-1 and the co-repressor complex; CIR directly contacts CBF-1, SKIP and SAP30 (Hsieh et al., 1999; Zhou et al., 2000), while SMRT/N-CoR makes direct contacts with CBF-1, SKIP, Sin3A, SAP30 and possibly HDAC (Nagy et al., 1997; Kao et al., 1998; Laherty et al., 1998; Zhou et al., 2000). SMRT and N-CoR may be more than simple scaffolds in this co-repressor complex, since recent studies have shown that these proteins directly activate the deacetylase activity of the class I enzyme HDAC3 through one of the SANT domains (Guenther et al., 2001; Zhang et al., 2002). SMRT and N-CoR are capable of interacting with multiple class I and class II HDAC enzymes, and the repertoire of HDACs involved in CBF-1-mediated repression is not yet known. Nevertheless, the ultimate involvement of HDACs in CBF-1-mediated repression is clear, as the class I enzyme HDAC1 co-immunoprecipitates with CBF-1 in tissue culture cells, and expression of a Xenopus CBF-1 target gene is potentiated in the presence of the HDAC inhibitor trichostatin A (TSA; Kao et al., 1998).
Several lines of evidence indicate that CtBP proteins function at least in part by recruiting HDAC activity. In support of this model, mammalian CtBP binds directly to multiple class II HDAC enzymes via PXDLR motifs (Zhang et al., 2001), and at least some modes of CtBP-mediated repression can be relieved using TSA (Criqui-Filipe et al., 1999). The class II enzyme Drosophila HDAC4 also contains a possible CtBP binding site (PVNLS), although an interaction has not yet been specifically tested. Since dCtBP self-associates (Poortinga et al., 1998), dCtBP dimers may form a bridge between the Su(H)–H complex and HDAC (Figure 1B).
Drosophila Gro displays genetic and physical interactions with the class I HDAC Rpd3, indicating that Gro also directly recruits deacetylase activity (Figure 1C; Chen et al., 1999). Since the active form of Gro seems to be a tetramer (Chen et al., 1998), Gro could in principle mediate the formation of higher-order HDAC-containing complexes. Finally, experiments in cultured human cells have shown that vertebrate Gro is present in a complex that includes not only HDAC1, but also Sin3 and RbAp48 (Choi et al., 1999). These data suggest that Gro and SMRT/N-CoR may have similar functions in recruiting a Sin3/HDAC complex to mediate repression by CSL proteins.
It is worth noting that some aspects of repression mediated by CSL or its co-repressors may be HDAC independent. For example, interactions of CBF-1 with the transcriptional activators dTAFII110 and TFIIA during repression of the pI adenovirus promoter suggest that CBF-1 can directly interfere with co-activator recruitment (Olave et al., 1998). Similarly, it has been suggested that Gro family members may also interact directly with and inhibit the basal transcription machinery (reviewed in Courey and Jia, 2001). Finally, some modes of CtBP-mediated repression are not sensitive to TSA, suggesting that CtBP does not always exert its effects through HDAC (reviewed in Turner and Crossley, 2001; Chinnadurai, 2002). Intriguingly, CtBP proteins bear significant similarity to acid dehydrogenases, and a rat homolog has been shown to possess acyl transferase activity, suggesting that CtBP might exert some of its effect through an intrinsic enzymatic activity. Taken together, the accumulated data suggest that recruitment of HDAC activity represents a major, although probably not exclusive, strategy for transcriptional repression by CSL proteins.
Several recent studies indicate that the opposing reaction of histone acetylation is an intrinsic mechanistic feature of transcriptional activation by the CSL/NIC complex (Figure 1D). In fact, mammalian NIC is capable of associating with multiple histone acetyltransferases (HATs), including p300/CBP (Oswald et al., 2001) and PCAF/GCN5 (Kurooka and Honjo, 2000). The latter study showed that mammalian PCAF/GCN5 also interacts with Drosophila NIC, suggesting that this interaction is a conserved feature of the CSL co-activator complex. The precise mechanisms of HAT recruitment are not fully resolved; the aforementioned studies suggest a direct physical interaction with NIC, while a more recent study suggests that HAT activity is recruited to NIC via Mastermind (Mam; Fryer et al., 2002). It is also not known whether multiple HATs are ever simultaneously recruited to the CSL co-activator complex or whether they are utilized individually in different settings. Regardless, these results collectively suggest that repression and activation by CSL proteins are coordinately linked to histone hypo- and hyper-acetylation, respectively.
Conclusions and speculations
As the central transcription factors in the N pathway, it may seem self-evident that CSL proteins should promote target gene expression in response to N activation. But why should so much machinery be dedicated to allowing CSL proteins to function as repressors as well? Many detailed genetic analyses in Drosophila have elegantly shown that cell-fate decisions mediated by the N pathway are highly sensitive to small differences in pathway activity. The ability of CSL proteins to actively repress transcription in the absence of N pathway activity sharpens the cellular response to pathway activation, since it eliminates stray gene expression prior to signaling and terminates gene expression following completion of signaling. Notably, a similar dual strategy of gene regulation is employed by the central transcription factors in many other widely utilized signaling pathways, including the nuclear receptor, Hedgehog, Wingless/WNT and Decapentaplegic/TGF-β pathways. Thus, repression of target gene expression in the absence of signaling is likely to be the rule rather than the exception (reviewed in Barolo et al., 2002). N is also subject to negative post-translational regulation; for example, multiple ubiquitin ligases target the activated N receptor for degradation (reviewed in Lai, 2002). The combination of strong negative regulatory mechanisms that function both pre- and post-signaling ensures a cellular response to N activation that is both temporally and quantitatively sensitive.
It remains to be determined whether these different co-repressor complexes evolved independently or are used throughout metazoan phyla. For example, the lack of an obvious vertebrate H ortholog could reflect either a specific role for this novel protein in Diptera or primary sequence divergence precluding its identification in other genomes. A Drosophila ortholog of CIR exists, but its function has not yet been examined. SMRTER, a probable fly homolog of SMRT, has recently been found to associate with Su(H), and it has been suggested that SMRTER may participate in Su(H)-mediated repression during eye development (Tsuda et al., 2002). However, genetic evidence for this is still required. Finally, it remains to be seen whether the nematode CSL ortholog, Lag-1, participates in transcriptional repression, and, if so, what cofactors might be involved; the Caenhorhabditis elegans proteome includes orthologs of CIR, Gro and SKIP, but does not contain recognizable orthologs of H and SMRT.
The dual activity of CSL proteins in the N pathway also begs the question of what their ancestral function may have been. Although CSL-encoding genes are generally absent from plants and lower eukaryotes, the genome of the yeast Schizosaccharomyces pombe encodes two distant CSL homologs. This unicellular organism does not possess most components of the N pathway, but does utilize many components of the mammalian Sin3a/HDAC repression complex. Thus, it is conceivable that CSL proteins in this yeast are linked to an HDAC complex and that repression was the original CSL activity. In this scenario, a signal-dependent transcriptional activation mode may well have been a subsequent multicellular adaptation. These possibilities await future studies.

Eric C. Lai
Acknowledgments
Acknowledgements
The author would like to thank Adina Bailey, Albert Courey and Francois Schweisguth for useful discussions, and Scott Barolo, James Posakony, Raghavendra Nagaraj and Utpal Banerjee for communicating data prior to publication. He is supported by Gerald Rubin and the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation, DRG 1632.
References
- Artavanis-Tsakonas S., Rand, M.D. and Lake, R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776. [DOI] [PubMed] [Google Scholar]
- Bailey A.M. and Posakony, J.W. (1995) Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes Dev., 9, 2609–2622. [DOI] [PubMed] [Google Scholar]
- Bang A.G., Bailey, A.M. and Posakony, J.W. (1995) Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway. Dev. Biol., 172, 479–494. [DOI] [PubMed] [Google Scholar]
- Barolo S., Walker, R., Polyanovsky, A., Freschi, G., Keil, T. and Posakony, J.W. (2000) A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell, 103, 957–969. [DOI] [PubMed] [Google Scholar]
- Barolo S., Stone, T., Bang, A.G. and Posakony, J.W. (2002) Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev., 16, 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C., Logeat, F., Lecourtois, M., Vandekerckhove, J., Kourilsky, P., Schweisguth, F. and Israel, A. (1994) Inhibition of the DNA-binding activity of Drosophila Suppressor of Hairless and of its human homolog, KBF2/RBP-Jκ, by direct protein–protein interaction with Drosophila Hairless. Genes Dev., 8, 2491–2503. [DOI] [PubMed] [Google Scholar]
- Chen G., Nguyen, P.H. and Courey, A.J. (1998) A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol., 18, 7259–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Fernandez, J., Mische, S. and Courey, A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. and Evans, R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. (2002) CtBP, and uncoventional transcriptional corepressor in development and oncogenesis. Mol. Cell, 9, 213–224. [DOI] [PubMed] [Google Scholar]
- Choi C.Y., Kim, Y.H., Kwon, H.J. and Kim, Y. (1999) The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem., 274, 33194–33197. [DOI] [PubMed] [Google Scholar]
- Courey A.J. and Jia, S. (2001) Transcriptional repression: the long and the short of it. Genes Dev., 15, 2786–2796. [DOI] [PubMed] [Google Scholar]
- Criqui-Filipe P., Ducret, C., Maira, S. and Wasylyk, B. (1999) Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J., 18, 3392–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou S., Zeng, X., Cortes, P., Erdjument-Bromage, H., Tempst, P., Honjo, T. and Vales, L.D. (1994) The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol. Cell. Biol., 14, 3310–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M.E. and Artavanis-Tsakonas, S. (1994) The Suppressor of Hairless protein participates in Notch receptor signaling. Cell, 79, 273–282. [DOI] [PubMed] [Google Scholar]
- Fryer C.J., Lamar, E., Turbachova, I., Kintner, C. and Jones, K.A. (2002) Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev., 16, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M. and Bray, S. (2000) Dissecting the mechanisms of Suppressor of Hairless function. Dev. Biol., 227, 520–532. [DOI] [PubMed] [Google Scholar]
- Furriols M. and Bray, S. (2001) A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol., 11, 60–64. [DOI] [PubMed] [Google Scholar]
- Furukawa T., Kobayakawa, Y., Tamura, K., Kimura, K., Kawaichi, M., Tanimura, T. and Honjo, T. (1995) Suppressor of Hairless, the Drosophila homologue of RBP-Jκ, transactivates the neurogenic gene E(spl)m8. Jpn J. Genet., 70, 505–524. [DOI] [PubMed] [Google Scholar]
- Glass C. and Rosenfeld, M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Grossman S.R., Johannsen, E., Tong, X., Yalamanchili, R. and Kieff, E. (1994) The Epstein–Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl Acad. Sci. USA, 91, 7568–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M., Barak, O. and Lazar, M. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol., 21, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P., Muller, B., Basler, K. and Paroush, Z. (2001) Brinker requires two corepressors for maximal and versatile repression in Dpp signaling. EMBO J., 20, 5725–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T., Ling, P.D., Hayward, S.D. and Peterson, M.G. (1994) Mediation of Epstein–Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science, 265, 92–95. [DOI] [PubMed] [Google Scholar]
- Horlein A. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Hsieh J.J., Henkel, T., Salmon, P., Robey, E., Peterson, M.G. and Hayward, S.D. (1996) Truncated mammalian Notch1 activates CBF-1/RBPJκ-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol. Cell. Biol., 16, 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J.J., Zhou, S., Chen, L., Young, D.B. and Hayward, S.D. (1999) CIR, a corepressor linking the DNA binding factor CBF-1 to the histone deacetylase complex. Proc. Natl Acad. Sci. USA, 96, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J.J.-D. and Hayward, S.D. (1995) Masking of the CBF-1/RBPJκ transcriptional repression domain by Epstein–Barr virus EBNA2. Science, 268, 560–563. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Ordentlich, P., Koyano-Nakagawa, N., Tang, Z., Downes, M., Kintner, C.R., Evans, R.M. and Kadesch, T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev., 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Oyama, T., Kawashima, T., Yedvobnick, B., Kumar, A., Matsuno, K. and Harigaya, K. (2001) A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol., 21, 4337–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T., Seugnet, L., Haenlin, M. and Arias, A.M. (2000) Two different activities of Suppressor of Hairless during wing development in Drosophila. Development, 127, 3553–3566. [DOI] [PubMed] [Google Scholar]
- Kurooka H. and Honjo, T. (2000) Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem., 275, 17211–17220. [DOI] [PubMed] [Google Scholar]
- Laherty C. et al. (1998) SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell, 2, 33–42. [DOI] [PubMed] [Google Scholar]
- Lai E.C. (2002) Protein degradation: four E3s for the Notch pathway. Curr. Biol., 12, R74–R78. [DOI] [PubMed] [Google Scholar]
- Lecourtois M. and Schweisguth, F. (1995) The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split complex genes triggered by Notch signaling. Genes Dev., 9, 2598–2608. [DOI] [PubMed] [Google Scholar]
- Matsunami N., Hamaguchi, Y., Yamamoto, Y., Kuze, K., Kangawa, K., Matsuo, H., Kawaichi, M. and Honjo, T. (1989) A protein binding to the Jκ recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature, 342, 934–937. [DOI] [PubMed] [Google Scholar]
- Morel V. and Schweisguth, F. (2000) Repression by Suppressor of Hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev., 14, 377–388. [PMC free article] [PubMed] [Google Scholar]
- Morel V., Lecourtois, M., Massiani, O., Maier, D., Preiss, A. and Schweisguth, F. (2001) Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless–dCtBP complex in Drosophila. Curr. Biol., 11, 789–792. [DOI] [PubMed] [Google Scholar]
- Mumm J.S. and Kopan, R. (2000) Notch signaling: from the outside in. Dev. Biol., 228, 151–165. [DOI] [PubMed] [Google Scholar]
- Nagy L., Kao, H.Y., Chakravarti, D., Lin, R.J., Hassig, C.A., Ayer, D.E., Schreiber, S.L. and Evans, R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Olave I., Reinberg, D. and Vales, L.D. (1998) The mammalian transcriptional repressor RBP (CBF-1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev., 12, 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F., Tauber, B., Dobner, T., Bourtelle, S., Kostezka, U., Adler, G., Liptay, S. and Schmid, R. (2001) p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol., 21, 7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z., Finley, R.L. Jr, Kidd, T., Wainwright, S.M., Ingham, P.W., Brent, R. and Ish-Horowicz, D. (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell, 79, 805–815. [DOI] [PubMed] [Google Scholar]
- Petcherski A. and Kimble, J. (2000) LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature, 405, 364–368. [DOI] [PubMed] [Google Scholar]
- Phippen T., Sweigart, A., Moniwa, M., Krumm, A., Davie, J. and Parkhurst, S.M. (2000) Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both Mad and Groucho transcriptional repression. J. Biol. Chem., 275, 37628–37637. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Watanabe, M. and Parkhurst, S. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. and Posakony, J.W. (1992) Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell, 69, 1199–1212. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. and Posakony, J.W. (1994) Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development, 120, 1433–1441. [DOI] [PubMed] [Google Scholar]
- Tsuda L., Nagaraj, R., Zipursky, S.L. and Bannerjee, U. (2002) An EGFR/Ebi/Sno pathway promotes Delta expression by inactiviting Su(H)/SMRTER expression during Notch signaling. Cell, in press. [DOI] [PubMed] [Google Scholar]
- Tun T., Hamaguchi, Y., Matsunami, N., Furukawa, T., Honjo, T. and Kawaichi, M. (1994) Recognition sequence of a highly conserved DNA binding protein RBP-Jκ. Nucleic Acids Res., 22, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. and Crossley, M. (2001) The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays, 23, 683–690. [DOI] [PubMed] [Google Scholar]
- Waltzer L., Bourillot, P.Y., Sergeant, A. and Manet, E. (1995) RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein–Barr virus transcription factor EBNA2. Nucleic Acids Res., 23, 4939–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Aster, J., Blacklow, S., Lake, R., Artavanis-Tsakonas, S. and Griffin, J. (2000) MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet., 26, 484–489. [DOI] [PubMed] [Google Scholar]
- Zhang C., McKinsey, T., Lu, J. and Olson, E.N. (2001) Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem., 276, 35–39. [DOI] [PubMed] [Google Scholar]
- Zhang H. and Levine, M. (1999) Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kalkum, M., Chait, B. and Roeder, R. (2002) The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell, 9, 611–623. [DOI] [PubMed] [Google Scholar]
- Zhou S. and Hayward, S.D. (2001) Nuclear localization of CBF-1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol., 21, 6222–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Fujimuro, M., Hsieh, J., Chen, L., Miyamoto, A., Weinmaster, G. and Hayward, S.D. (2000) SKIP, a CBF-1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol., 20, 2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]