Abstract
Snu114p is a yeast U5 snRNP protein homologous to the ribosomal elongation factor EF-2. Snu114p exhibits the same domain structure as EF-2, including the G-domain, but with an additional N-terminal domain. To test whether Snu114p in the spliceosome is involved in rearranging RNA secondary structures (by analogy to EF-2 in the ribosome), we created conditionally lethal mutants. Deletion of this N-terminal domain (snu114ΔN) leads to a temperature-sensitive phenotype at 37°C and a pre-mRNA splicing defect in vivo. Heat treatment of snu114ΔN extracts blocked splicing in vitro before the first step. The snu114ΔN still associates with the tri-snRNP, and the stability of this particle is not significantly impaired by thermal inactivation. Heat treatment of snu114ΔN extracts resulted in accumulation of arrested spliceosomes in which the U4 RNA was not efficiently released, and we show that U4 is still base paired with the U6 RNA. This suggests that Snu114p is involved, directly or indirectly, in the U4/U6 unwinding, an essential step towards spliceosome activation.
INTRODUCTION
Splicing of mRNA precursors (pre-mRNA) proceeds via two consecutive transesterification steps that are catalysed by the spliceosome. The spliceosome is formed by the ordered interaction of the U1, U2, U5 and U4/U6 snRNPs (in which the U4 and U6 RNAs are base paired) and of several splicing factors with the pre-mRNA (Burge et al., 1999). The splicing complex is formed by the initial interaction of the U1 snRNP with the 5′ splice site (ss), whereafter the U2 snRNP recognizes the branch site to form the pre-spliceosome. Spliceosome assembly is completed by the subsequent association of the U4/U6 and U5 snRNPs as an [U4/U6.U5] tri-snRNP complex. Activation of the assembled spliceosome into a catalytically active machine requires a dramatic reorganization of the components and remains idle until a ‘switch is flipped’. Thus, during spliceosome activation, base pairing between U4 and U6 is disrupted and a new base-pairing interaction between U2 and U6 is formed (reviewed in Nilsen, 1998). Concomitant with these events, base pairing of U1 to the 5′ ss is exchanged for base pairing between U6 and the 5′ ss (Kandels-Lewis and Séraphin, 1993; Lesser and Guthrie, 1993). After these rearrangements, U1 and U4 snRNPs are released from the spliceosome prior to catalysis. The conserved loop 1 of U5 RNA contacts exonic sequences at the 5′ and 3′ ss, while the splicing reaction proceeds (Sontheimer and Steitz, 1993). The U5 snRNP plays a central role in both spliceosome assembly and splicing. Studies of the human 20S U5 snRNP have shed light on its composition and organization. It contains, in addition to the seven common Sm proteins, several particle-specific proteins (Will and Lührmann, 2001).
The U4/U6 unwinding is an important step towards the activation of the spliceosome and likely depends on the function of a large number of protein factors, several of which are associated with the U5 snRNP. One such factor is Prp8p (in man 220K), which has been shown to regulate the unwinding of U4 from U6 by way of protein–protein interactions (Kuhn et al., 1999). Indeed, within the U5 snRNP, the human orthologue of Prp8p forms a stable RNA-free complex with several U5 proteins. These include the ATP-dependent RNA unwindase 200K/Brr2p, which has been shown to unwind the base pairing between U4 and U6 in vitro, and the 116K/Snu114p, which is homologous to the elongation factor EF-2, a GTPase that catalyses the rearrangement of the ribosome during the translocation step (Fabrizio et al., 1997; Achsel et al., 1998; Laggerbauer et al., 1998; Raghunathan and Guthrie, 1998). In yeast, the sequential recognition of the 5′ ss by U1 and subsequently by U6 has been shown to be mediated by another ATP-dependent RNA helicase associated with the U5 snRNP: the Prp28p (Staley and Guthrie, 1999; Chen et al., 2001; Stevens et al., 2001). This raises the possibility that the sequence of multiple structural rearrangements during spliceosome activation is facilitated by several ATPases associated with the U5 snRNP.
We have identified Snu114p (a Saccharomyces cerevisiae protein essential for splicing in vivo) and its human orthologue 116K as close homologues of the ribosomal elongation factor EF-2 (Fabrizio et al., 1997). Aside from an N-terminal acidic domain that is not present in EF-2, Snu114p/116K exhibits the same domain structure as EF-2, including the G-domain. This suggests that the two proteins may also share some functional properties. Indeed, several amino acid substitutions in the P loop of the GTP-binding domain of Snu114p are lethal, suggesting that GTP binding and hydrolysis are important for its function. To investigate whether Snu114p, as predicted by its homology to EF-2, is involved in remodelling the spliceosomal RNP structure, we generated conditionally lethal mutations. One such mutant was obtained by deleting the N-terminal 128 amino acids (snu114ΔN). We show that this deletion leads to a temperature-sensitive phenotype and an in vivo and in vitro splicing defect at 37°C. Interestingly, the thermal inactivation results in accumulation of arrested spliceosomes in which U4 is still base paired with the U6 RNA, indicating that Snu114p is involved in the dissociation of the U4/U6 duplex.
RESULTS AND DISCUSSION
The N-terminal domain of Snu114p is evolutionarily conserved and its deletion leads to a temperature-sensitive phenotype
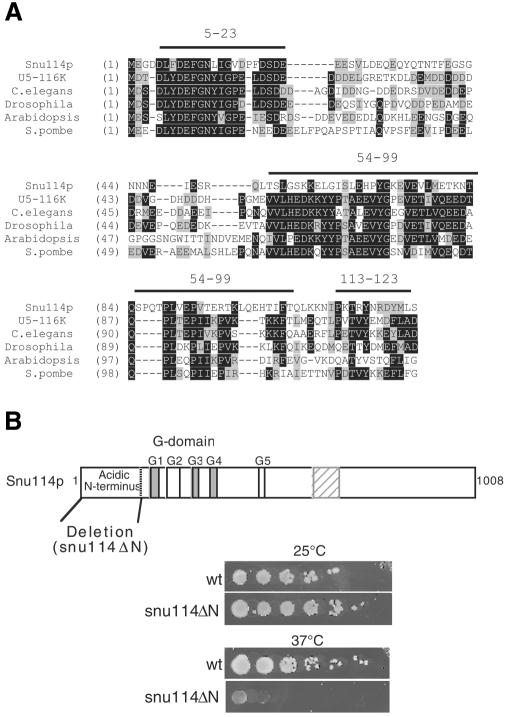
We have shown previously that Snu114p/116K exhibits the same domain structure as EF-2, aside from an acidic N-terminal domain not present in EF-2 (Fabrizio et al., 1997). A new database search now identified several more proteins with this conserved N-terminal domain followed by the overall domain structure of EF-2. The N-terminal extension makes them a separate group of orthologues, distinct from elongation factors. Figure 1A shows an alignment of ∼125 amino acids of the N-terminal domain of putative orthologues of Snu114p. This domain is evolutionarily conserved between budding yeast (Snu114p), man (U5-116K), nematodes, flies, plants and fission yeast (23–32% identity and 45–51% similarity with Snu114p). Three blocks of conserved amino acids, which are rich in hydrophobic residues, are observed and, with the exception of the first block (5–23), are more clearly conserved among the other proteins than in Snu114p. This evolutionary conservation suggests that this domain is important for spliceosomal-specific functions.
Fig. 1. The N-terminal domain of Snu114p is evolutionarily conserved. (A) Alignment of ∼125 amino acids of the N-terminal domains of potential orthologues of Snu114p from S. cerevisiae and other proteins that display sequence similarity. Lines above the sequences indicate evolutionarily conserved hydrophobic blocks (the numbering follows the Snu114p sequence). Identical residues are highlighted in black and similar residues in grey. (B) Block diagram of the primary structure of Snu114p. G1 to G5 are the conserved motifs of the G-domain. Below the diagram, yeast cells carrying the wild type (wt) and the temperature-sensitive deletion mutant snu114ΔN are shown after growth at 25 and 37°C, respectively.
To investigate whether Snu114p is involved in remodelling RNA structures required for splicing, we searched for conditionally lethal mutants. The N-terminal domain deletion mutant (snu114ΔN) was obtained by removing the first 128 amino acids from wild-type Snu114p. As shown in Figure 1B, this deletion results in a temperature-sensitive phenotype at 37°C. This indicates that the N-terminal domain, as predicted by its evolutionary conservation, has an important function so that cells lacking it grow at 25°C but die at 37°C.
snu114ΔN leads to a temperature-dependent pre-mRNA splicing defect in vivo and in vitro
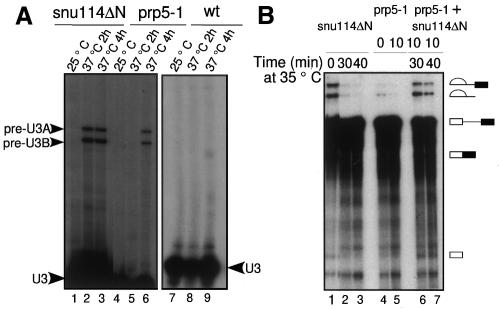
Next, we investigated whether the lethality at 37°C of cells carrying snu114ΔN was due to a splicing defect. The involvement of snu114ΔN in pre-mRNA splicing in vivo was studied using primer extension analysis of U3 transcripts isolated from snu114ΔN and wild-type cells grown at the permissive and non-permissive temperatures (Figure 2A). Indeed, the snu114ΔN temperature-sensitive deletion leads to a splicing defect that is seen in vivo after the strain has been incubated at the non-permissive temperature for 2 or 4 h (lanes 2 and 3). Unspliced U3A and U3B precursors accumulate, as can also be seen with the well-characterized prp5-1 mutant used as a control (lane 6). With the wild-type strain, no accumulation of unspliced U3A and U3B precursors was seen at either 25 or 37°C (lanes 7–9).
Fig. 2. Deletion of the N-terminal domain (snu114ΔN) leads to a splicing defect in vivo and in vitro. (A) Total RNA was extracted from snu114ΔN as well as wild-type (wt) cells grown at 25°C (lanes 1 and 7) and after the switch to 37°C for 2 h (lane 2 and 8) and 4 h (lanes 3 and 9). Primer extension was performed to measure the levels of unspliced pre-U3A/U3B transcripts. (B) In vitro splicing reactions were performed at 25°C using snu114ΔN and prp5-1 extracts inactivated at 35°C for the time indicated above each lane. Splicing activity complementation was obtained by mixing equal volumes of the two heat-inactivated extracts prior to the splicing reaction (lanes 6 and 7). The identity of the 32P-labeled RNA species is indicated (from top to bottom): intron-lariat-exon 2 intermediate, excised lariat-intron, pre-mRNA, mature mRNA and cleaved exon 1 intermediate.
To investigate whether snu114ΔN leads to a splicing defect in vitro, we prepared splicing extracts from the corresponding strain and analysed splicing activity in vitro before and after thermal inactivation (Figure 2B). We inactivated splicing extracts at 35°C in the presence of high KPO4, prior to the addition of the pre-mRNA, and withdrew aliquots of the reaction mixture at the times indicated in Figure 2B. Splicing was performed by adding the actin pre-mRNA and incubating the reaction for 25 min at 25°C. After treatment at the non-permissive temperature, the system with snu114ΔN showed a significant decrease of splicing prior to the first step (lanes 2 and 3). To determine whether this inhibition can be ascribed to the specific inactivation of snu114ΔN, we tested whether splicing activity could be restored by complementing two heat-inactivated extracts from the two mutants of Figure 2A. The complementation efficiently reversed the splicing defect (Figure 2B, lanes 6 and 7). We conclude that the splicing defect in vitro is due to the inactivation of snu114ΔN (and/or to the weak interactions resulting from this inactivation) but not unspecific random inactivation of other factors.
A splicing defect was also observed by incubating the splicing reaction, including the pre-mRNA, directly at 30°C. The temperature-induced splicing defect obtained with these conditions was removed by cooling to 25°C (data not shown).
Tri-snRNP particles are present in snu114ΔN extracts heat inactivated in vitro
Snu114p is associated with U5 and [U4/U6.U5] snRNP particles (Fabrizio et al., 1997; Gottschalk et al., 1999; Stevens and Abelson, 1999). To understand whether the splicing defect observed with snu114ΔN extracts is due to loss of the mutant protein from tri-snRNP particles and/or insufficient levels of properly formed tri-snRNP, we investigated wild-type and mutant tri-snRNPs by immunoprecipitation and glycerol gradient centrifugation. The immunoprecipitation revealed that snu114ΔN, although lacking ∼130 amino acids, associates with the tri-snRNP particle (data not shown). Glycerol gradient centrifugation verified that, even after heat treatment (35°C), plenty of tri-snRNP particles were present in the mutant extract (∼50–80%, when compared with the wild type) (Figure 3). However, either before or after inactivation, the levels of the U5 snRNP declined to a certain extent. A U5 snRNP with a reduced sedimentation velocity (∼10–12S) appears in the mutant, the majority being approximately in fractions 6–11 instead of fractions 8–13. This indicates that snu114ΔN may lead to a reduced stability of the U5 snRNP complex, so that some of its associated factors are less stably associated in the mutant. This implies that the N-terminus of Snu114p may be required to connect Snu114p with other proteins within the U5. It appears that the amount of U4/U6 snRNPs increases concomitantly, especially after heat treatment. This may be due to partial dissociation of the tri-snRNP particles or to a weaker association of U5 with U4/U6 snRNPs. Whether this effect also appears in vivo is not known. However, since the tri-snRNP build up, we can hypothesize that in vivo, at least at the permissive temperature, plenty of U5 snRNP is available to form a functional tri-snRNP particle. In conclusion, the splicing defect seen in vitro must be due to the inhibition of a subsequent step.
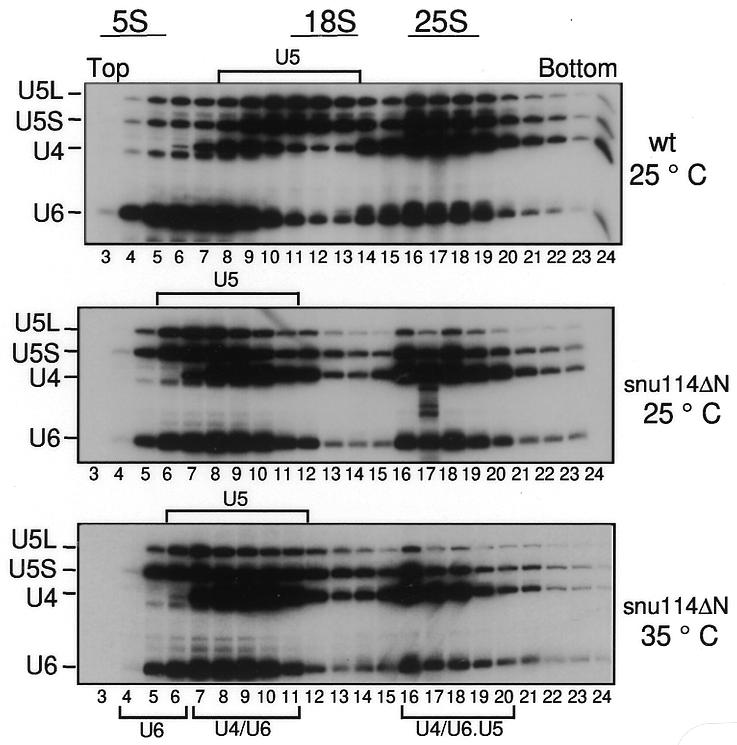
Fig. 3. Tri-snRNP particles are present in snu114ΔN extracts heat inactivated in vitro. Wild-type (wt) and snu114ΔN extracts were treated at 25 or 35°C for 35 min and then sedimented on a 10–30% glycerol gradient. Fractions were analysed for RNA contents by northern blotting.
Snu114p participates in spliceosome activation
To investigate whether tri-snRNP particles carrying snu114ΔN are integrated into mature spliceosomes, we used an affinity-purification technique. The extract was first heat inactivated at 35°C; afterwards, biotinylated pre-mRNA was added and the mixture was incubated at 25°C under splicing conditions for the time indicated (Figure 4A). Splicing reactions were performed using either wild-type (lanes 1–5) or snu114ΔN (lanes 6–10) extracts. Splicing complexes associated with the biotinylated pre-mRNA were subsequently isolated using streptavidin beads, and their RNA moieties were analysed by northern blot analysis. Figure 4A (lanes 1 and 6) shows parallel reactions using non-biotinylated substrates. At the earliest time point (1.5 min), tri-snRNP particles join the pre-spliceosome, here represented by the U1 and U2 RNAs (lanes 2 and 7). In the wild-type extract, the spliceosome is activated for catalysis after binding of the tri-snRNP particle. The activation sign is the characteristic unwinding and release of U4 RNA from the yeast spliceosomal complex prior to the first pre-mRNA cleavage-ligation step. This release is reflected in the time-dependent decrease in the yield of spliceosomal U4 RNA relative to U5 and U6 RNAs (Figure 4A, lanes 2–5). Furthermore, the U1/5′ ss interaction is disrupted and the U1 snRNP is less stably bound to the spliceosome. Consistently, the level of the U1 RNA decreases between 25 and 65 min (lane 4 and 5) (see Supplementary data available at EMBO reports Online for a lighter exposure). Quantifying the U4 and U6 RNA bands by phosphorimager analysis showed that the U4/U6 signal ratio decreased approximately from 0.9 after 1.5 min to 0.4 after 65 min of incubation (see Figure 4B for a graphical representation of the U4 RNA release).
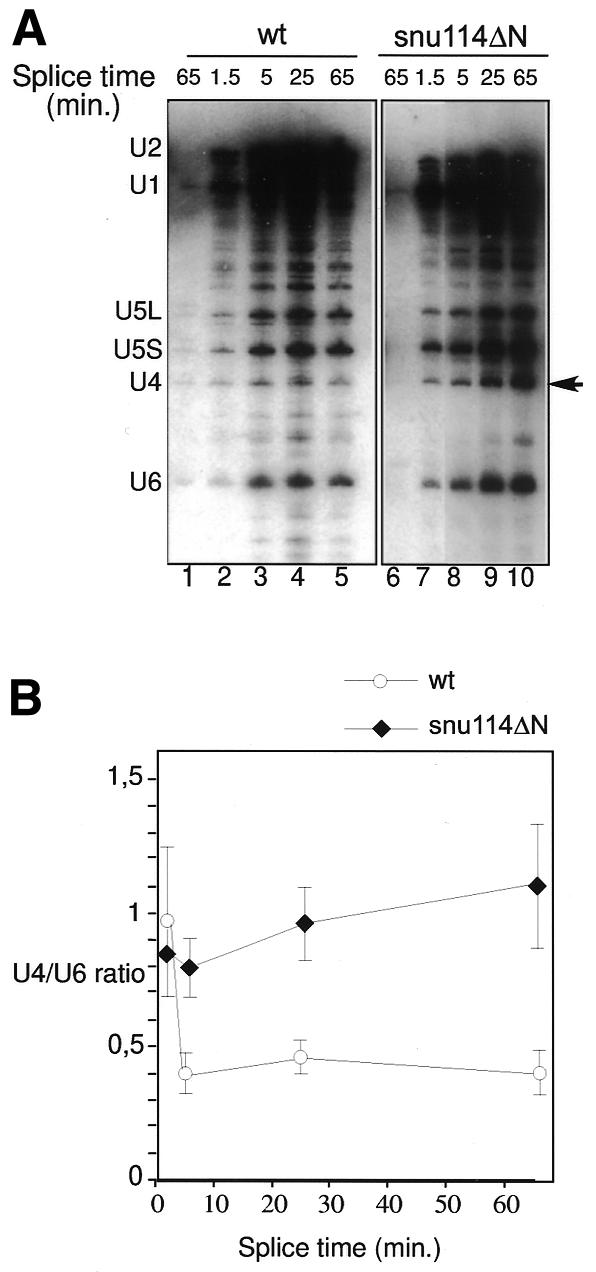
Fig. 4. Snu114p participates in spliceosome activation. (A) Extracts were heat inactivated for 25 min at 35°C, and then biotinylated pre-mRNA was added and incubated at 25°C under splicing conditions for the time indicated (splice time). Splicing reactions were performed using either wild-type (wt; lanes 1–5) or snu114ΔN (lanes 6–10) extracts. Splicing complexes were isolated using streptavidin beads, and their RNA content was analysed by northern blotting. Lanes 1 and 6 are control reactions using non-biotinylated substrates. The arrow points to the U4 RNA accumulating in the snu114ΔN mutant. (B) Plot of the U4 RNA release in the wild-type (wt) and snu114ΔN spliceosomes. The U4 and U6 RNAs bands were quantified by phosphorimager analysis and the U4/U6 ratio was plotted. The graph was obtained from five independent experiments.
The pattern of tri-snRNP addition to pre-spliceosomes in the mutant snu114ΔN was similar to that seen in the splicing-competent wild-type extract (Figure 4A, lane 7), with one striking distinction. With snu114ΔN, the U4 RNA is no longer efficiently released from the spliceosome and a time-dependent increase in both U4 and U1 RNAs is seen. The U4/U6 ratio increases from 0.83 after 1.5 min (Figure 4B and A, lane 7) to 1.1 after 65 min (Figure 4B and A, lane 10). These results suggest that Snu114p is required for mediating the dissociation of the U4 RNA and the concomitant release of U1 RNA, which are crucial rearrangements towards spliceosome activation.
U4 RNA remains bound to U6 RNA in snu114ΔN heat-inactivated spliceosomes
The observed retention of U4 RNA within the snu114ΔN spliceosome at elevated temperatures may be due to an insufficient unwinding of the U4/U6 RNA duplex or, alternatively, to hyperstabilization of U4 RNA within the spliceosome after its unwinding from U6 RNA. To distinguish between these two possibilities, wild-type and snu114ΔN spliceosomes were assembled in the presence of biotinylated pre-mRNA at the non-permissive temperature (30°C). Using these conditions, an in vitro splicing defect was also observed (data not shown). Aliquots were removed at the indicated time points and spliceosomes affinity purified as described above. Spliceosomal RNAs were isolated and fractionated on an SDS gel under conditions that resolve the U4/U6 RNA hybrid from free U4 and U6 RNAs. RNAs were analysed by hybridizing a northern blot with U4- and U6-specific probes (Figure 5). In the wild type, the U4/U6 duplex can be clearly observed after 1.5 min of incubation (Figure 5, lane 4). However, after only 25 min, the amount of U4/U6 duplex decreases, suggesting that U4 has been unwound from U6 RNA and released (Figure 5A, lane 5). This is consistent with the data presented for the wild-type strain in Figure 4A. In contrast, heat-inactivated snu114ΔN spliceosomes retain a paired U4/U6 duplex even after 75 min of incubation (Figure 5, lane 9). This indicates that the decreased dissociation of U4 RNA from the spliceosome (Figure 4A, lane 10) is mainly due to a reduced unwinding of U4 from U6 RNA. This suggests that the thermosensitivity of the snu114ΔN mutant is possibly due to an aberrant conformation at 30–35°C that affects the interaction with other factors involved in U4/U6 unwinding. As a consequence, spliceosomes retain a paired U4/U6 duplex. The N-terminal domain may be required to stabilize these protein–protein interactions at high temperatures.
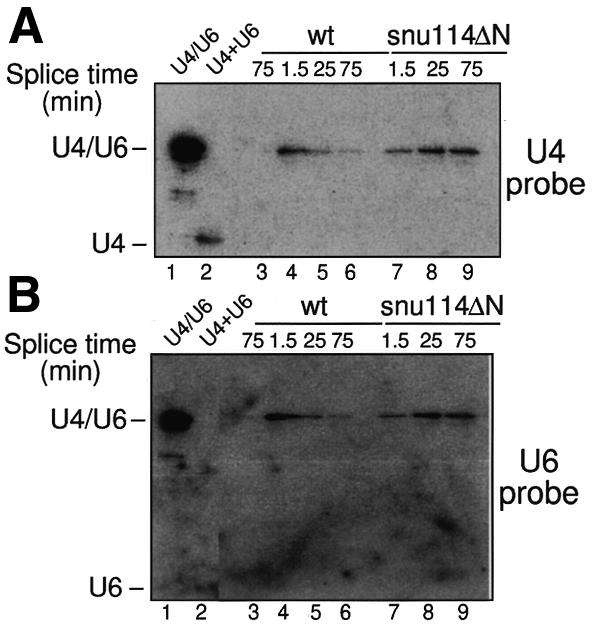
Fig. 5. U4 RNA remains bound to U6 RNA in snu114ΔN heat-inactivated spliceosomes. Wild-type (wt; lanes 4–6) and snu114ΔN (lanes 7–9) spliceosomes were assembled at non-permissive temperature (30°C) using a biotinylated actin pre-mRNA and were affinity purified on streptavidin beads. Spliceosomal RNAs were fractionated on an SDS gel. RNAs were analysed by northern blotting after probing the membrane with U4 (A) and U6 (B) probes. Lane 3 is a control reaction using non-biotinylated substrate. The U4/U6 duplex (lane 1) was obtained after phenol–chloroform extraction at 4°C of the reaction shown in lane 6, before affinity purification. In lane 2, the U4/U6 duplex was dissociated by heating the sample for 5 min at 95°C. (The free U6 RNA is only weakly visible.)
In addition to the putative U4/U6 helicase Brr2p, four protein splicing factors have previously been implicated in U4 RNA release during spliceosome activation: the U4/U6 snRNP protein Prp4p (Ayadi et al., 1997), the non-snRNP protein Prp19p (Tarn et al., 1993), the tri-snRNP protein Prp38p and the U5 snRNP protein Prp8p (Xie et al., 1998; Kuhn et al., 1999). The splicing arrest due to the loss of Prp38p function and to the mutant U4-cs1 (suppressed by prp8-201) has been characterized in part for what concern these proteins involvement in the U4/U6 unwinding. The effect of snu114ΔN in the U4/U6 rearrangement seems to be similar. Inactivation of either Prp38p or U4-cs1 results in the accumulation of arrested spliceosomes in which U4 RNA is still paired with U6 RNA (Xie et al., 1998; Kuhn et al., 1999). A link between Snu114p, Prp8p and the putative U4/U6 helicase Brr2p is indicated by several lines of evidence. The most direct of these is the strong RNA-free protein–protein interaction that has been detected in the human system between the 220K/Prp8p, the 200K/Brr2p and the 116K/Snu114p after dissociation from the U5 snRNP in high-salt buffer (Achsel et al., 1998). In yeast, cross-linking studies have shown that Snu114p is in contact with the U5 RNA 5′ internal loop 1 and is therefore in very close proximity to Prp8p, which also cross-links to this loop (Dix et al., 1998). Recently, an interaction between Prp8p and Brr2p was also detected by the yeast two-hybrid system (van Nues and Beggs, 2001). When bound at the 5′ ss, Prp8p may sequentially coordinate and recruit not only Brr2p but also at least one additional protein that catalyses rearrangements required for spliceosome activation: the 100K/Prp28p. In addition, Prp8p is the predominant interacting factor of Snu114p in two-hybrid screens (Fromont-Racine et al., 1997; Dix et al., 1998), and biochemical analyses demonstrated that the 220K protein binds strongly and directly to the 116K protein (Achsel et al., 1998). It is possible, therefore, that the G protein Snu114p is a pivotal factor that acts downstream to promote the function of Prp8p and the Brr2p/200K RNA unwindase in this intricate network of factors involved in spliceosome activation. Since deletion of the N-terminal region of Snu114p led to the temperature-sensitive phenotype, this further suggests that this part of Snu114p may be a critical element for the communication between Snu114p and other U5 proteins required for spliceosome activation. Future work will be aimed at defining the details of the reaction that Snu114p catalyses in concert with other factors and whether binding and hydrolysis of GTP by Snu114p are required for spliceosome activation.
METHODS
Splicing, affinity purification and glycerol gradients.
YPF36 cells (see Supplementary data) were grown at 25°C, and splicing extracts were prepared. For thermal inactivation in vitro, we used two protocols; these gave similar results. In protocol I, the extracts were incubated in high KPO4 (120 mM) at 35°C prior to the splicing reaction for the times indicated in Figure 2B, or else they were incubated for 35 min at 35°C (Figures 3 and 4A). Subsequently, the splicing reactions were performed for 25 min at 25°C under standard splicing conditions. In the experiment shown in Figure 5, heat inactivation was performed by incubating the splicing reaction (protocol II) at 30°C instead of 25°C. In vitro splicing reactions, affinity purification of spliceosomes, glycerol gradients and primer extensions were performed as described previously (Gottschalk et al., 1999). For U4/U6 duplex analysis, a phenol–chloroform extraction was performed at 4°C. The duplex formation was analysed at 4°C on an 11% SDS gel, which was followed by northern blot analysis.
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Rauhut for critical comments, D. Tetzlaff for initial experiments, J. Beggs for the gift of anti-Prp8p antibodies, and D. Meyer and M. Killian for technical assistance. This work was supported by a Deutsche Forschungsgemeinschaft grant (to P.F. and R.L).
REFERENCES
- Achsel T., Ahrens, K., Brahms, H., Teigelkamp, S. and Lührmann, R. (1998) The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol., 18, 6756–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi L., Miller, M. and Banroques, J. (1997) Mutations within the yeast U4/U6 snRNP protein Prp4 affect a late stage of spliceosome assembly. RNA, 3, 197–209. [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl, T. and Sharp, P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland, R.F. and Atkins, J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Chen J.Y., Stands, L., Staley, J.P., Jackups, R.R., Jr, Latus, L.J. and Chang, T.H. (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell, 7, 227–232. [DOI] [PubMed] [Google Scholar]
- Dix I., Russell, C.S., O’Keefe, R.T., Newman, A.J. and Beggs, J.D. (1998) Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA, 4, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Laggerbauer, B., Lauber, J., Lane, W.S. and Lührmann, R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J., 16, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain, J.C. and Legrain, P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer, G., Banroques, J., Mann, M., Lührmann, R. and Fabrizio, P. (1999) Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J., 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis S. and Séraphin, B. (1993) Involvement of U6 snRNA in 5′ splice site selection. Science, 262, 2035–2039. [DOI] [PubMed] [Google Scholar]
- Kuhn A.N., Li, Z. and Brow, D.A. (1999) Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell, 3, 65–75. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B., Achsel, T. and Lührmann, R. (1998) The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA, 95, 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser C.F. and Guthrie, C. (1993) Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science, 262, 1982–1988. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In Simons, R. and Grundber-Manago, M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 279–307.
- Raghunathan P.L. and Guthrie, C. (1998) RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol., 8, 847–855. [DOI] [PubMed] [Google Scholar]
- Sontheimer E.J. and Steitz, J.A. (1993) The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science, 262, 1989–1996. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie, C. (1999) An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell, 3, 55–64. [DOI] [PubMed] [Google Scholar]
- Stevens S.W. and Abelson, J. (1999) Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA, 96, 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W., Barta, I., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D. and Abelson, J. (2001) Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA, 7, 1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Tarn W.Y., Lee, K.R. and Cheng, S.C. (1993) Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl Acad. Sci. USA, 90, 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues R.W. and Beggs, J.D. (2001) Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics, 157, 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann, R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Xie J., Beickman, K., Otte, E. and Rymond, B.C. (1998) Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J., 17, 2938–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.