Abstract
Splicing and 3′-end processing (including cleavage and polyadenylation) of vertebrate pre-mRNAs are tightly coupled events that contribute to the extensive molecular network that coordinates gene expression. Sequences within the terminal intron of genes are essential to stimulate pre-mRNA 3′-end processing, although the factors mediating this effect are unknown. Here, we show that the pyrimidine tract of the last splice acceptor site of the human β-globin gene is necessary to stimulate mRNA 3′-end formation in vivo and binds the U2AF 65 splicing factor. Naturally occurring β-thalassaemia-causing mutations within the pyrimidine tract reduces both U2AF 65 binding and 3′-end cleavage efficiency. Significantly, a fusion protein containing U2AF 65, when tethered upstream of a cleavage/polyadenylation site, increases 3′-end cleavage efficiency in vitro and in vivo. Therefore, we propose that U2AF 65 promotes 3′-end processing, which contributes to 3′-terminal exon definition.
INTRODUCTION
Pre-mRNA 3′-end processing does not occur independently of the other steps of gene expression but contributes to a general coordination of gene expression (Maniatis and Reed, 2002; Proudfoot et al., 2002). Recent studies, for example, have provided a link between the cleavage/polyadenylation step and the removal of introns from pre-mRNAs. A direct functional influence of introns on the efficiency of 3′-end processing was shown in vitro by using chimeric splicing/polyadenylation precursor RNAs in which the presence of a 3′ splice site increased the rate of cleavage at a downstream poly(A)-addition site (Niwa et al., 1990; Cooke et al., 1999). A physical link for this coupling is also supported by various in vitro studies showing interactions between splicing and polyadenylation factors. The U1A protein of U1 snRNP bound to the SV40 late pre-mRNA was shown to interact with the 160 kDa subunit of cleavage/polyadenylation specificity factor to increase polyadenylation activity (Lutz et al., 1996), whereas the U1A protein bound to its own transcript interacts with poly(A) polymerase (PAP) to inhibit its activity (Gunderson et al., 1994). The U1 70K protein of U1 snRNP, bound to a 5′ splice site in the vicinity of the 3′-end processing signals of the bovine popilloma virus late pre-mRNA, interacts with PAP to inhibit polyadenylation (Gunderson et al., 1998). Finally, PAP is also able to stimulate splicing through its interaction with the splicing factor U2AF 65 (Gunderson et al., 1997; Vagner et al., 2000). Evidence from in vivo studies have also shown that the last intron of the human triosephosphate isomerase transcript is involved in 3′-end formation (Nesic et al., 1993; Nesic and Maquat, 1994). Transient (Liu and Mertz, 1996) and stable (Antoniou et al., 1998) transfection assays of various mutant human β-globin genes have shown that the 3′ splice site region of the second and last intron (βIVS-II) is a main determinant of the efficiency of 3′-end formation. All these data demonstrate that there is a relation between the last splice acceptor and the cleavage/polyadenylation sites of a pre-mRNA to define the last exon. However, the nature of the protein factor(s) that would mediate the stimulatory effect of the 3′ splice site on 3′-end formation has remained unknown.
The absolute requirement for the splice acceptor site region of the second and last β-globin intron (βIVS-II) to promote efficient 3′-end formation (Antoniou et al., 1998), combined with the occurrence of β-thalassaemia-causing mutations within the βIVS-II pyrimidine (PYR) tract in patients (Antoniou, 1995), led us to study the mechanism by which splicing signals influence 3′-end processing using the human β-globin gene as a model system.
RESULTS AND DISCUSSION
Mutations to the PYR tract of the β-globin last acceptor splice site reduce the efficiency of 3′-end formation
We began this investigation by constructing human β-globin genes possessing the following β-thalassaemia-causing mutations within the βIVS-II PYR tract. Single-mutant pyrimidine (SM-PYR) consisted of a single (C→A) mutation at –3 from the 3′ splice acceptor site; double-mutant pyrimidine (DM-PYR) harboured –7 C→G and –8 U→G substitutions; triple-mutant pyrimidine (TM-PYR) combined all three of the above point mutations. These mutant β-globin genes were stably transfected into murine erythroleukaemia (MEL) cells and expression analysed in cytoplasmic and nuclear RNA fractions after induced erythroid differentiation using an S1-nuclease protection assay. The human β-globin, end-labelled DNA probe employed allows the simultaneous detection of 3′ cleaved and uncleaved transcripts (Figure 1A). The ratio of the nuclear steady-state levels of 3′ cleaved (C) and uncleaved (U) products was used as a measure of the efficiency of 3′-end formation. Analysis of nuclear RNA from MEL cells transfected with β-globin genes harbouring the SM-PYR and DM-PYR mutations reduced the 3′ cleaved-to-uncleaved (C/U) ratio to ∼2 (from 7 with the wild-type RNA), implying an efficiency of 3′-end formation of ∼20% of the wild type (Figure 1B). The TM-PYR mutant reduced this still further to 0.5, implying an efficiency of 3′-end formation of 7% of the wild type (Figure 1B). Therefore, these β-thalassaemia-causing mutations appear to exert their effect not only by impairment of U2 snRNP binding to the lariat branch site (Ruskin et al., 1988) and subsequent inhibition of splicing (Sebillon et al., 1995) but also through a reduction in the efficiency of 3′-end formation.
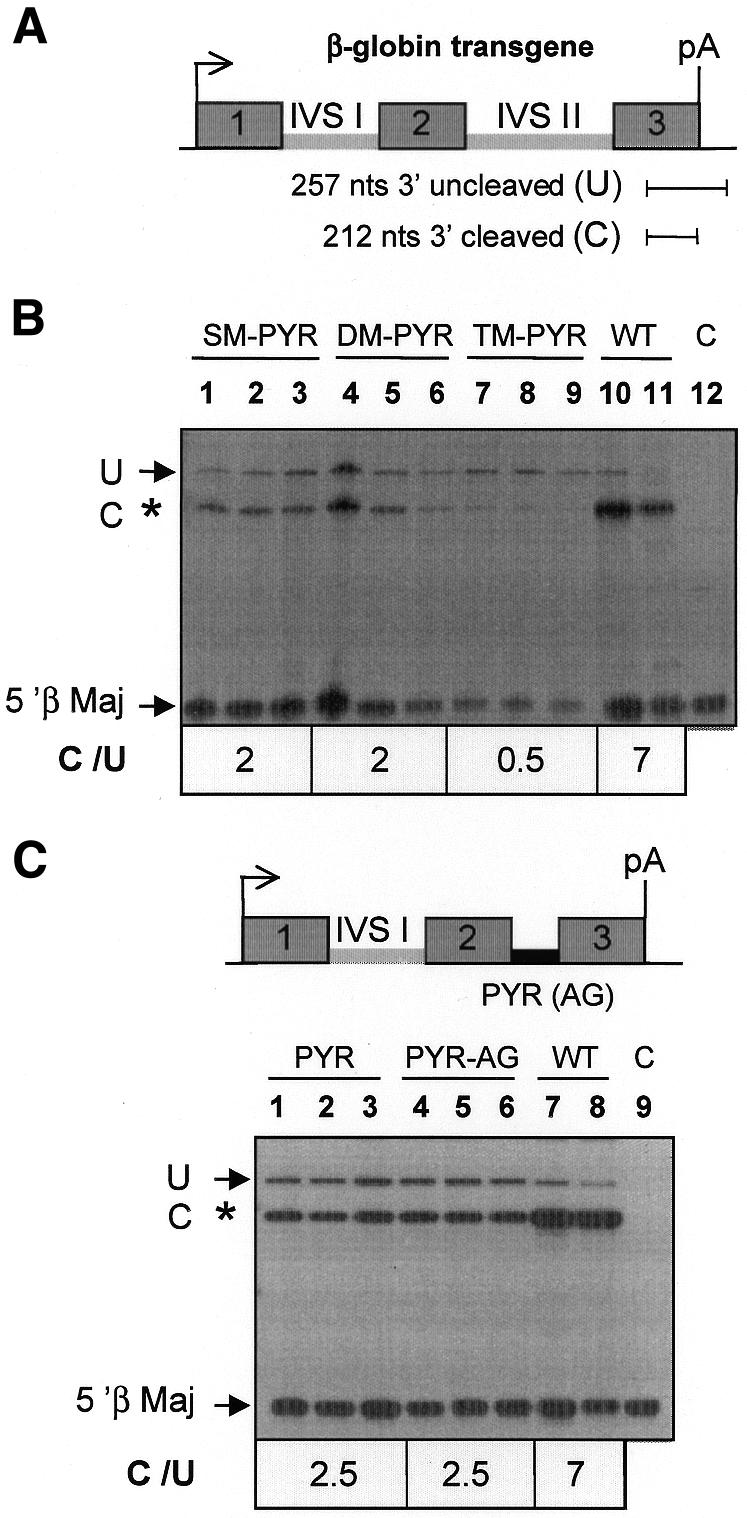
Fig. 1. The polypyrimidine tract of βIVS-II induces efficient 3′-end formation. Nuclear RNAs (5 µg) from MEL cells transfected with human β-globin genes harbouring mutations of βIVS-II were analysed for transgene expression by an S1-nuclease protection assay after 4 days of induced erythroid differentiation. (A) The probe used for S1-nuclease protection assay detects transcripts that have not undergone correct 3′ cleavage as a 257-nucleotide product (U) and correctly cleaved RNA as 212-nucleotide fragments (C). Simultaneous probing for mouse β-major globin (5′ β Maj) mRNA acted as an internal reference control. (B) Lanes 1–3, SM-PYR, single (C→A) mutation at –3 from the 3′ splice acceptor site; lanes 4–6, DM-PYR, double PYR-tract mutation at –7 C→G and –8 U→G; lanes 7–9, TM-PYR, triple mutant combining all three of the point mutations of SM-PYR and DM-PYR. Lanes 10–11, WT, wild-type β-globin gene; lane C, total RNA from untransfected MEL cells. C/U represents the 3′ cleaved-to-uncleaved ratio. (C) S1-nuclease protection analysis with human β-globin genes in which the βIVS-II intron was replaced by a 21-nucleotide-long sequence corresponding to the PYR tract of this intron either with a wild-type (PYR-AG) or a mutant (PYR) 3′ AG splice acceptor site.
The terminal PYR tract alone supports efficient 3′-end formation
We next assessed the potential role of the PYR tract alone in promoting pre-mRNA 3′ cleavage by analysing mutant β-globin genes containing a 21-nucleotide sequence corresponding to the PYR tract either with (PYR-AG) or without (PYR) the AG 3′ splice site (Figure 1C) inserted in the βIVS-II position, thereby replacing the normal intron. The results show that both mutants give a C/U ratio of 2.5, giving an efficiency of 3′-end formation of 35% of the wild type (Figure 1C). Interestingly, both mutants gave cytoplasmic mRNA levels that were also ∼30% of the wild-type gene. The cytoplasmic PYR-AG and PYR mutant mRNAs were, as expected, not spliced at the βIVS-II position and therefore contained the inserted 21 bases oligonucleotide PYR tract sequences (data not shown). These data further demonstrate that the βIVS-II PYR tract alone is able to promote efficient mRNA 3′-end formation.
The terminal PYR tract binds U2AF 65
UV cross-linking and immunoprecipitation experiments were then conducted to address the nature of the protein factor(s) that bind to and may mediate the effect of the βIVS-II PYR tract in promoting efficient 3′-end formation. UV cross-linking using a 32P-end-labelled RNA oligonucleotide corresponding to the βIVS-II PYR tract in the presence of MEL nuclear extract gives a single species of ∼65 kDa (Figure 2A). A similar band was observed with a PYR tract triple-mutant oligonucleotide (Figure 2A). UV cross-linking followed by immunoprecipitation with the monoclonal antibody MC3 specific for U2AF 65 (Gama-Carvalho et al., 1997) gave an immunoprecipitated complex with both wild-type PYR and TM-PYR (Figure 2B). Interestingly, the efficiency of U2AF 65 binding to TM-PYR was only ∼30% of wild-type PYR, which correlates with its reduced ability to promote 3′-end formation. Similar results were obtained with HeLa nuclear extracts (data not shown). Furthermore, antibodies against the other known PYR-tract-binding proteins PTB (Garcia-Blanco et al., 1989) and hnRNP C (Swanson and Dreyfuss, 1988) failed to give an immunoprecipitated product (data not shown). These data indicate that U2AF 65 binds to the βIVS-II PYR tract. Moreover, β-thalassaemia-causing mutations in the PYR tract reduces this binding.
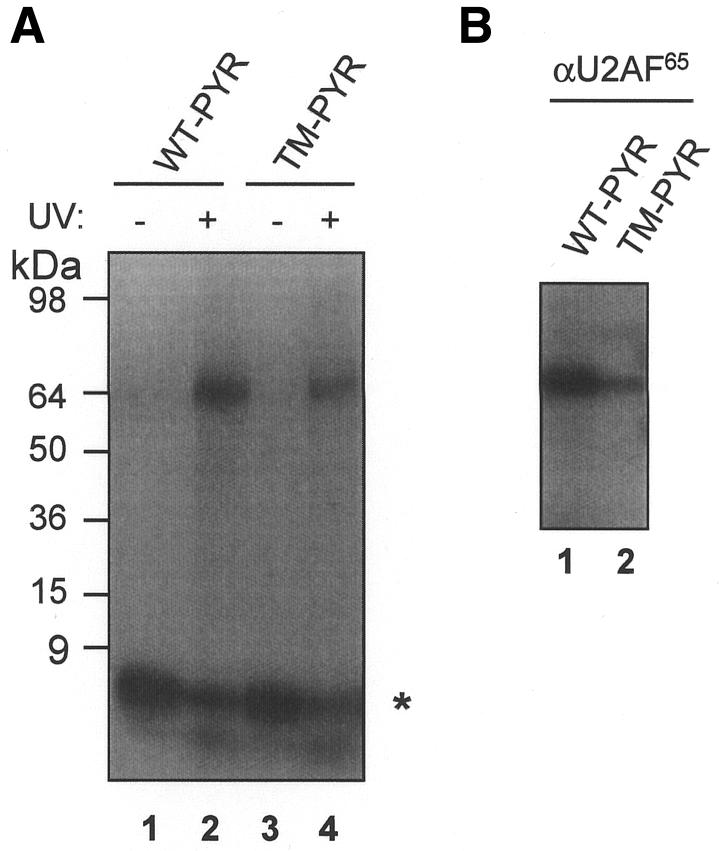
Fig. 2. U2AF 65 binding to the wild-type (WT) or triple-mutant (TM) human β-globin IVS-II PYR tract. (A) UV cross-linking of 5′ 32P-labelled RNA oligonucleotides and MEL cell nuclear extracts and resolution by SDS–PAGE. Lanes 2 and 4, UV-irradiated samples; lanes 1 and 3, non-UV-irradiated controls. The migration of the free RNA oligonucleotides (asterisk) and of the protein size markers are indicated. (B) UV cross-linking/immunoprecipitation. After incubation of the two RNA oligonucleotides in MEL cell extracts and UV irradiation, immunoprecipitation was performed with the MC3 monoclonal antibody against U2AF 65 followed by analysis by SDS–PAGE.
U2AF 65 stimulates in vivo and in vitro 3′-end processing
In order to demonstrate the role of U2AF on 3′-end cleavage, we wished to tether this splicing factor to the RNA upstream of a cleavage/polyadenylation site and to examine its effect on the efficiency of 3′-end formation. Human U2AF (Zamore and Green, 1989) is composed of two subunits of 65 kDa (U2AF 65) and 35 kDa (U2AF 35), but only the large subunit U2AF 65 contacts the polypyrimidine tract directly. U2AF 65 or U2AF 35 were therefore fused to an RNA-binding domain that could be used to position it on a pre-mRNA devoid of the polypyrimidine tract. The R17 phage coat protein, which binds a 72-nucleotide-long RNA sequence with high affinity and specificity (Bardwell and Wickens, 1990), was chosen to tether U2AF 65 or U2AF 35 to the RNA. A human β-globin gene harbouring a R17-binding site in place of βIVS-II (β-R17; Figure 3A) was stably transfected into MEL cells alone or together with plasmids encoding either R17–U2AF 65 or R17–U2AF 35 fusion proteins. Transgene expression was analysed in nuclear RNA fractions after induced erythroid differentiation using the S1-nuclease protection assay as before (Figure 1). The 3′ C/U ratio for this RNA was very low (∼0.4; Figure 3A and B) and comparable to that of an RNA that does not contain βIVS-II (see lane IVS-I in Figure 3C). However, in MEL cells expressing both the β-R17 reporter gene and R17–U2AF 65, the C/U ratio was increased to 2, implying a 4- to 5-fold increase in cleavage efficiency (Figure 3A and B). Conversely, the C/U ratio was not changed in cells expressing R17–U2AF 65 and a β-globin reporter gene devoid of R17 sequences (Figure 3C), showing that tethering R17–U2AF 65 to the RNA is required to promote an increase in 3′-end cleavage. No stimulatory effect on 3′-end formation was observed upon tethering R17–U2AF 35 to the β-R17 reporter gene (Figure 3B), suggesting that only the 65 kDa subunit of U2AF is capable of interfacing with the 3′ cleavage/polyadenylation machinery. However, negative results with R17–U2AF 35 could also reflect inactivity of the fusion protein.
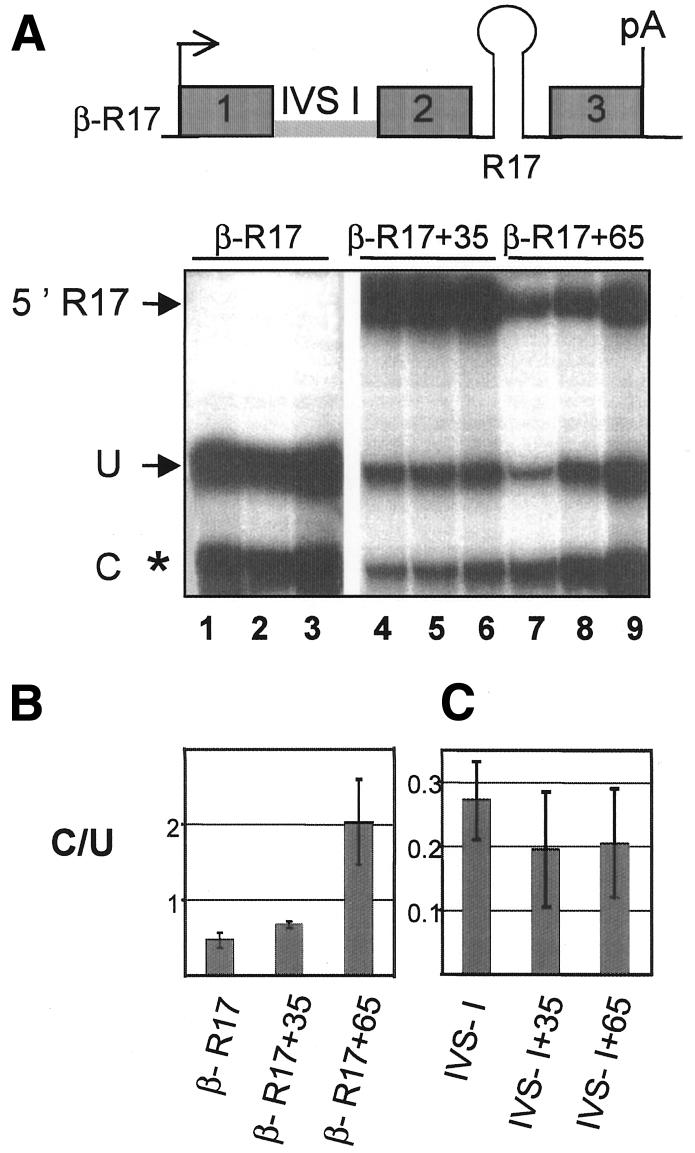
Fig. 3. Tethering U2AF 65 to β-globin pre-mRNA devoid of βIVS-II promotes 3′-end processing. (A) S1-nuclease protection analysis with human β-globin genes containing a replacement of βIVS-II with a 72 bp sequence corresponding to the phage R17-binding site (β-R17). Experiments were performed as in Figure 1, with MEL cells expressing the β-R17 reporter gene either alone (lanes 1–3) or co-transfected with the R17–U2AF 35 fusion protein expression vector (lanes 4–6) or with the R17–U2AF 65 fusion protein expression vector (lanes 7–9). The R17 phage coat protein is fused at the N-terminus of U2AF 65 and U2AF 35. 5′ R17 shows the 5′ end of the mRNA from the R17–U2AF 35 gene (lanes 4–6) or the R17–U2AF 65 gene (lanes 7–9). (B) C/U represents the 3′ cleaved-to-uncleaved ratio for results in (A). Error bars are SD values between the three samples within each set. (C) Same as in (B), but using a β-globin gene devoid of βIVS-II as a reporter either alone (lane IVS-I) or co-transfected with the R17–U2AF 35 fusion protein expression vector (lane IVS-I+35) or the R17–U2AF 65 fusion protein expression vector (lane IVS-I+65).
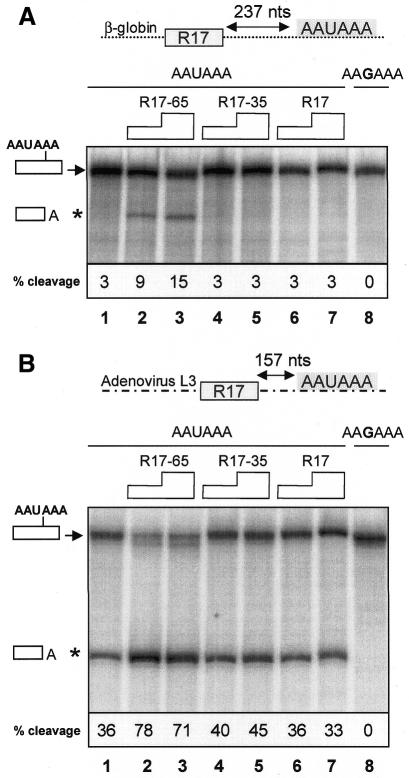
In order to more generally assess the role of U2AF 65, we performed in vitro cleavage reactions with HeLa nuclear extracts either alone or in combination with R17–U2AF 65, R17–U2AF 35 or R17 proteins expressed in and purified from Escherichia coli. The RNA cleavage substrate contained a R17-binding site, in place of the PYR tract located upstream of the β-globin cleavage/polyadenylation signal (Figure 4A, top line). A comparison of the input uncleaved RNA (upper band) and cleaved product (lower band) showed that the efficiency of 3′ cleavage was ∼3% (Figure 4A, lane 1). No cleavage product was observed with an RNA substrate containing an AAUAAA to AAGAAA mutation in the β-globin cleavage/polyadenylation site (Figure 4A, lane 8). Addition of R17–U2AF 65 (Figure 4A, lanes 2 and 3) but neither R17 (Figure 4A, lanes 6 and 7) nor R17–U2AF 35 (Figure 4A, lanes 4 and 5) to HeLa nuclear extracts stimulated cleavage efficiency by up to 5-fold. Similar experiments were carried out with an RNA substrate consisting of a R17-binding site upstream of the very efficient cleavage/polyadenylation signal of the adenovirus L3 pre-mRNA. In this case, the addition of R17–U2AF 65 specifically stimulated 3′-end cleavage efficiency by up to 2-fold (Figure 4B, lanes 2 and 3). Taken together, these results demonstrate that an R17–U2AF 65 fusion protein, when tethered to various RNA cleavage substrates, is able to stimulate 3′-end processing in both stably transfected MEL cells in vivo and in cell-free HeLa nuclear extracts in vitro.
Fig. 4. U2AF 65 stimulates in vitro pre-mRNA 3′-end cleavage. (A) In vitro cleavage reactions using RNA substrates containing an R17-binding site located upstream of the wild-type (AAUAAA; lanes 1–7) or mutant (AAGAAA; lane 8) β-globin cleavage/polyadenylation site. 32P-labelled RNA substrate was incubated with HeLa nuclear extracts in the presence of increasing amounts of E. coli produced GST–R17–U2AF 65 (lane 2, 0.25 µg; lane 3, 0.5 µg), GST–R17–U2AF 35 (lane 4, 0.25 µg; lane 5, 0.5 µg) or GST–R17 (lane 6, 0.25 µg; lane 7, 0.5 µg) recombinant fusion proteins. Quantification of cleavage reactions after resolution on polyacrylamide gels was by PhosphorImager analysis and phosphor screen exposure times of 1 h (Molecular Dynamics). Cleavage activity was calculated by dividing the amount of upstream cleavage product by the sum of upstream cleavage plus precursor products. (B) Same as (A), with RNA substrates containing an R17-binding site located upstream of the wild-type (AAUAAA; lanes 1–7) or mutant (AAGAAA; lane 8) adenovirus L3 cleavage/polyadenylation site.
The exon definition model of mammalian genes (Berget, 1995) postulates that splicing factors binding to the 3′ splice site of an upstream intron promotes the binding of splicing factors to the 5′ splice site of the immediate downstream intron, and vice versa. This model, however, does not account for the definition of the last exon of a gene. The coupling of splicing and 3′-end processing of the terminal intron are thought to define the last exon with the poly(A)-addition site replacing the 5′ splice site downstream of the last intron splice acceptor site. In support of this model, we have shown previously that PAP, in association with a poly(A)-addition recognition site, stimulates the binding of U2AF 65 to a PYR tract within an upstream intron, thereby promoting splicing (Vagner et al., 2000). This report addresses the reciprocal question of whether U2AF 65 is responsible for the enhancing effect of the splicing machinery on 3′-end cleavage. Our results demonstrate that U2AF 65 bound to the PYR tract of the splice acceptor site stimulates 3′-end processing. Because PAP is required for the cleavage step of some pre-mRNAs, the interaction of U2AF 65 and PAP could be involved in the stimulatory effect of U2AF 65 on 3′-end cleavage. However, it will be of value to investigate other possible interactions between U2AF 65 and the cleavage/polyadenylation factors. For instance, since U2AF partially co-purifies with the cleavage factors CF Im and CF IIAm (de Vries et al., 2000), it is still possible that some direct physical interactions between U2AF 65 and the protein subunits of these cleavage complexes participate in the U2AF 65 role in 3′-end cleavage. Much work remains in order to describe physical and functional interactions between U2AF 65 and the cleavage/polyadenylation machinery. In conclusion, our findings point to a novel function for U2AF 65 as a 3′-end formation-promoting factor in addition to its role in early recognition of splice acceptor signals. U2AF 65 is therefore one of the prime determinants for the coupling of splicing and 3′-end formation that in turn defines the terminal exon of a gene.
METHODS
Tissue culture. The maintenance, stable transfection by electroporation and induction to terminal erythroid differentiation (with 2% DMSO for 4 days) of MEL cells were all as described previously (Antoniou et al., 1998). Stable pools of transfected cells were selected with 800 µg/ml G418 either alone or in combination with 2.5 µg/ml puromycin (both from Sigma) in co-transfection experiments. All plasmids were linearized with PvuI prior to transfection.
Plasmid DNA constructs and generation of βIVS-II mutants. The mutant series of βIVS-II (SM-PYR, DM-PYR, TM-PYR, PYR, PYR-AG, β-R17; see Figures 1 and 3) were produced within a pBluescript (Stratagene, La Jolla, CA) subclone of the β-globin gene harbouring a fully functional 89 bp deletion variant (Δ89) of this intron as described previously (Antoniou et al., 1998). Genes were cloned between the ClaI and KpnI sites of the βLCR expression vector that also carries a neomycin (G418) resistance gene (Collis et al., 1990). The R17–U2AF 65 and R17–U2AF 35 fusion protein genes were generated by linking in-frame the cDNA coding sequences of these polypeptides and inserting them into the βLCR expression vector pEV modified to contain a puromycin resistance gene in place of the neomycin resistance gene (Needham et al., 1995). This allowed the stable co-transfection into MEL cells of either the β-R17 or βIVS-I reporter genes and the R17–U2AF 65 or R17–U2AF 35 fusion protein pEV-puro expression vectors by double drug selection with G418 and puromycin. The βLCR confers physiological levels of expression that are directly proportional to transgene copy number once these constructs are stable transfected in MEL cells (Antoniou et al., 1998).
Extraction of RNA and analysis by S1-nuclease protection assays. The preparation of total, nuclear and cytoplasmic RNA from MEL cells was as described previously (Antoniou et al., 1998). Human β-globin and mouse βmaj-globin mRNA was quantified in these RNA fractions by an S1-nuclease protection assay using double-stranded, end-labelled DNA probes as before. The 3′ human β-globin probe is derived of an intronless human β-globin gene and is illustrated in Figure 1A. This probe detects 3′ uncleaved and cleaved transcripts from the human β-globin transgenes respectively as 257- and 212-nucleotide S1-nuclease protected products (Figure 1C). The mouse βmaj-globin probe is a 700 bp HindIII–NcoI fragment from the 5′ half of this gene and gives a 96-nucleotide S1-nuclease protected product derived from the 5′ half of exon II. The products from the S1-nuclease digestion were resolved on a 6% polyacrylamide gel in the presence of 8 M urea, dried, quantified by PhosphorImager (Molecular Dynamics) analysis and exposed to Kodak XAR 5 or Fuji medical X-ray film to obtain a hard copy of the results.
In vitro cleavage reactions. Protein extracts were prepared from HeLa cell nuclei (Dignam et al., 1983). Substrate RNAs were transcribed by T7 or T3 RNA polymerase in the presence of 32P-UTP and ApppG capped. Cleavage reactions were as described previously (Boelens et al., 1993).
UV cross-linking/immunoprecipitation. MEL or HeLa cell nuclear protein extracts were incubated for 15 min with either 32P-end-labelled RNA oligonucleotide probes (wild-type PYR: CCUCUUAUCUUCCUCCCACAG; TM-PYR: CCUCUUAUCUUCCGGCCAAAG) under cleavage conditions. Reactions were performed as described previously (Vagner et al., 2000).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Maria Carmo-Fonseca for the MC3 monoclonal antibody and Sophie Bonnal for the recombinant GST–R17 protein. This work was supported by the European Union Human Capital and Mobility, Biomed-2, Biotechnology and Framework IV programmes (to M.A.), INSERM and the French Ministry of Research (ACI ‘Jeunes chercheurs’) (to S.V.). S.M. is a recipient of an EMBO long-term postdoctoral fellowship.
REFERENCES
- Antoniou M. (1995) Clinical defects in pre-mRNA processing. In Lamond, A.I. (ed.), Pre-mRNA Processing. R.G. Landes Company, Austin, TX, pp. 187–201.
- Antoniou M., Geraghty, F., Hurst, J. and Grosveld, F. (1998) Efficient 3′-end formation of human β-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res., 26, 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell V.J. and Wickens, M. (1990) Purification of RNA and RNA–protein complexes by an R17 coat protein affinity method. Nucleic Acids Res., 18, 6587–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S.M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- Boelens W.C., Jansen, E.J., van Venrooij, W.J., Stripecke, R., Mattaj, I.W. and Gunderson, S.I. (1993) The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell, 72, 881–892. [DOI] [PubMed] [Google Scholar]
- Collis P., Antoniou, M. and Grosveld, F. (1990) Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J., 9, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C., Hans, H. and Alwine, J.C. (1999) Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol., 19, 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H., Ruegsegger, U., Hubner, W., Friedlein, A., Langen, H. and Keller, W. (2000) Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J., 19, 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz, R.M. and Roeder, R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M., Krauss, R.D., Chiang, L., Valcarcel, J., Green, M.R. and Carmo-Fonseca, M. (1997) Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol., 137, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco M.A., Jamison, S.F. and Sharp, P.A. (1989) Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev., 3, 1874–1886. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Beyer, K., Martin, G., Keller, W., Boelens, W.C. and Mattaj, I.W. (1994) The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell, 76, 531–541. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Vagner, S., Polycarpou-Schwarz, M. and Mattaj, I.W. (1997) Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev., 11, 761–773. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Polycarpou-Schwarz, M. and Mattaj, I.W. (1998) U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell, 1, 255–264. [DOI] [PubMed] [Google Scholar]
- Liu X. and Mertz, J.E. (1996) Sequence of the polypyrimidine tract of the 3′-terminal 3′ splicing signal can affect intron-dependent pre-mRNA processing in vivo. Nucleic Acids Res., 24, 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C.S., Murthy, K.G., Schek, N., O’Connor, J.P., Manley, J.L. and Alwine, J.C. (1996) Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev., 10, 325–337. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. (2002) An extensive network of coupling among gene expression machines. Nature, 416, 499–506. [DOI] [PubMed] [Google Scholar]
- Needham, M.et al. (1995) Further development of the locus control region/murine erythroleukemia expression system: high level expression and characterization of recombinant human calcitonin receptor. Protein Expr. Purif., 6, 124–131. [DOI] [PubMed] [Google Scholar]
- Nesic D. and Maquat, L.E. (1994) Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev., 8, 363–375. [DOI] [PubMed] [Google Scholar]
- Nesic D., Cheng, J. and Maquat, L.E. (1993) Sequences within the last intron function in RNA 3′-end formation in cultured cells. Mol. Cell. Biol., 13, 3359–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M., Rose, S.D. and Berget, S.M. (1990) In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev., 4, 1552–1559. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger, A. and Dye, M.J. (2002) Integrating mRNA processing with transcription. Cell, 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore, P.D. and Green, M.R. (1988) A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell, 52, 207–219. [DOI] [PubMed] [Google Scholar]
- Sebillon P., Beldjord, C., Kaplan, J.C., Brody, E. and Marie, J. (1995) A T to G mutation in the polypyrimidine tract of the second intron of the human β-globin gene reduces in vitro splicing efficiency: evidence for an increased hnRNP C interaction. Nucleic Acids Res., 23, 3419–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.S. and Dreyfuss, G. (1988) RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J., 7, 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S., Vagner, C. and Mattaj, I.W. (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev., 14, 403–413. [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D. and Green, M.R. (1989) Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl Acad. Sci. USA, 86, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]