Abstract
In eukaryotic ribosomes, termination of translation is triggered by class 1 polypeptide release factor, eRF1. In organisms with a universal code, eRF1 responds to three stop codons, whereas, in ciliates with variant codes, only one or two codon(s) remain(s) as stop signals. By mutagenesis of the Y–C–F minidomain of the N domain, we converted an omnipotent human eRF1 recognizing all three stop codons into a unipotent ‘ciliate-like’ UGA-only eRF1. The conserved Cys127 located in the Y–C–F minidomain plays a critical role in stop codon recognition. The UGA-only response has also been achieved by concomitant substitutions of four other amino acids located at the Y–C–F and NIKS minidomains of eRF1. We suggest that for eRF1 the stop codon decoding is of a non-linear (non-protein-anticodon) type and explores a combination of positive and negative determinants. We assume that stop codon recognition is profoundly different by eukaryotic and prokaryotic class 1 RFs.
INTRODUCTION
In the standard (‘universal’) genetic code, 61 sense codons are decoded by cognate transfer RNAs aminoacylated by 20 natural amino acids. The remaining three codons, UAA, UGA and UGA, are thought to be recognized by proteins called class 1 polypeptide release factors (RFs) that are intimately involved in triggering termination of protein synthesis (reviewed by Kisselev and Buckingham, 2000; Poole and Tate, 2000).
Termination of translation in eukaryotes is governed by a single release factor, eRF1, when it occupies the ribosomal A site. Cleavage of the last peptidyl-tRNA (termination reaction) takes place if one of the three stop codons is located at the same site within the ribosome. Specificity of stop codon recognition is dictated by the eRF1 itself and not by the ribosome (Kervestin et al., 2001; Ito et al., 2002). This means that some features of the eRF1 protein molecule determine stop codon recognition.
The eRF1 protein is composed of three domains: N (or 1), M (or 2) and C (or 3) (Song et al., 2000). The C domain is not involved in stop codon recognition but binds to the second termination factor, eRF3 (Frolova et al., 2000 and references therein). The M domain, containing an ubiquitous GGQ motif, was supposed to be located at the peptidyl transferase centre or nearby and involved in triggering the peptidyl-tRNA cleavage (Frolova et al., 1999; Song et al., 2000).
Recent data indicate that the structural basis of decoding specificity resides in the N domain of eRF1. Point mutations of the amino acids located in the N domain cause alteration in stop codon recognition in vivo (Bertram et al., 2000). Substitutions of the amino acid residues in the NIKS motif of human eRF1 (the N domain, positions 61–64) cause changes in the stop codon responses in an in vitro RF assay (Frolova et al., 2002). Sequence analysis of eRF1s from a wide variety of organisms also points to the N domain as a potential stop codon decoding site (Lozupone et al., 2001; Inagaki et al., 2002).
Several hypotheses, mostly based on indirect evidence, on the precise location of the decoding site have been proposed. ‘Linear’ models point to TAS (Nakamura et al., 2000), ‘wobble’ TAS (Muramatsu et al., 2001) or NIKS (Knight and Landweber, 2000) motifs as potential sites of stop codon recognition. ‘Non-linear’ models (Bertram et al., 2000; Inagaki et al., 2002) rely on single amino acid residues associated with structurally different parts of the N domain and spread between positions 51 and 132.
In universal-code eukaryotes, a single factor, eRF1, responds to all three stop codons, whereas, in variant-code organisms, one or two of the stop codons are reassigned for sense codons (reviewed in Knight et al., 2001; Lozupone et al., 2001). For example, in ciliate Euplotes, UGA is reassigned for cysteine, and in vitro Euplotes eRF1 is blind towards UGA remaining active towards UAA and UAG (Kervestin et al., 2001). The eRF1s from variant-code organisms are restricted in the recognition of certain codons, probably due to constraints imposed on their eRF1 amino acid sequences during the evolution from standard to variant genetic codes. Thus, one may anticipate that omnipotent eRF1s from standard-code organisms can, in principle, be converted into unipotent, ciliate-like eRF1s by alteration of the eRF1 structure.
The aim of this work was to mimic omnipotent-to-unipotent conversion by the substitution of some critically essential amino acids in the N domain of eRF1.
RESULTS AND DISCUSSION
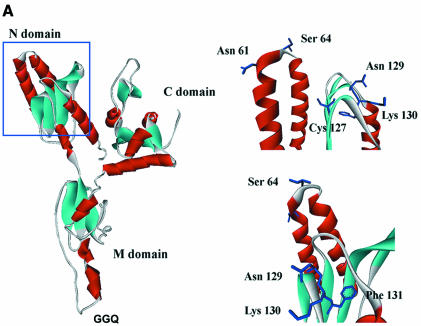
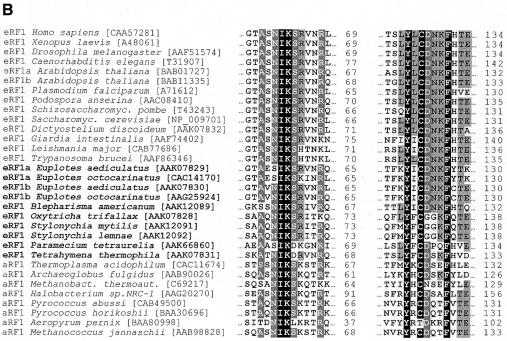
From the three-dimensional (3D) structure of the N domain of human eRF1 (Figure 1A), two minidomains are clearly visible: one that contains a NIKS motif invariant in eukaryotes with a universal genetic code (Kisselev et al., 2000; Song et al., 2000; Frolova et al., 2002); and one that contains Tyr125, Cys127 and Phe131, common for the eRF1/aRF1 structural family (Figure 1B).
Fig. 1. (A) The ribbon diagram of the whole human eRF1 (left) and its N-terminal domain (right, view from two sides) derived from crystallographic data (Song et al., 2000) by the WebLab ViewerLite program version 4.0 (Molecular Simulations). (B) Alignments of the eRF1 and aRF1 amino acid sequences for the NIKS and Y–C–F minidomains. Positions are numbered for human eRF1. Accession numbers in brackets are from the NCBI-Entrez-Proteins database. Amino acids are shaded according to their identity percentage (white letters, black shading, –95%; white letters, dark grey shading, –85%; black letters, light grey shading, –35%). Variant-code ciliates are in bold.
Here, we focused on the Tyr–Cys–Phe (Y–C–F) minidomain that is of particular interest for several reasons: (i) it is composed of semi-conserved and invariant amino acid residues as anticipated for the stop codon recognition site, since in all three stop codons the first U is invariant and is therefore probably recognized by an invariant amino acid(s), whereas positions 2 and 3 are occupied by either an A or a G and presumably can be recognized by conserved, semi-conserved or variant amino acids; (ii) it is located near the NIKS minidomain, the essential functional role of which has been demonstrated already (Frolova et al., 2002); (iii) the distance between the Y–C–F minidomain and the GGQ motif is ∼80 Å (Song et al., 2000; Inagaki et al., 2002), close to the distance between the anticodon and the CCA end of tRNA (75 Å), and it can be placed near to the stop codon at the A site if the GGQ motif is located at the peptidyl transferase centre near the CCA end of peptidyl-tRNA; and (iv) Tyr125, Cys127, Phe131 and some other amino acids from this region are the slowest evolving in the N domain (Inagaki et al., 2002), and therefore these residues could be involved in the formation of the stop codon decoding site.
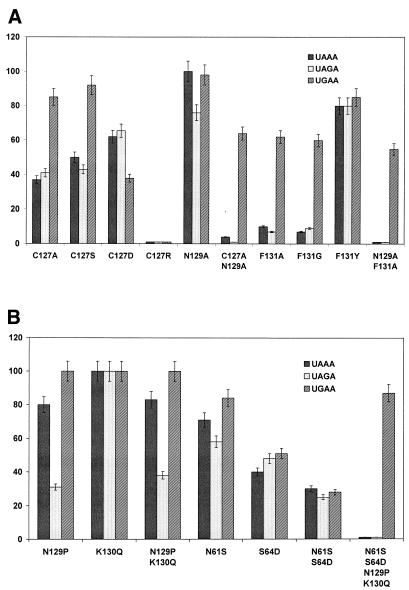
Figure 2A shows the RF activity in vitro of human eRF1 mutagenized at positions 127, 129 and 131. The Cys127Ser and Cys127Ala mutants retain their ability to respond to the UGA stop codon but possess a reduced capacity to respond to UAA/UAG. The Cys127Asp mutant differs from the former two mutants, as the response to UAA and UGA is more strongly affected than the UAG response. The almost total abolishment of the RF activity towards all three stop codons observed for the Cys127Arg mutant (Figure 2A) is not caused by the loss of eRF1 binding to the ribosome. This mutant, as well as the other mutants, completely retained their ability to stimulate eRF3 GTPase activity (data not shown), known to be a measure of eRF1 binding ability towards the ribosome (Seit-Nebi et al., 2001; Frolova et al., 2002). Collectively, the RF activity of the Cys127 mutants clearly points to a very essential role of this amino acid in stop codon recognition.
Fig. 2. The in vitro RF activity of the human eRF1 mutants. (A) Mutations at positions 127, 129 and 131. (B) Multiple mutations at two different minidomains. The RF activity of the wild-type eRF1 was equal to 100%. In each case, the average from three independent measurements is presented. The standard deviation of the measurements was 11%.
The Phe131 is also a functionally important residue, since even mild Phe→Tyr substitution reduces the RF activity towards all three stop codons, whereas elimination of the aromatic ring in Phe131Ala and Phe131Gly mutants almost entirely abolishes two out of the three stop codon responses (Figure 2A).
The Asn129 located between the invariant Cys127 and Phe131 is conserved in universal-code eukaryotes but varies in Eukarea with variant genetic codes and in Archaea (Figure 1B). Therefore, it seems reasonable to mutagenize this position. Two out of the four Asn129 mutants, Asn129Pro and Asn129Ser, were selected because of the presence of these amino acid residues in other eRF1s (Figure 1B), whereas two others, Asn129Ala and Asn129Asp, were chosen to broaden the spectrum of mutations of amino acid residues with different chemical properties. Conversion of Asn129 into Asp shows no effect and Asn→Ser substitution has a weak effect (data not shown), whereas in the Asn129Pro and Asn129Ala mutants the UAG response is more sensitive (Figure 2) than the other stop codon responses. This behaviour may indicate that Asn129 is implicated in the recognition of G in the 3d position of the stop codon.
To confirm and extend the above data, we combined two mutations in the same protein molecule and followed the RF activity (Figure 2A). In the double mutants Cys127Ala+Asn129Ala and Asn129Ala+Phe131Ala, the UAA and UAG responses are entirely abolished, whereas the UGA response is reduced only ∼2-fold (Figure 2A).
Consequently, we have shown for the first time that the invariant Cys127 and Phe131 of the human eRF1 located at the Y–C–F minidomain of the N domain (Figure 1A) play a pivotal role in stop codon decoding by eRF1. Remarkably, these positions are near the positions 126, 128 and 132 mentioned earlier as potentially involved in stop codon recognition (Bertram et al., 2000; Inagaki et al., 2002). Although in these works other approaches have been applied to study the stop codon recognition problem, collectively, all these data point to the critical role of the 126–132 region in stop codon discrimination.
Earlier, the important functional role of the NIKS tetrapeptide and neighbouring amino acid residues in human eRF1 (Frolova et al., 2002; Ito et al., 2002) has been demonstrated and, therefore, it has been reasonable to convert NIKS tetrapeptide of human eRF1 into SIKD tetrapeptide typical for Paramecium eRF1, the organism with a variant genetic code where the UGA is the only stop codon (see Kervestin et al., 2001), together with additional substitutions of Asn129 and Lys130 of human eRF1 for Pro129 and Gln130 present in Paramecium eRF1 (Figure 1B). The double mutant Asn61Ser+Ser64Asn diminishes the RF activity for all three stop codons, at least partly due to the decrease in ribosome binding (Frolova et al., 2002), whereas Asn129Pro+Lys130Gln leads to damaged response only towards the UAG stop codon (Figure 2B). The four-site mutant Asn61Ser+Ser64Asp+Asn129Pro+Lys130Gln, which mimics the structure of Paramecium eRF1 in these two minidomains, responds only to UGA (Figure 2B). Surprisingly, although the SIKD mutant possesses 3-fold reduced UGA-dependent activity, the quadruple mutant exhibits 80% of UGA response (Figure 2B). This phenomenon requires further analysis.
The demonstration of the profound changes in the stop codon decoding pattern after simultaneous multiple substitutions in two minidomains and the failure to induce the same transformation by separate mutations in each minidomain (Figure 2B) implies that specific stop codon recognition depends on two structurally distinct minidomains of the same N domain located in space close to each other (Figure 1A).
Since the recognition pattern of this four-site Paramecium-like eRF1 mutant is virtually identical to the Phe131Ala, Phe131Gly, Cys127Ala+Asn129Ala and Asn129Ala+Phe131Ala mutants (Figure 2A), we conclude that the same alteration of the recognition pattern can be achieved by entirely different ways, namely by double-site mutagenesis in the Y–C–F minidomain and by multiple-site mutagenesis of semi-conserved and variable amino acids located within two different minidomains (NIKS and Y–C–F) of the N domain (Figure 1A).
The most striking result is a non-predicted and surprising effect of the alteration of two invariant amino acids, Cys127 and Phe131, on the stop codon recognition pattern of mutant eRF1s (Figure 2A). Retention of the UGA response for Phe131 mutants implies that this position is not critical for the maintenance of the overall 3D structure of the N domain. Even for the Cys127Arg mutant where the RF activity was completely abolished, ribosome binding is completely retained (data not shown), in accordance with the idea that Cys127 is also not critical for the conservation of the 3D structure of eRF1. It has been suggested (Inagaki et al., 2002) that Cys127 is involved in the recognition of the first base of the stop codon, a universal U. All four substitutions of Cys127 (Figure 2A) cause dramatic functional effects, although they are different towards different stop codons. This is a strong evidence in favour of the assumption that this residue is critical for the maintenance of the recognition pattern towards all stop codons. The UAA and UAG responses for the Phe131Ala and Phe131Gly mutants point to the role of Phe131 in the recognition of A in the second position of the stop codon. However, this assumption is inconsistent with the data showing that Phe131 is also present in UGA-only ciliate eRF1s (Figure 1B). In these variant-code organisms, G, not A, occupies the second position of the stop codon. This apparent controversy may be resolved by assuming that in UGA-only ciliate eRF1s the influence of Phe131 on the second A recognition is hindered by some negative determinants (yet unidentified) surrounding in space these residues, similar to what is known for tRNA recognition by aminoacyl-tRNA synthetases (Giege et al., 1993). In the N domains of eRF1s from universal-code and variant-code organisms, there are numerous amino acids differences (see Inagaki and Doolittle, 2001; Kervestin et al., 2001; Lozupone et al., 2001; Inagaki et al., 2002) that may serve as negative determinants.
If the negative-determinant hypothesis is correct, then variant-code eRF1s can be converted into universal-code eRF1s by replacements of these putative negative elements. It remains unknown whether Cys127 and/or Phe131 are in physical contact with the stop codon triplet or their influence is mediated by other structural features of eRF1 or by rRNAs known to affect translation termination in eukaryotes (Velichutina et al., 2001 and references therein).
It seems unlikely that prokaryotes and eukaryotes share a common mechanism for stop codon discrimination, due to profound differences in primary, secondary and tertiary structures between eukaryotic and prokaryotic class 1 RFs (Frolova et al., 1994; Kisselev et al., 2000; Song et al., 2000; Vestergaard et al., 2001; Kisselev, 2002) and in the location and structure of the functionally important sites (Ito et al., 2000; Frolova et al., 2002; the present work).
Our data (see also Bertram et al., 2000; Inagaki et al., 2002) disfavour for eukaryotes the ‘linear’ or the ‘protein-anticodon’ model suggested for prokaryotes, in which PAT and SPF tripeptides of RF1 and RF2 interact directly with UAA/UAG and UAA/UGA, respectively (Ito et al., 2000).
Thus, we postulate the decoding mechanism, in eukaryotic class 1 RFs, with the following features, not discussed earlier: (i) the involvement of two distinct minidomains, Y–C–F and NIKS, of the N domain in stop codon recognition by eRF1; (ii) the existence of the negative determinants (constraints) that restrict the recognition of some stop codons by eRF1s from organisms with variant genetic codes; and (iii) the critical role of invariant amino acids of the Y–C–F minidomain in the eRF1 decoding function but not in peptidyl-tRNA hydrolysis or in the conservation of the 3D structure.
Successful omnipotent-to-unipotent conversion of eRF1 (Figure 2) provides the structural basis for the hypothesis on the evolution of variant-code organisms from universal-code organisms (Knight et al., 2001; Lozupone et al., 2001; Inagaki et al., 2002). The data described in the present work open a new avenue to decipher the decoding mechanism in eukaryotes by detailed structural/functional analysis of the two minidomains in eukaryotic class 1 RFs.
The obvious controversy between the recognition models mentioned above and our data points to the fact that we still have to do much more (sequencing of new eRF1s, assay of stop codon specificity for mutant eRF1s, crystallization of eRF1–ribosome complexes and eRF1 mutants, etc.) before the genuine mechanism of stop codon recognition will be resolved unambiguously.
METHODS
Cloning and mutagenesis of human eRF1. The full-length cDNA encoding eRF1 with the C-terminal His-tag fusion (pERF4B construct) was cloned as described previously (Seit-Nebi et al., 2001; Frolova et al., 2002). The mutagenesis procedure was performed according to the PCR-based ‘megaprimer’ method (Sarkar and Sommer, 1990). The PCR primers used for the generation of eRF1 mutants are listed in Table I. The direct primer, one of those mentioned in Table I (except Asn61Ser, reverse primer), and the reverse primer (RFBst) 5′-CCATTCTTAAGCGGGCAAAACGCAAGG-3′ (Bst98I site underlined), were used at the first step of PCR. The resulting 400-bp PCR product was used as the reverse ‘megaprimer’ together with the direct primer (RFNde), 5′-GAGATATACATATGGCGGACGACCC-3′ (NdeI site underlined) at the second step of PCR. The resulting 590-bp PCR product was hydrolyzed with NdeI and Bst98I and ligated with pERF4B plasmid (Seit-Nebi et al., 2001; Frolova et al., 2002) treated with the same endonucleases. The ligated mixture was transformed into Escherichia coli strain JM109. The cloned DNAs were sequenced, and appropriate clones were used for the expression of the mutant eRF1. DNA amplifications were carried out as described previously (Frolova et al., 2002).
Table I. PCR primers used to generate the constructs for bacterial expression of human eRF1 mutant proteins.
| eRF1 mutant | Primers |
|---|---|
| Cys127Ala | 5′-TCATTGTATTTGGCTGACAACAAATTCC-3′ |
| Cys127Arg | 5′- GTATTTGCGTGACAACAAATTCC -3′ |
| Cys127Asp | 5′- GTATTTGGATGACAACAAATTCC -3′ |
| Cys127Ser | 5′-GTATTTGTCTGACAACAAATTCC-3′ |
| Asn129Ala | 5′-GTGTGACGCAAAATTCCATACAGAGG-3′ |
| Cys127Ala+Asn129Ala | 5′-TTGTATTTGGCTGACGCCAAATTCCATACAG-3′ |
| Phe131Ala | 5′-GACAACAAAGCCCATACAGAGGCTC-3′ |
| Phe131Gly | 5′-GACAACAAAGGCCATACAGAGGCTC-3′ |
| Phe131Tyr | 5′-GACAACAAATATCATACAGAGGCTC-3′ |
| Asn129Ala+Phe131Ala | 5′-TTGTGTGACGCCAAAGCCCATACAGAGG-3′ |
| Asn129Pro | 5′-GTGTGACCCTAAATTCCATACAGAGG-3′ |
| Lys130Gln | 5′-GTGTGACAACCAGTTCCATACAGAGG-3′ |
| Asn129Pro+Lys130Gln | 5′-TTGTATTTGTGTGACCCACAATTCCATACAGAGGC-3′ |
| Asn61Ser | 5′-CTTAATTGAAGATGCAGTTCCAAACTCATCCG-3′ |
| Ser64Asp | 5′-CTGCATCTAACATTAAGGACCGAGTAAACCG-3′ |
The double mutant Asn61Ser+Ser64Asp was obtained as described previously (Frolova et al., 2002). The quadruple mutant Ans61Ser+Ser64Asp+Asn129Pro+Lys130Gln was obtained by the insertion of a NdeI–BsrGI DNA fragment of pERF4B containing Asn61Ser64+Ser64Asp into the same construct containing Asn129Pro+Lys130Gln and treated by the same endonucleases.
Expression and purification of human eRF1. Wild-type human eRF1 and its mutants containing His-tag at the C terminus were expressed in E. coli strain BL21(DE3) and purified using Ni-NTA resin (Superflow; Qiagen) as described previously (Frolova et al., 1994, 2000).
Ribosomes. Rabbit reticulocyte 80S ribosomes washed with 0.5 M KCl were treated with puromycin and GTP for dissociation into subunits, which were subsequently resolved by centrifugation in a 10–25% (w/v) sucrose gradient containing 0.3 M KCl, 3 mM MgCl2, 1 mM DTT and 20 mM Tris–HCl pH 7.6. Before addition to the incubation mixtures, the subunits were combined in an equimolar ratio.
In vitro RF assay. The eRF1 activity was measured as described previously (Caskey et al., 1974; Frolova et al., 1994, 1998) at saturating levels (50 µM) of one out of the three stop-codon-containing tetraplets. The incubation mixture (25 µl) contained 20 mM Tris–HCl pH 7.5, 15 mM MgCl2, 8 mM NH4Cl, 1.5 pmol of f[35S]Met-tRNAfMet-AUG-ribosome complex and 4 pmol of eRF1. The reaction was run at 20°C for 20 min. The background was measured without tetraplets and subtracted from all values. The amount of f[35S]Met released without stop codons was 500–800 c.p.m. AUG and ribotetraplets were synthesized by A. Veniaminova and M. Ryabkova (Institute of Bioorganic Chemistry, Novosibirsk, Russia).
Acknowledgments
ACKNOWLEDGEMENTS
We thank V. Dubovaya for the eRF1 (Cys127Arg) mutant. This work was supported by the Human Frontier Science Program (grant 96-032), INTAS grant 00-0041, the Russian Foundation for Basic Research (grant to L.F.) and the Program of Support for Scientific Schools (grant to L.K.).
REFERENCES
- Bertram G., Bell, H.A., Ritchie, D.W., Fullerton, G. and Stansfield, I. (2000) Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA, 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey C.T., Beaudet, A.L. and Tate, W.P. (1974) Mammalian release factor: in vitro assay and purification. Methods Enzymol., 30, 293–303. [DOI] [PubMed] [Google Scholar]
- Frolova L. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- Frolova L.Y., Simonsen, J.L., Merkulova, T.I., Litvinov, D.Y., Martensen, P.M., Rechinsky, V.O., Camonis, J.H., Kisselev, L.L. and Justesen, J. (1998) Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem., 256, 36–44. [DOI] [PubMed] [Google Scholar]
- Frolova L.Yu., Tsivkovskii, R., Sivolobova, G., Oparina, N., Serpinsky, O., Blinov, V., Tatkov, S. and Kisselev, L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Yu., Merkulova, T.I. and Kisselev, L.L. (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of two functionally and structurally distinct domains. RNA, 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L., Seit-Nebi, A. and Kisselev, L. (2002) Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA, 8, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege R.J., Puglisi, J.D. and Florentz, C. (1993) tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol., 45, 129–206. [DOI] [PubMed] [Google Scholar]
- Inagaki Y. and Doolittle, W.F. (2001) Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res., 29, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y., Blouin, C., Doolittle, W.F. and Roger, A.J. (2002) Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: evolution of stop codon specificity. Nucleic Acids Res., 30, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno, M. and Nakamura, Y. (2000) A tripeptide anticodon deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Ito K., Frolova, L., Seit-Nebi, A., Karamyshev, A., Kisselev, L. and Nakamura, Y. (2002) Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within the domain 1. Proc. Natl Acad. Sci. USA, 99, 8494–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S., Frolova, L., Kisselev, L. and Jean-Jean, O. (2001) Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO rep., 2, 680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev L. (2002) Polypeptide release factors in prokaryotes and eukaryotes: same function, different structure. Structure, 10, 8–9. [DOI] [PubMed] [Google Scholar]
- Kisselev L.L. and Buckingham, R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- Kisselev L.L., Oparina, N.Yu. and Frolova, L.Yu. (2000) Class-1 translation termination factors are structurally and functionally similar to suppressor tRNAs and belong to distinct structural/functional families (prokaryotes/mitochondria and eukaryotes/archaea). Mol. Biol. (Moscow), 34, 427–442. [PubMed] [Google Scholar]
- Knight R.D. and Landweber, L.F. (2000) The early evolution of the genetic code. Cell, 101, 569–572. [DOI] [PubMed] [Google Scholar]
- Knight R.D., Freeland, S.J. and Landweber, L.F. (2001) Rewiring the keyboard: evolvability of genetic code. Nat. Rev. Genet., 2, 49–58. [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Knight, R.D. and Landweber, L.F. (2001) The molecular basis of nuclear genetic code change in ciliates. Curr. Biol., 11, 65–74. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Heckmann, K., Kitanaka, C. and Kuchino, Y. (2001) Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett., 488, 105–109. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito, K. and Ehrenberg, M. (2000) Mimicry grasps reality in translation termination. Cell, 101, 349–352. [DOI] [PubMed] [Google Scholar]
- Poole E. and Tate, W. (2000) Release factors and their role as decoding proteins: specificity and fidelity for termination of protein synthesis. Biochim. Biophys. Acta, 1493, 1–11. [DOI] [PubMed] [Google Scholar]
- Sarkar G. and Sommer, S.S. (1990) The ‘megaprimer’ method of site-directed mutagenesis. Biotechniques, 8, 404–407. [PubMed] [Google Scholar]
- Seit-Nebi A., Frolova, L., Justesen, J. and Kisselev, L. (2001) Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res., 29, 3982–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Mugnier, P., Webb, H.M., Evans, D.R., Tuite, M.F., Hemmings, B.A. and Barford, D. (2000) The crystal structure of human eukaryotic release factors eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Velichutina I.V., Hong, J.Y., Mesecar, A.D., Chernoff, Y.O. and Liebman, S.W. (2001) Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18S rRNA. J. Mol. Biol., 305, 715–727. [DOI] [PubMed] [Google Scholar]
- Vestergaard B., Van, L.B., Andersen, G.R., Nyborg, J., Buckingham, R.H. and Kjeldgaard, M. (2001) Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell, 8, 1375–1382. [DOI] [PubMed] [Google Scholar]