Abstract
To develop a reliable strategy for cell-specific delivery of retroviral vectors, we genetically modified the envelope (Env) protein of the ecotropic Moloney murine leukemia virus. We found a site in the variable region A, where the insertion of ligands, epidermal growth factor (EGF) and stromal-derived factor-1α (SDF-1α), was possible without abolishing virion incorporation of the Env protein and its ecotropic entry function. The vector containing the EGF–Env did not show the EGF receptor-dependent transduction. The vector containing the SDF-1α–Env, however, specifically transduced human cells expressing CXCR4, the receptor for SDF-1α, at titers of 103–104 c.f.u./ml. Further experiments showed that the CXCR4-dependent transduction was based on the specific interaction between the SDF-1α moiety of the SDF-1α–Env and CXCR4 and was independent of the ecotropic entry function. The direct targeting of the retroviral vector may be possible if the proper chimeric Env structure and the appropriate ligand–receptor system are employed.
INTRODUCTION
Retroviral vectors are valuable tools for the transfer of genes into animal cells. Compared with other methods of gene transfer, they have the distinct ability to efficiently and stably integrate genes into the chromosomes of the cells and persistently express the transferred genes. The representative retroviral vectors are derived from murine leukemia viruses (MLVs). Extensive studies have established the safety of these vectors in human applications (Anderson, 1998). MLVs are classified into five subgroups according to their host ranges. This host range is determined mainly by the interaction between the viral envelope (Env) protein and the cellular receptor for infection. Recently, receptors for MLVs were identified as multiple membrane-spanning proteins involved in the transport of solutes (Sommerfelt, 1999). The main determinant of infectability is appropriate receptor expression; therefore, in an animal, the infection of different types of cells occurs because of the ubiquitous expression of the receptor in the body. While this underlies the broad applicability of MLV vectors provided that the recipient cells are dividing, it limits their usefulness, as direct injection into the body and the transfer of genes to specific target cells cannot be achieved.
Various attempts have been made to obtain retroviral vectors that specifically deliver genes to desired types of cells (Russell and Cosset, 1999). The most thoroughly examined approach is to change the delivery tropism of the vector by incorporating a segment of DNA that encodes a peptide ligand (or a single-chain antibody) into the env gene. The MLV Env is synthesized as a precursor and proteolytically cleaved into surface (SU) and transmembrane (TM) subunits. They appear as trimers of SU–TM heterodimers in the envelope. SU and TM proteins are involved in receptor binding and envelope–cell membrane fusion, respectively. The ligand is either inserted into the SU (Cosset et al., 1995a; Yajima et al., 1998; Benedict et al., 1999; Zhao et al., 1999; Gollan and Green, 2002a,b) or it substitutes as a part of the SU (Kasahara et al., 1994). In the direct targeting strategy for the change of the delivery tropism, the ligand moiety binds to its cognate receptor, and this induces the chimeric Env protein to mediate the membrane fusion that is required for the transfer of genes (Russell and Cosset, 1999). In most cases so far, however, the chimeric Env protein binds to the cognate receptor, but membrane fusion and subsequent cellular entry of the vector occur very inefficiently or not at all.
In this study, we investigated the factors that would affect the direct targeting of MLV vectors. We reasoned that, to achieve efficient coupling of binding to the targeted receptor with membrane fusion, the ligand should be in the native receptor-binding surface of the SU, which is mainly composed of the variable region A (VRA) of the receptor-binding domain (RBD) (Fass et al., 1997). We searched for sites in the VRA of the ecotropic (mouse and rat cell-tropic) Moloney (Mo) MLV Env, into which the insertion of a ligand would not affect the incorporation of Env into virus particles. Epidermal growth factor (EGF) and stromal-derived factor-1α (SDF-1α) were used as ligands to test an idea that the topology of membrane proteins is related to their potential for functioning as receptors for MLV entry.
RESULTS
Search for sites in the VRA that tolerate ligand insertion
The effects of the insertion of the human EGF sequence into six different sites in the VRA (Figure 1) on the incorporation of chimeric Env proteins into viral particles were examined. The EGF sequence with linkers encoding five amino acids at each end was inserted into different sites of the env gene, and each expression construct was transfected into TELCeB6 cells, which produce the Env-negative lacZ retroviral vector (Cosset et al., 1995b). A significant amount of SU protein was detected only in the virions from cells stably transfected with the site 3 EGF–chimeric env (E3) plasmid (see Supplementary data available at EMBO reports Online). Its chimeric nature was confirmed by the increased molecular size and the reactivity with an anti-EGF antibody. No SU proteins were detected in the virions from cells transfected with other EGF–chimeric env plasmids. Analysis of the cell lysates showed that all of the EGF–chimeric Env proteins were expressed at similar levels, but that only the E3 Env precursor was proteolytically processed to the SU and TM proteins (see Supplementary data).
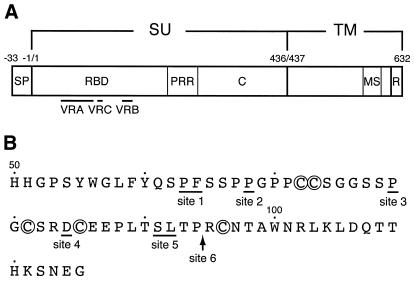
Fig. 1. Location of the ligand insertion sites in the Mo MLV Env. (A) The structure of the Env precursor is schematically shown. SU, surface subunit; TM, transmembrane subunit; SP, signal peptide; RBD, receptor-binding domain; PRR, proline-rich region; C, C-terminal domain; MS, membrane-spanning segment; R, R peptide. Variable regions (VRA, VRB and VRC) are indicated. (B) Ligand insertion sites are depicted under the amino acid sequence of VRA. These sites are indicated by underlines or an arrow. Cysteine residues involved in intrachain disulfide bond formation (Linder et al., 1992) are circled.
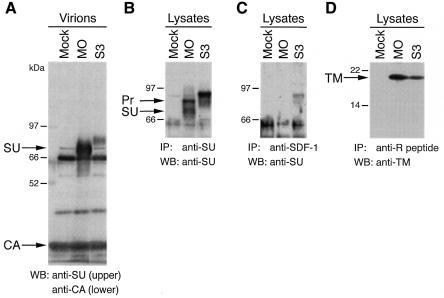
We then examined the insertion of the human SDF-1α (CXCL12), a CXC chemokine, as a ligand at site 3. A chimeric env (S3) plasmid, which contained the SDF-1α sequence without a linker, was constructed. Virions were pelleted from the culture supernatant of the stably transfected TELCeB6 cells. An SU protein was detected in the virions, although its amount was at a lower level relative to the virions from cells transfected with the wild-type env plasmid (Figure 2A, lane MO). It had a larger molecular size than the wild-type SU. Analysis of the cell lysates by immunoprecipitation and subsequent western blotting revealed that the S3 Env precursor could be immunoprecipitated by an anti-SDF-1 antibody (Figure 2B and C). It also showed the presence of the TM protein in the S3 env-transfected cells (Figure 2D), indicating an occurrence of cleavage of the Env precursor to the SU and TM proteins. Consequently, we found a single site (site 3) in the VRA into which peptide ligands can be inserted without abolishing cellular processing and viral incorporation of the chimeric Env proteins.
Fig. 2. Viral incorporation and cellular processing of the site 3 SDF-1α–chimeric Env protein (S3). (A) Viral incorporation was analyzed by western blotting (WB) of the virions. (B–D) The presence of the SDF-1α sequence in the S3 Env protein and its cellular processing were examined by immunoprecipitation (IP) and subsequent western blotting of the cell lysate. MO, S3 and Mock indicate either virions or lysates from TELCeB6 cells transfected with the wild-type env plasmid, the S3 env plasmid and without a plasmid, respectively, and cultured at 32°C. For (A)–(C) 8% SDS–polyacrylamide gels were used, and for (D) a 12% gel was used. The lower portion of the blot for (A) was used for detection of the Gag capsid protein (CA). Pr, Env precursor.
Delivery tropism of the vectors pseudotyped by the site 3 ligand–chimeric Env proteins
The transduction of cell lines by the lacZ retroviral vectors pseudotyped by the E3 and S3 Env proteins was examined. The E3 vector efficiently transduced mouse NIH 3T3 cells through the ecotropic MLV receptor, mCAT1 (Albritton et al., 1989) (see Supplementary data). However, it did not show the transduction of human cell lines irrespective of the expression of the EGF receptor (EGF-R), and its transduction through mCAT1 was specifically inhibited in EGF-R-overexpressing NIH 3T3 cells (see Supplementary data).
Like the E3 vector, the S3 vector efficiently transduced NIH 3T3 cells (Table I, 3T3.T4). Interference with the ecotropic Env protein (3T3.T4-E) indicated that the transduction occurs through the mCAT1 receptor. This was also shown by efficient transduction of HOS.mCAT1 cells by the S3 vector. This cell line was derived from the human osteosarcoma HOS cells (HOS.pBABE-puro) after stable transfection with the mCAT1 receptor cDNA. The cognate receptor for SDF-1α is CXCR4, which is a protein with seven membrane-spanning domains (Feng et al., 1996) in contrast to EGF-R. Remarkably, the S3 vector could transduce the HOS cells when they expressed CXCR4 (HOS.pBABE-puro versus HOS.CXCR4) and attained titers of 103 to 104 c.f.u./ml (Table I). The dependence of transduction by the S3 vector on the expression of CXCR4 was then characterized using human breast cancer cell lines naturally expressing CXCR4 at different levels (Müller et al., 2001) (Table I). Of the three cell lines examined, only the DU-4475 cell line, which was found to express the highest level of CXCR4 mRNA, was susceptible to transduction by the S3 vector. Collectively, the S3 vector showed an expanded tropism for the CXCR4-expressing cells. The wild-type ecotropic (MO) vector was unable to transduce the human cell lines except for the HOS.mCAT1 line, while the amphotropic (AM) vector efficiently transduced all these cell lines (Table I).
Table I. Transduction titers of the S3 vector for various cell lines.
| Enva |
Transduction titer (c.f.u./ml) for the cell line |
|||||||
|---|---|---|---|---|---|---|---|---|
| 3T3.T4 | 3T3.T4-E | HOS.pBABE-puro | HOS.mCAT1 | HOS.CXCR4b | MDA-MB-468c | MDA-MB-361c | DU-4475c | |
| S3 | 1.1 × 107 | <10 | <10 | 3.9 × 106 | 1.1 × 104 | <10 | 1.3 × 101 | 2.5 × 103 |
| MO | 4.3 × 107 | <10 | <10 | 9.3 × 106 | <10 | <10 | <10 | <10 |
| AM | —d | — | 5.5 × 106 | — | 6.0 × 106 | 4.3 × 106 | 1.7 × 106 | 3.6 × 105 |
aEnv protein that pseudotyped the lacZ vector.
bTwo other preparations of the S3 vector had transduction titers of 5.6 and 2.4 × 103 c.f.u./ml on this cell line.
cHuman breast cancer cell lines. Relative levels of CXCR4 mRNA expression were determined. They were 0 for MDA-MB-468, 1 for MDA-MB-361 and 12 for DU-4475.
dNot determined.
Evidence for the direct targeting by the S3 vector
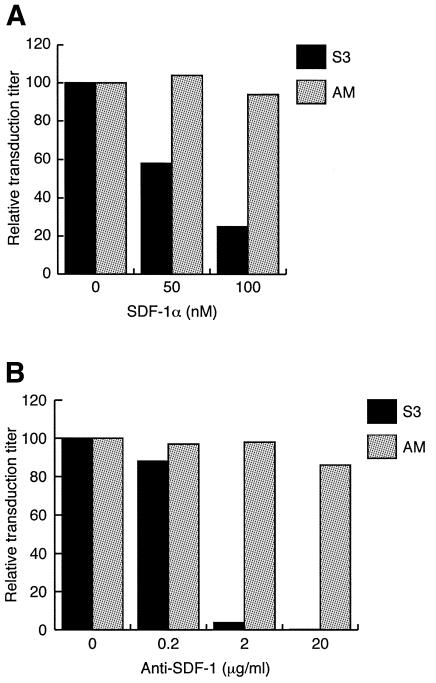
To know whether the CXCR4-dependent transduction by the S3 vector is based on the specific interaction between the SDF-1α moiety of the S3 Env protein and CXCR4, we examined the effects of soluble SDF-1α and an anti-SDF-1 antibody on transduction. As shown in Figure 3A, increasing concentrations of SDF-1α reduced the transduction of HOS.CXCR4 cells by the S3 vector to 25% of that obtained without the addition of SDF-1α. The inhibition of transduction by the anti-SDF-1 antibody was also observed (Figure 3B). Pretreatment of the S3 vector by the antibody at 2 µg/ml almost completely blocked the transduction of HOS.CXCR4 cells. SDF-1α and anti-SDF-1 had no effect on the transduction of the same cells by the AM vector. These results indicate that the SDF-1α sequence is displayed on the S3 vector envelope and that its specific interaction with CXCR4 is required for transduction.
Fig. 3. Inhibition of the CXCR4-dependent transduction of HOS.CXCR4 cells by the S3 vector with (A) soluble SDF-1α and (B) an anti-SDF-1 antibody. Transduction assays were performed using the S3 (black bars) and AM (gray bars) vectors in the presence of the indicated concentrations of SDF-1α or after pretreatment of the vector with the indicated concentrations of the antibody. Percent transduction titers relative to those obtained in the absence of the inhibitors are shown: S3, 6.1 and 4.6 × 103 c.f.u./ml for (A) and (B), respectively; and AM, 6.6 and 6.9 × 106 c.f.u./ml for (A) and (B), respectively.
Since the S3 vector retained the ability to transduce cells through the mCAT1 receptor, we considered the possibility that the CXCR4-dependent transduction of human cells might involve a weak interaction between the S3 Env protein and a human CAT1 protein. This hypothetical interaction could activate the membrane fusion potential of the S3 Env protein under the circumstance where the S3 vectors are adsorbed on the cell surface through specific binding to CXCR4. To examine this possibility, the transduction of HOS.CXCR4 cells expressing the ecotropic Env protein (HOS.CXCR4-E) by the S3 vector was determined. The S3 vector transduced HOS.CXCR4-E cells with a similar titer as that for HOS.CXCR4 cells (Table II), showing that the above possibility is unlikely. That the ecotropic entry function is not required for the CXCR4-dependent transduction by the S3 vector was also indicated using the NIH 3T3 cells expressing CXCR4. The expression of the ecotropic Env protein in the 3T3.T4.CXCR4 cells (3T3.T4.CXCR4-E) abolished the transduction by the MO vector, whereas it did not completely block the transduction by the S3 vector (Table II). This contrasted with the complete inhibition of transduction observed in the 3T3.T4-E cells (Table I). Taken together, these results indicate that the binding to CXCR4 by itself most likely activates the membrane fusion potential of the S3 Env protein.
Table II. Lack of contribution of the ecotropic entry function of the S3 vector to the CXCR4-dependent transduction.
| Enva |
Transduction titer (c.f.u./ml) for the cell line |
|||
|---|---|---|---|---|
| HOS.CXCR4 | HOS.CXCR4-E | 3T3.T4.CXCR4 | 3T3.T4.CXCR4-Eb | |
| S3 | 1.1 × 104 | 1.0 × 104 | 1.2 × 107 | 1.6 × 103 |
| MO | —c | — | 3.4 × 107 | <10 |
aEnv protein that pseudotyped the lacZ vector.
bTwo other preparations of the S3 vector had transduction titers of 1.3 and 1.2 × 103 c.f.u./ml on this cell line.
cNot determined.
DISCUSSION
Site 3 corresponds to Pro79 of the Mo MLV Env (Figure 1B). It is identical to, or very close to, the sites that have recently been shown to tolerate the insertion of a 15mer peptide and a single-chain antibody variable domain (Lorimer and Lavictoire, 2000; Wu et al., 2000). Pro79 is located in a small disulfide-bonded loop involving Cys73 and Cys81 and is probably exposed on the surface of the RBD, as predicted by the crystal structure of the related RBD of the Friend MLV Env protein (Fass et al., 1997).
The inability of the E3 vector to transduce cells through EGF-R could be due to the failure in one or several post-receptor-binding steps necessary for transduction. These include coupling the receptor binding with the induction of a fusogenic state of Env and placing the virions close to the cell membranes. In addition, the internalization property of EGF-R may not be suitable for transduction.
The S3 vector showed an expanded delivery tropism, namely it could transduce human cells at titers of 103 to 104 c.f.u./ml when these cells expressed a certain level of CXCR4 (Table I). Interference experiments (Table II) indicated that the ecotropism of the S3 vector does not contribute to the CXCR4-dependent transduction of cells. We obtained the S3 vector without coexpression of the wild-type Mo MLV Env. The present study thus indicates that the S3 vector utilizes CXCR4 as a receptor for the transduction of cells. The efficiency of transduction through CXCR4 by the S3 vector, however, is still low. The titer of the S3 vector for transduction through CXCR4 (HOS.CXCR4) was at least two orders of magnitude lower than that through mCAT1 (Table I, HOS.mCAT1). Further study is necessary to improve the efficiency of the targeted transduction by the S3 vector before its practical applications.
Our results are consistent with the idea that only proteins having multiple membrane-spanning domains can act as receptors for MLV vectors. It is possible that these membrane proteins, by virtue of their topology, are favored for placing the virions close to the cell membranes. CXCR4 has not been known as a receptor for infection by any subgroup of MLVs, but it has been known to function as a coreceptor for infection by human immunodeficiency virus type 1 (HIV-1) (Feng et al., 1996). It would be interesting to know whether the range of proteins that can be utilized as receptors for MLV vectors extend to the coreceptors for HIVs, members of the chemokine receptors or any protein with multiple membrane-spanning domains.
SDF-1α is known to inhibit the infection of HIV-1. Its efficacy as an inhibitor of HIV-1 entry is much greater than that as an inhibitor of transduction of the S3 vector. The concentration of SDF-1α for 50% inhibition of the HIV-1 entry was ∼3 nM (Amara et al., 1997), whereas it was >50 nM for 50% inhibition of the S3 vector transduction (Figure 3A). This may reflect a difference of the cell entry point between HIV-1 and ecotropic MLVs.
The present study suggests that a functional non-viral recognition module can be incorporated into the Env protein to lead to the direct targeting of MLV vectors, provided that the structures of essential Env regions are not significantly affected and that the targeted membrane protein has the appropriate topology and internalization property. Recently, it was reported that the HIV-1 vector with replacement of the V3 region of Env by a portion of SDF-1a could transduce cells through CXCR4 and CD4 (Yonezawa et al., 2001). The S3 vector is distinct in that it transduces cells through a membrane protein unrelated to the known MLV receptors and that its transduction is independent of CD4.
METHODS
Cell lines.
TELCeB6 was provided by F.-L. Cosset (ENS de Lyon, France). HOS.pBABE-puro (HOS cells transduced by the BABE-puro retroviral vector), HOS.CXCR4 (Deng et al., 1996), 3T3.T4 and 3T3.T4.CXCR4 (Deng et al., 1997) were obtained through the AIDS Research and Reference Reagent Program, NIH. Human breast cancer cell lines MDA-MB-468, MDA-MB-361 and DU-4475 were obtained from the ATCC. Relative amounts of CXCR4 mRNA in these cells were determined by the LightCycler-based real-time quantitative RT–PCR method using the PCR primers 5′-GACCACAATCATCCCCATCC-3′ and 5′-CCTCGGTGATGGAAATCCAC-3′. HOS.mCAT1 was prepared by stable transfection of HOS.pBABE-puro cells with the pIRESneo–mCAT1 plasmid, which was constructed by inserting the mCAT1 cDNA (provided by J.M. Cunningham, Harvard Medical School) into the EcoRI site of the pIRESneo2 vector (Clontech). 3T3.T4-E and 3T3.T4.CXCR4-E were prepared by the infection of 3T3.T4 and 3T3.T4.CXCR4, respectively, with Mo MLV. HOS.CXCR4-E was prepared by stable transfection of HOS.CXCR4 with pCE4.1. Unless otherwise indicated, all the cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and an appropriate selection drug at 37°C in 5% CO2.
Construction of Env expression plasmids.
The Mo MLV env sequence was amplified by PCR using pML48 (Bacheler and Fan, 1981) as a template, cloned into pTargeT (Promega) and sequenced (pCE4.1). For construction of EGF–Env expression plasmids, the 0.8-kb BamHI fragment of pCE4.1 encoding the N-terminal half of the SU was subcloned into pKF18k (Takara) (pKF18-B0.8). Six plasmids were produced, each containing a SacII recognition sequence at different sites in the region of pKF18-B0.8 corresponding to the VRA, using site-directed mutagenesis (Mutan-super express Km, Takara). Mutagenic oligonucleotides and the coordinates (Shinnick et al., 1981) of the SacII sites are as follows: site 1, 5′-pGCTAGAATATCAATCCGCGGTTTCTTCTCC-3′ (6060–6065); site 2, 5′-pCAATCCCCTTTTTCTTCTCCGCGGGGG-3′ (6074–6079); site 3, 5′-pGGCAGCAGCCGCGGCTGTTCCAG-3′ (6109–6114); site 4, 5′-pCCCAGGCTGTTCCCGCGGCTGCGAAGAACC-3′ (6121–6126); site 5, 5′-pAAGAACCTTTAACCGCGGTCACCCCTCGGTGC-3′ (6144–6149); and site 6, 5′-pCCCTCACCCCGCGGTGCAACAC-3′ (6155–6160). Three sets of sense and antisense primers (for cassette 1, 5′-CCGCGGGGCACTATTCCGTAGGAAATAGTGACTCTG-3′ and 5′-CCGCGGCCCCGTGGCCAGCGTGGCCCAGTTCCCACC-3′; cassette 2, 5′-CCGCGGCACTATTCCGTAGGAAATAGTGACTCTG-3′ and 5′-CCGCGGCCCGTGGCCAGCGTGGCCCAGTTCCCACC-3′; and cassette 3, 5′-CCGCGGCCACTATTCCGTAGGAAATAGTGACTCTG-3′ and 5′-CCGCGGGCCCCGTGGCCAGCGTGGCCCAGTTCCCACC-3′) were used to amplify the 53-amino-acid human EGF sequence by PCR using the λ EGF116 phage DNA (ATCC 59957) as a template. Each primer set was designed so that (i) sequences encoding five amino acids flanking the mature EGF sequence were also amplified, (ii) single Arg residues at one amino acid N-terminal side to the N-terminus of EGF and at the C-terminus of EGF were changed to Gly residues and (iii) SacII sites were created at both ends. The PCR product was cloned into pTargeT and sequenced. One of these EGF cassettes was chosen, based on correct reading frame (cassette 1 for sites 1 and 5, cassette 2 for sites 2 and 6, and cassette 3 for sites 3 and 4) and inserted into the unique SacII site created in pKF18-B0.8 to make the pKBS-E plasmids. Finally, the 1.0-kb BamHI insert of pKBS-E was used to replace the corresponding 0.8-kb fragment of pCE4.1, resulting in six different EGF–Env expression plasmids.
For construction of the S3 Env expression plasmid, the sequence encoding the 68-amino-acid segment of SDF-1α from Lys22 was amplified by PCR using the plasmid containing human SDF-1α cDNA (Shirozu et al., 1995; provided by T. Honjo, Kyoto University, Japan) as a template and the primers 5′-CCGCGGGAAGCCCGTCAGCCTGAGC-3′ and 5′-CCGCGGGACTTGTTTAAAGCTTTCTCCAG-3′. The PCR product containing SacII sites at both ends was cloned into pTargeT and sequenced. The SDF-1α cassette was then inserted into the SacII site created at site 3 in pKF18-B0.8 to result in the pKBS-S3 plasmid. The whole Env-encoding sequence was reconstituted as described above.
Retroviral vector production.
The Env expression plasmid was transfected by the modified calcium phosphate precipitation method into TELCeB6 cells, and the cells were selected for resistance to G418 (450 µg/ml). Drug-resistant cells were mixed as a stably transfected cell pool. The pooled cells were cultured at 37°C to near confluency and then transferred for culture at 32°C. Five days after transfer, the culture medium was replaced by fresh medium and the cells cultured for an additional 24 h. The culture supernatant containing a retroviral vector was collected, cleared by centrifugation at 500 g at 4°C for 10 min, filtered through a 0.45 µm filter (Millipore) and used immediately or stored frozen at –80°C until use. Vectors pseudotyped by the ecotropic and the amphotropic MLV Env were produced using pCE4.1 and pCMV4070A (provided by J.M. Heard, Institut Pasteur), respectively.
Detection of Env proteins.
Goat anti-R-MLV gp70 (anti-SU) and anti-R-MLV p30 (anti-CA) were obtained from ViroMed Biosafety Laboratories and goat anti-SDF-1 from Santa Cruz Biotechnology. Rabbit anti-R peptide (C-terminal 16 amino acids of the TM protein) and anti-MLV TM were provided by J.H. Elder (Scripps Res. Inst.) and A. Rein (NCI, NIH), respectively. Virions were pelleted from culture supernatants by the ultracentrifugation method (Januszeski et al., 1997). Virion proteins were separated on an 8% SDS–polyacrylamide gel and subjected to western blot analysis. Primary antibodies used were anti-SU (1:1300) and anti-CA (1:10 000). Horseradish peroxidase (HRP)-conjugated anti-goat IgG was used as a secondary antibody. Lysates were prepared from cells grown at 32°C using the lysis buffer consisting of PBS(-), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitors (Boehringer Mannheim) by incubation on ice for 1 h and centrifugation at 10 000 g for 10 min to pellet nuclei. Cell lysates were immunoprecipitated by the standard method using anti-SU, anti-SDF-1 and anti-R peptide. The immunoprecipitates were subjected to 8 or 12% SDS–PAGE and western blotting using anti-SU or anti-TM (1:500) as a primary antibody and HRP-conjugated protein G as a secondary reagent.
Transduction assay.
Assay cells (1–5 × 105) were seeded in 35-mm dishes. On the next day, the cells were exposed to 1 ml of the vector solution in the presence of 5 µg/ml polybrene at 37°C for 4 h. Fresh medium (1 ml) was added, and the cells were cultured for 48 h prior to X-Gal staining to detect cells expressing the Escherichia coli β-galactosidase. The number of colonies containing stained cells or the fraction of stained cells per total cells (for DU-4475 cell line) was measured microscopically. Titers (c.f.u./ml) were determined from the duplicate assays. For SDF-1α inhibition, cells were exposed to 1 ml of the vector solution containing the recombinant human SDF-1α (Sigma) and polybrene for 4 h. For inhibition by anti-SDF-1, vectors were pretreated with the antibody (R & D Systems) at 4°C for 1 h and used for transduction.
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Esser for establishing the NE13 cell line, T. Kawamura and M. Ogura for technical assistance, A. Sarai for helpful discussion, and H. Nakauchi, Y. Aoki, R. Ishimura, M. Sakaue, A. Ishimoto and Y. Kitamura for support and advice. This study was performed through Special Coordination Funds of the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government.
REFERENCES
- Albritton L.M., Tseng, L., Scadden, D. and Cunningham, J.M. (1989) A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell, 57, 659–666. [DOI] [PubMed] [Google Scholar]
- Amara A., Le Gall, S., Schwartz, O., Salamero, J., Montes, M., Loetscher, P., Baggiolini, M., Virelizier, J.-L. and Arenzana-Seisdedos, F. (1997) HIV coreceptor downregulation as antiviral principle: SDF-1α-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J. Exp. Med., 186, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W.F. (1998) Human gene therapy. Nature, 392 (Suppl.), 25–30. [DOI] [PubMed] [Google Scholar]
- Bacheler L. and Fan, H. (1981) Isolation of recombinant DNA clones carrying complete integrated proviruses of Moloney murine leukemia virus. J. Virol., 37, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C.A., Tun, R.Y.M., Rubinstein, D.B., Guillaume, T., Cannon, P.M. and Anderson, W.F. (1999) Targeting retroviral vectors to CD34-expressing cells: binding to CD34 does not catalyze virus–cell fusion. Hum. Gene Ther., 10, 545–557. [DOI] [PubMed] [Google Scholar]
- Cosset F.-L., Morling, F.J., Takeuchi, Y., Weiss, R.A., Collins, M.K.L. and Russell, S.J. (1995a) Retroviral retargeting by envelopes expressing an N-terminal binding domain. J. Virol., 69, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosset F.-L., Takeuchi, Y., Battini, J.L., Weiss, R.A. and Collins, M.K.L. (1995b) High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol., 69, 7430–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.K. et al. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature, 381, 661–666. [DOI] [PubMed] [Google Scholar]
- Deng H.K., Unutmaz, D., KewalRamani, V.N. and Littman, D.R. (1997) Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature, 388, 296–300. [DOI] [PubMed] [Google Scholar]
- Fass D., Davey, R.A., Hamson, C.A., Kim, P.S., Cunningham, J.M. and Berger, J.M. (1997) Structure of a murine leukemia virus receptor binding glycoprotein at 2.0 Å resolution. Science, 277, 1662–1666. [DOI] [PubMed] [Google Scholar]
- Feng Y., Broder, C.C., Kennedy, P.E. and Berger, E.A. (1996) HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science, 272, 872–877. [DOI] [PubMed] [Google Scholar]
- Gollan T.J. and Green, M.R. (2002a) Redirecting retroviral tropism by insertion of short, nondisruptive peptide ligands into envelope. J. Virol., 76, 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollan T.J. and Green, M.R. (2002b) Selective targeting and inducible destruction of human cancer cells by retroviruses with envelope proteins bearing short peptide ligands. J. Virol., 76, 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszeski M.M., Cannon, P.M., Chen, D., Rozenberg, Y. and Anderson, W.F. (1997) Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol., 71, 3613–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara N., Dozy, A.M. and Kan, Y.W. (1994) Tissue-specific targeting of retroviral vectors through ligand–receptor interactions. Science, 266, 1373–1376. [DOI] [PubMed] [Google Scholar]
- Linder M., Linder, D., Hahnen, J., Schott, H.H. and Stirm, S. (1992) Localization of the intrachain disufide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur. J. Biochem., 203, 65–73. [DOI] [PubMed] [Google Scholar]
- Lorimer I.A.J. and Lavictoire, S.J. (2000) Targeting retrovirus to cancer cells expressing a mutant EGF receptor by insertion of a single chain antibody variable domain in the envelope glycoprotein receptor binding lobe. J. Immunol. Methods, 237, 147–157. [DOI] [PubMed] [Google Scholar]
- Müller A. et al. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature, 410, 50–56. [DOI] [PubMed] [Google Scholar]
- Russell S.J. and Cosset, F.L. (1999) Modifying the host range properties of retroviral vectors. J. Gene Med., 1, 300–311. [DOI] [PubMed] [Google Scholar]
- Shinnick T.M., Lerner, R.A. and Sutcliiffe, J.G. (1981) Nucleotide sequence of Moloney murine leukemia virus. Nature, 293, 543–548. [DOI] [PubMed] [Google Scholar]
- Shirozu M., Nakano, T., Inazawa, J., Tashiro, K., Tada, H., Shinohara, T. and Honjo, T. (1995) Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics, 28, 495–500. [DOI] [PubMed] [Google Scholar]
- Sommerfelt M.A. (1999) Retrovirus receptors. J. Gen. Virol., 80, 3049–3064. [DOI] [PubMed] [Google Scholar]
- Wu B.W., Lu, J., Gallaher, T.K., Anderson, W.F. and Cannon, P.M. (2000) Identification of regions in the Moloney murine leukemia virus SU protein that tolerate the insertion of an integrin-binding peptide. Virology, 269, 7–17. [DOI] [PubMed] [Google Scholar]
- Yajima T., Kanda, T., Yoshiike, K. and Kitamura, Y. (1998) Retroviral vector targeting human cells via c-Kit–stem cell factor interaction. Hum. Gene Ther., 9, 779–787. [DOI] [PubMed] [Google Scholar]
- Yonezawa A., Hori, T., Takaori-Kondo, A., Morita, R. and Uchiyama, T. (2001) Replacement of the V3 region of gp120 with SDF-1 preserves the infectivity of T-cell line-tropic human immunodeficiency virus type 1. J. Virol., 75, 4258–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhu, L., Lee, S., Li, L., Chang, E., Soong, N.W., Douer, D. and Anderson, W.F. (1999) Identification of the block in targeted retroviral-mediated gene transfer. Proc. Natl Acad. Sci. USA, 96, 4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.