Abstract
Background:
Members of the sulfotransferase superfamily (SULT) influence the activity of a wide range of hormones, neurotransmitters, metabolites and xenobiotics. However, their roles in developmental processes are not well characterized even though they are expressed during embryogenesis. We previously found in a microarray screen that Six1 up-regulates LOC100037047, which encodes XB5850668.L, an uncharacterized sulfotransferase.
Results:
Since Six1 is required for patterning the embryonic ectoderm into its neural plate, neural crest, preplacodal and epidermal domains, we used loss- and gain-of function assays to characterize the role of XB5850668.L during this process. Knockdown of endogenous XB5850668.L resulted in the reduction of epidermal, neural crest, cranial placode and otic vesicle gene expression domains, concomitant with neural plate expansion. Increased levels had minimal effects, but infrequently expanded neural plate and neural crest gene domains, and infrequently reduced cranial placode and otic vesicle gene domains. Mutation of two key amino acids in the sulfotransferase catalytic domain required for PAPS binding and enzymatic activity tended to reduce the effects of overexpressing the wild-type protein.
Conclusions:
Our analyses indicates that XB5850668.L is a member of the SULT2 family that plays important roles in patterning the embryonic ectoderm. Some aspects of its influence likely depend on sulfotransferase activity.
Keywords: LOC100037047, otic vesicle, neural border, neural plate, Xenopus, preplacodal ectoderm, neural crest, Six1
INTRODUCTION
Members of the cytosolic sulfotransferase (SULT) superfamily catalyze the transfer of a sulfuryl (SO3) group from a donor to a substrate. The most common sulfate donor is 3’phosphoadenosine 5’phosphosulfate (PAPS), whereas substrates include many hormones, neurotransmitters, metabolites and xenobiotic compounds, such as environmental toxins, antibiotics and cancer promoting compounds.1, 2, 3, 4 Members of the SULT superfamily share considerable amino acid sequence and structural similarities. However, each family displays distinct but overlapping substrate specificities resulting in different biological functions.1, 5, 6 The biological functions and substrate specificities have been extensively studied in adult tissues where they play important roles in detoxifying compounds and metabolizing drugs.6, 7 Adding sulfuryl groups to endogenous and xenobiotic compounds is thought to increase their water solubility, thus increasing diffusion and excretion, and to modify their bioactivity. The role of SULTs in developmental processes, however, has not been studied in any detail. SULTs have been detected in embryonic tissues in nematode, fly, fish, frog and humans,4, 8, 9, 10, 11, 12 but aside from studies that identified potential substrates, little is known about how their activities affect developmental processes.
In a search for downstream targets of Six1, a homeodomain-containing transcription factor involved in patterning the embryonic ectoderm and development of cranial sensory organs,13, 14, 15 we performed a microarray screen to identify novel genes regulated by Six1 in ectodermal explants. We identified an mRNA designated LOC100037047 that was upregulated >3-fold by Six1 and whose embryonic expression required the presence of Six1.16 The provisional gene symbol for LOC100037047 is XB5850668.L, which encodes an uncharacterized sulfotransferase. XB5850668.L is expressed in many of the same embryonic tissues as Six1,17 including the cranial sensory placodes, neural crest-derived branchial arches, somites and nephric mesoderm.16 Herein we present a phylogenetic analysis that indicates XB5850668.L is a member of the SULT2 family.
Because the patterning of the cranial derivatives of the embryonic ectoderm is involved in many craniofacial syndromes, we investigated whether the establishment of the progenitor fields of the neural and non-neural domains of the embryonic ectoderm are altered after either loss-of-function or gain-of function of this uncharacterized sulfotransferase. Reducing the endogenous levels of XB5850668.L by microinjection of translation blocking antisense morpholino oligonucleotides (MOs) caused neural plate gene expression domains to expand, whereas the predominant phenotype for neural crest, preplacodal ectoderm (PPE) and epidermal gene domains was a reduced expression domain. At later stages, otic vesicle gene expression domains also were reduced. In contrast, increasing the levels of XB5850668.L by mRNA injections had no effect in the majority of embryos. A small percentage of embryos showed broader domains of neural plate and neural crest genes, whereas about 40% of embryos showed smaller domains of most PPE and all otic vesicle genes analyzed. To determine whether the overexpression effects required enzymatic activity, we mutated two key amino acids required for the sulfotransferase activity of other members of the SULT superfamily.18, 19 We found that for most genes, at least one of the mutated proteins caused less of an effect compared to that of the wild type protein. Together these results demonstrate that XB5850668.L plays a role in craniofacial development by balancing the proportion of the embryonic ectoderm that differentiates into neural plate versus neural crest and cranial placode structures. Because XB5850668.L is regulated by Six1,16 a gene that is mutated in Branchio-oto-renal (BOR; OMIM 608389) and in Deafness, autosomal dominant 23 (DFNA23; OMIM 605192) syndromes, we suggest that members of the SULT2 family should be investigated for potential involvement in with these syndromes.
RESULTS
LOC100037047-XB5850668.L encodes a sulfotransferase
LOC100037047 (NCBI accession number BC130199.1,20) was identified as an upregulated Six1 target by a microarray screen of Six1-treated ectodermal explants.16 Specifically, LOC100037047 was increased >3-fold by Six1 in the explants, and its expression domain was expanded in embryos by Six1 mRNA injection and reduced by Six1 translation-blocking antisense morpholino oligonucleotide (MO) injection. Sequencing of the commercially available full-length ORF clone (BG885936, Open Biosystems) identified this clone as a minor variant of Xenopus laevis LOC100037047 (Fig. 1). According to the Xenopus laevis gene assembly (v10.1) the provisional gene symbol for LOC100037047 is XB5850668.L (xenbase.org; NCBI Accession number NM_001097778), which is located on chromosome 7L; there is no homeolog on chromosome 7S (Fig. 2A). It has 88.16% identity with the Xenopus tropicalis ortholog, XB5850668 (NCBI Accession number NM_001126496.1). XB5850668.L likely functions as a sulfotransferase because it contains a highly conserved sulfotransferase domain between residues 30-276 (Fig. 1), including the 5’ phosphosulfate binding loop (PSB loop) (aa 38-45; aa 118-128) and the C-terminal PB-loop motif (aa 245-257) both of which are required for PAPS binding.7, 18, 19
Figure 1:

Clone BG88539 encodes a sulfotransferase. Amino acid sequence of BG88539 (red font) is identical (red dots) to that of XB5850668.L (black font, NM_001097778; and to LOC100037047, BC130199.1, not shown) except at amino acids #17, #151, #219 and #229. The highly conserved sulfotransferase domain extends from residues 30-276 and includes signature sequences (red bars) for the 5’ phosphosulfate binding loop (PSB loop; aa 38-45; aa 118-128) and a C-terminal P-loop motif (aa 245-257) that are required for PAPS binding. Red arrows denote the amino acids (K40, H96) that were experimentally altered.
Figure 2:
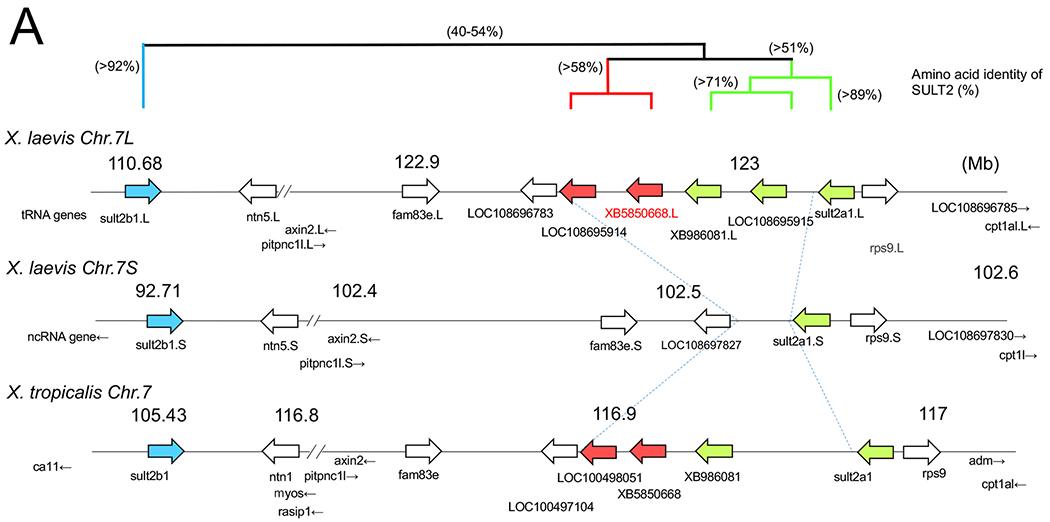
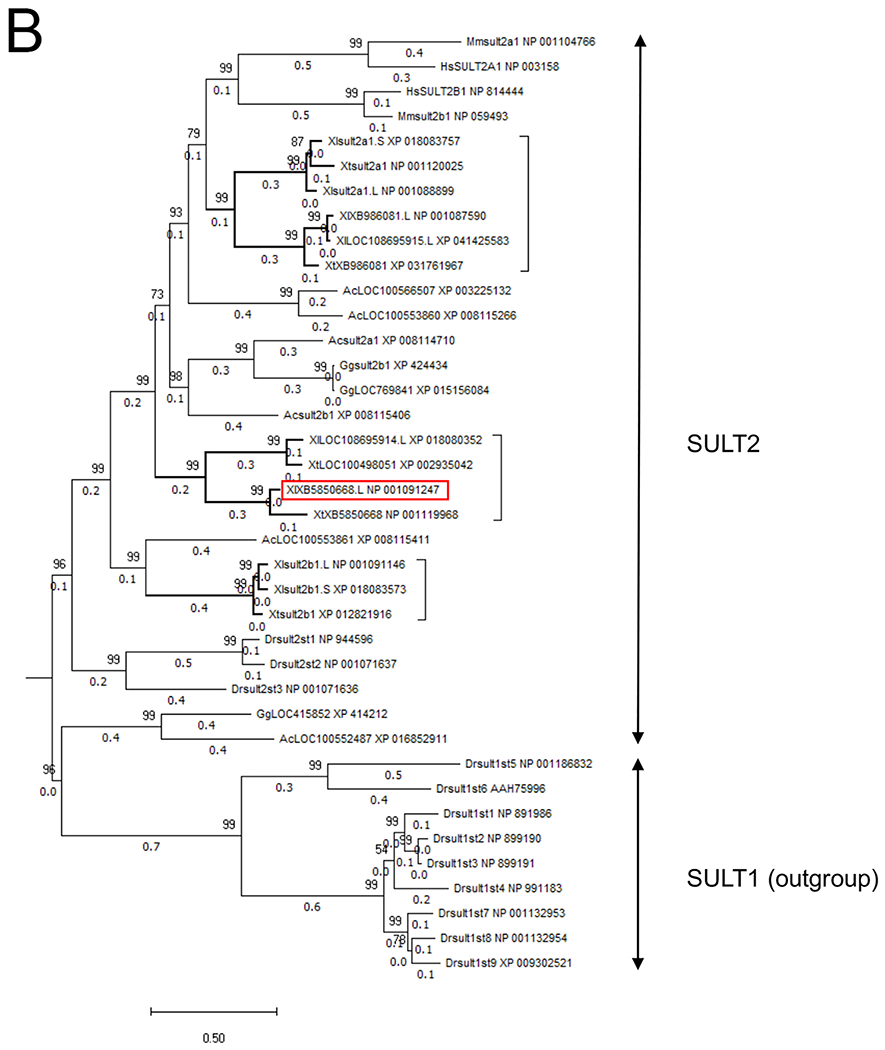
Chromosomal localization of the members of SULT2 gene family including XB5850668.L in Xenopus laevis and Xenopus tropicalis, and their phylogenetic relationships.
A: The loci of SULT2 family genes on Xenopus chromosome 7. The gene cluster consists of 6 members on the L-subgenome and 2 members on the S-subgenome in the allotetraploid X. laevis, and 5 members in the diploid X. tropicalis. Four members were lost from the S-subgenome in X. laevis. This chromosomal region showed shared synteny between X. laevis and X. tropicalis (see also Table 1), but little synteny across species from mammals to a teleost (see also Table 2). The three groups in the Xenopus SULT2 gene family are shown by bold arrows in blue, red, and green, and their phylogenetic relationships are depicted with amino acid identity (%) above the chromosomal maps.
B: Phylogenetic tree of the vertebrate SULT2 gene family. The tree was constructed with the maximum likelihood method from eight X. laevis (Xl), five X. tropicalis (Xt) and 16 other vertebrate SULT2 amino acid sequences and with nine Danio rerio (Dr) SULT1 amino acid sequences as an outgroup. Node values represent the Chi-square-based parametric values returned by the approximate likelihood ratio test. Gene and protein IDs, and annotations and other information for each member are shown in Table 1. The first two letters of the gene names denote the species: Ac, Anolis carolinensis (green anole); Dr, Danio rerio (zebrafish); Gg, Gallus gallus (chicken); Hs, Homo sapiens (human); Mm, Mus musculus (house mouse); Xl, Xenopus laevis (African clawed frog); and Xt, Xenopus tropicalis (western clawed frog). The last letter (S or L) of the gene names for Xenopus laevis indicate homeologs derived from either the S- or L-subgenome. The three groups in the Xenopus SULT2 gene family are shown by bold branches and a bracket, and the XB5850668.L gene is outlined by a red rectangle.
In comparison to LOC100037047 (BC130199.1) and XB5850668.L (NM_001097778), BG885936 has four amino acid (aa) substitutions (Fig. 1). At aa 17, which lies outside the sulfotransferase domain, lysine (K) is substituted by asparagine (N); this amino acid also is K in X. tropicalis XB5850668 (NM_001126496.1). At aa 151, leucine (L) is substituted by isoleucine (I); this amino acid is L in X. tropicalis XB5850668. At aa 219, threonine (T) is substituted to isoleucine (I); this amino acid also is I in X. tropicalis XB5850668. At aa 229, serine (S) is substituted to a threonine (T) that is not predicted to be phosphorylated (NetPhos2.0); this amino acid also is S in X. tropicalis XB5850668. These minor variants fall outside the key regions for PAPS binding (Fig. 1) and therefore are not expected to have an effect on sulfotransferase activity. However, amino acid changes within the sulfotransferase domain potentially could influence substrate specificity.5
Phylogenetic analysis of the vertebrate SULT2 gene family and identification of Xenopus members
A BLAST search analysis of X. laevis XB5850668.L identified 8 members of the X. laevis SULT2 gene family. In the allotetraploid X. laevis, 5 members were derived from chromosome 7 of the L-subgenome and 2 of the S-subgenome (Fig. 2A). This chromosomal region showed shared synteny between X. laevis and X. tropicalis (Table 1), but little synteny across species from mammals to a teleost (Table 2). Annotations of these genes in public databases are still currently incomplete. Therefore, to clarify their identification, we conducted a maximum likelihood (ML) phylogenetic tree (Fig. 2B) with the SULT2 gene family members of various animal groups from fish to mammals. Our ML tree of SULT2 genes did not correspond to a phylogenetic tree of vertebrates, suggesting multiple events of gene duplication and subsequent deletion in SULT2 loci during vertebrate evolution. In the vertebrate SULT2 gene family, our ML tree revealed the presence of at least eight major classes with more than 47% amino acid identities. Among them, three SULT2 classes were detected in Xenopus. These are: (1) SULT2A1 and its related genes (green in Fig. 2A), which are most related to the mammalian SULT2A/2B classes; (2) SULT2B1 class (blue in Fig. 2A), which did not form a monophyletic clade with any of the mammalian, avian or reptilian SULT2B1 classes; and (3) the intermediate class (red in Fig. 2A), to which X. laevis XB5850668.L belongs. Homeologous genes of XB5850668.L and related gene (LOC 108695914.L) were not detected in the S-subgenome in X. laevis (Fig. 2A). The deduced amino acid sequences of the Xenopus SULT2 and related genes showed >51%, >58% and 92% identities within the classes (1), (2) and (3), respectively, and 40-54% identities among the three classes.
Table 1.
Amino acid identity percents of Xenopus SULT2 family members
| Xlsult2a1.L_NP_001088899_ | Xlsult2a1.S_XP_018083757_ | Xtsult2a1_NP_001120025_ | XlXB986081.L_NP_001087590_ | XlLOC108695915.L_XP_041425583_ | XtXB986081_XP_031761967_ | XlLOC108695914.L_XP_018080352_ | XtLOC100498051_XP_002935042_ | XlXB5850668.L_NP_001091247_ | XtXB5850668_NP_001119968_ | Xlsult2b1.L_NP_001091146_ | Xlsult2b1.S_XP_018083573_ | Xtsult2b1_XP_012821916_ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xlsult2a1.L_NP_001088899_ | ID | 0.923 | 0.891 | 0.603 | 0.62 | 0.526 | 0.503 | 0.506 | 0.498 | 0.475 | 0.434 | 0.441 | 0.434 |
| Xlsult2a1.S_XP_018083757_ | 0.923 | ID | 0.902 | 0.586 | 0.602 | 0.519 | 0.496 | 0.5 | 0.501 | 0.475 | 0.431 | 0.434 | 0.441 |
| Xtsult2a1_NP_001120025_ | 0.891 | 0.902 | ID | 0.593 | 0.609 | 0.515 | 0.496 | 0.5 | 0.515 | 0.496 | 0.431 | 0.434 | 0.438 |
| XlXB986081.L_NP_001087590_ | 0.603 | 0.586 | 0.593 | ID | 0.962 | 0.71 | 0.501 | 0.501 | 0.52 | 0.493 | 0.461 | 0.464 | 0.464 |
| XlLOC108695915.L_XP_041425583_ | 0.62 | 0.602 | 0.609 | 0.962 | ID | 0.728 | 0.513 | 0.513 | 0.539 | 0.506 | 0.462 | 0.465 | 0.465 |
| XtXB986081_XP_031761967_ | 0.526 | 0.519 | 0.515 | 0.71 | 0.728 | ID | 0.435 | 0.432 | 0.489 | 0.451 | 0.4 | 0.397 | 0.4 |
| XlLOC108695914.L_XP_018080352_ | 0.503 | 0.496 | 0.496 | 0.501 | 0.513 | 0.435 | ID | 0.902 | 0.604 | 0.581 | 0.45 | 0.45 | 0.447 |
| XtLOC100498051_XP_002935042_ | 0.506 | 0.5 | 0.5 | 0.501 | 0.513 | 0.432 | 0.902 | ID | 0.621 | 0.588 | 0.453 | 0.453 | 0.45 |
| XlXB5850668.L_NP_001091247_ | 0.498 | 0.501 | 0.515 | 0.52 | 0.539 | 0.489 | 0.604 | 0.621 | ID | 0.853 | 0.479 | 0.472 | 0.469 |
| XtXB5850668_NP_001119968_ | 0.475 | 0.475 | 0.496 | 0.493 | 0.506 | 0.451 | 0.581 | 0.588 | 0.853 | ID | 0.461 | 0.447 | 0.454 |
| Xlsult2b1.L_NP_001091146_ | 0.434 | 0.431 | 0.431 | 0.461 | 0.462 | 0.4 | 0.45 | 0.453 | 0.479 | 0.461 | ID | 0.937 | 0.93 |
| Xlsult2b1.S_XP_018083573_ | 0.441 | 0.434 | 0.434 | 0.464 | 0.465 | 0.397 | 0.45 | 0.453 | 0.472 | 0.447 | 0.937 | ID | 0.92 |
| Xtsult2b1_XP_012821916_ | 0.434 | 0.441 | 0.438 | 0.464 | 0.465 | 0.4 | 0.447 | 0.45 | 0.469 | 0.454 | 0.93 | 0.92 | ID |
Table 2.
Vertebrate sulfotransferase 2 (SULT2) members
| Species | Gene name used in Fig. 1 | Gene ID | protein ID | Amino acid residue | Annotation and other information | Linked genes |
|---|---|---|---|---|---|---|
| Danio rerio (Zebrafish ) | ||||||
| Drsult1st1 | LOC323424 | AAO64983, NP_891986.1 | 299 | Chr. 8, partial start | ||
| Drsult1st2 | LOC791732 | AAO64984, NP_899190.2 | 301 | Chr. 8 | ||
| Drsult1st3 | LOC368270 | AAP55637, NP_899191.2 | 301 | Chr. 8 | ||
| Drsult1st4 | LOC402915 | AAP55638, NP_991183.1 | 304 | Chr. 8 | ||
| Drsult1st5 | LOC619193 | AAY47051, NP_001186832.1 | 293 | Chr.23 | ||
| Drsult1st6 | LOC436872 | AAX59994, NP_001002599.1 | 308 | Chr. 12 | ||
| Drsult1st7 | LOC100196920 | ACA81603, XP_017213027.1 | 301 | Chr. 8 | ||
| Drsult1st8 | LOC100499180 | ACA81604. NP_001132954.1 | 301 | Chr. 8 | ||
| Drsult1st9 | LOC100196921 | AFC93291, XP_009302521.1 | 300 | Chr. 8, partial | ||
| Drsult2st1 | LOC338214 | NP_944596 | 287 | Chr.12 | wu:fj29h11, bsk146, abcc3 | |
| Drsult2st2 | LOC777793 | NP_001071637 | 287 | Chr. 12 | ||
| Drsult2st3 | LOC777792 | NP_001071636 | 288 | Chr. 16 | ncam3, cd22, tmem147, gapdhs | |
| Xenopus laevis (African clawed frog) | ||||||
| Xlsult2a1.L | LOC496246 | NP_001088899 | 287 | sult1a1 L homeolog; Chr. 7L | rps9,cpt1 | |
| Xlsult2b1.L | LOC100036899 | NP_001091146 | 288 | sult2b1 L homeolog; Chr. 7L | ntn5, | |
| XlLOC108695914.L | LOC108695914 | XP_018080352 | 287 | sult2b1; Chr. 7L | fam83e, axin2, pitpnc1 | |
| XlXB5850668.L | LOC100037047 | NP_001091247 | 284 | uncharacterized XB5850668 L homeolog; Chr. 7L | ||
| XlXB986081.L | LOC447414 | NP_001087590 | 290 | provisional ortholog of sult2b1 L homeolog; Chr. 7L | ||
| XlLOC108695915.L | LOC108695915 | XP_041425583 | 287 | sult2b1-like; Chr. 7L | ||
| Xlsult2a1.S | LOC108697829 | XP_018083757 | 287 | sult2a1 S homeolog; Chr. 7S | rps9, cpt1,adm | |
| Xlsult2b1.S | LOC432283 | NP_018083573 | 288 | sult2b1 S homeolog; STD2; Chr.7S | ntn5, | |
| Xenopus tropicalis (Tropical clawed frog) | ||||||
| Xtsult2a1 | LOC100144988 | NP_001120025 | 287 | sult2a1; Chr.7 | rps9, adm, cpt1 | |
| Xtsult2b1 | LOC493303 | XP_012821916 | 288 | sult2b1X1; Chr.7 | ca11, ntn1 | |
| XtLOC100498051 | LOC100498051 | XP_002935042 | 287 | sult2b1; Chr. 7 | pitpnc1, axin2, fam83e | |
| XtXB5850668 | LOC100144919 | NP_001119968 | 286 | uncharacterized XB5850668; Chr. 7 | ||
| XtXB986081 | LOC100497264 | XP_031761967 | 238 | provisional ortholog of sult2b1; Chr. 7 | 短い | |
| Anolis carolinensis (Green anole) | ||||||
| Acsult2a1 | LOC100563874 | XP_008114710.1 | 299 | sult2a1X2; NW_003338800.1 | lig1, atf5, arhgef40, znf219 | |
| AcLOC100553860 | LOC100553860 | XP_008115266.1 | 281 | bile salt sulfotransferase; NW_003338823.1 | hpn, prg4, fxyd3, lgi4 | |
| Acsult2b1 | LOC100562572 | XP_008115406.1 | 287 | sult2b1X2; Chr. NW_003338826.1 | sphk2, rpl18, ca11, | |
| AcLOC100553861 | LOC100553861 | XP_008115411.1 | 307 | sult2b1; Chr. NW_003338826.1 LOW QUALITY | ntn1, mamstr, rasip1 | |
| AcLOC100566507 | LOC100566507 | XP_003225132.1 | 287 | sult2b1; NW_003338828.1 | lim2, galectin 7, nkg7, etfb | |
| AcLOC100552487 | LOC100552487 | XP_016852911.1 | 287 | sult2b1; NW_003339034.1 | cpne7, mtnr1b, dpep1, chmp1a | |
| Gallus gallus (chicken) | ||||||
| GgLOC415852 | LOC415852 | XP_414212 | 288 | sult2b1-likelX2; Chr.11 | rpl13, cpne7, dpep1,chmp1a | |
| Ggsult2b1 | LOC426825 | XP_424434.3 | 295 | sult2b1X1; Chr.31 | dmbt1l, pit54, cd163l | |
| GgLOC769841 | LOC769841 | XP_015156084.2 | 295 | sult2b1-likeX2; Chr.31 | dmbt1, znf2, | |
| Mus musculus (mouse) | ||||||
| Mmsult2a1 | LOC20859 | NP_001104766 | 285 | dehydroepiandrosterone (DHEA)-preferring; Chr. 7 | Cabp5, Bsph2, Vmn1r90, Obox7 | |
| Mmsult2b1 | LOC54200 | NP_059493 | 338 | ST2B1; SULT2B, Chr. 7 | Fam83e, Ca11, Ntn5, SEC1, Fut2 | |
| Homo sapiens (human) | ||||||
| HsSULT2A1 | LOC6822 | NP_003158.2 | 285 | SULT2A1, bile salt SULT, STD; Chr.19 | CRX, TPRX2, BSPH1, ELSPBP1 | |
| HsSULT2B1 | LOC6820 | NP_814444.1 | 365 | SULT2B1, Chr.19 | FAM83E, RPL18, CA11, NTN5 |
Loss of endogenous XB5850668.L expands neural plate and reduces neural crest, PPE and epidermal gene expression domains
To determine whether XB5850668.L is involved in separating the embryonic ectoderm into its four primary domains – neural plate, neural crest, cranial placodes and epidermis – we injected an equimolar mixture of two translation-blocking MOs (Fig. 3A) into blastomeres that are the major contributors to the cranial neural crest and PPE.21 Western blot analysis showed that an equimolar combination of MO1 and MO2 effectively blocked XB5850668.L translation (Fig. 3B) and when the mixture was injected into embryos there was a higher frequency of gene expression phenotypes than obtained by each MO alone (Fig. 3C). Expression of an MO-insensitive version of XB5850668.L mRNA allowed translation in the presence of both MOs (Fig. 3B) and rescued MO phenotypes in embryos (Fig. 3D, E). Therefore, for all embryo experiments described below an equimolar mixture of MO1 and MO2 was used to effectively reduce endogenous XB5850668.L.
Figure 3:
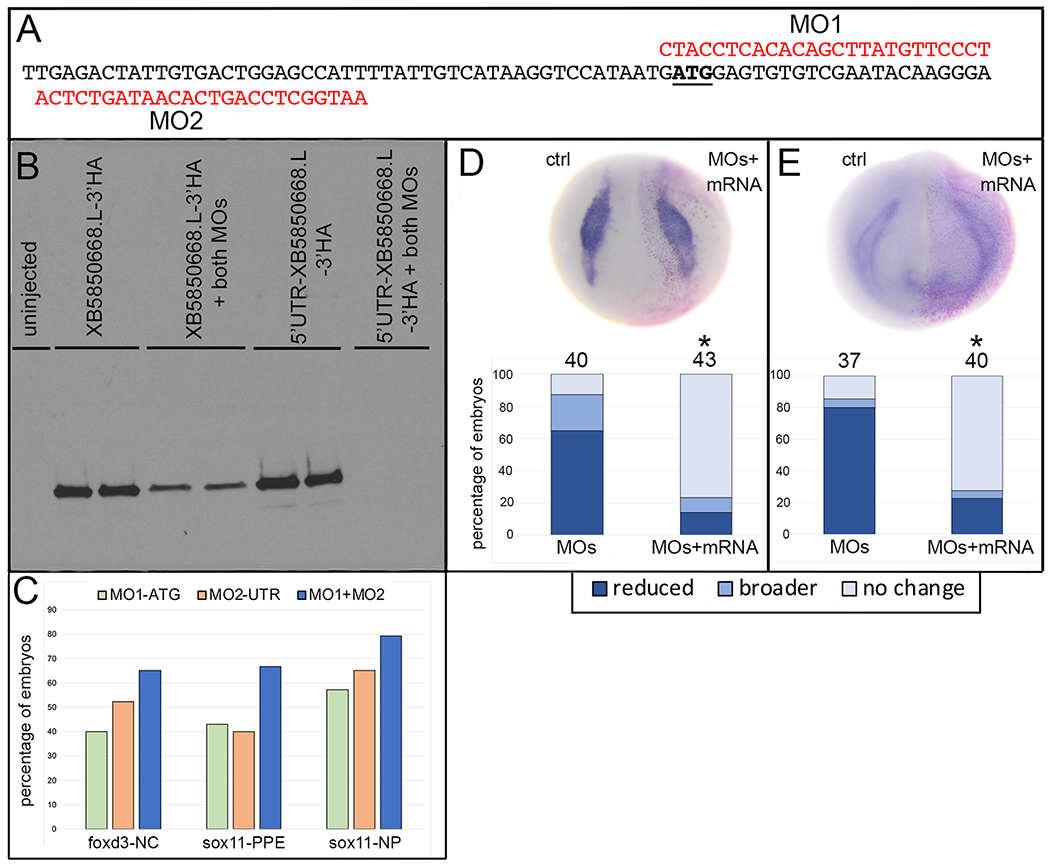
Validation of efficacy and specificity of the XB5850668.L antisense morpholino oligonucleotides.
A: Sequence of a portion of the 5’UTR (to the left of the translational start ATG [underlined]) and the coding region (to the right of the ATG) of XB5850668.L mRNA. In red are the two antisense morpholino oligonucleotides (MO1, MO2) that were used to block translation of endogenous XB5850668.L mRNA. These MOs bind with no mismatches to LOC100037047 and BG88539 mRNAs, but not to other closely related variants (e.g., XM_041568315, XM_018224208).
B: Western blot of 1-egg equivalent lysates from oocytes injected with HA-tagged mRNAs containing (5’UTR-XB5850889.L-3’HA) or not containing (XB5850889.L-3’HA) the 5’UTR to which MO2 binds. Both mRNAs are efficiently translated in the absence of an equimolar mixture of MO1+MO2. In the presence of both MOs, 5’UTR-XB5850889.L-3’HA translation is blocked, whereas XB5850889.L-3’HA translation is detected due to lack of MO2 binding but reduced due to binding of MO1. Two lanes per condition were run on each blot and assays were run in duplicate two independent times. Uninjected = lysate from oocytes that were not injected with either mRNA.
C: The percentage of embryos that showed reduced foxd3 expression in the neural crest (foxd3-NC), reduced sox11 expression in the preplacodal ectoderm (sox11-PPE) or broader expression of sox11 in the neural plate (sox11-NP). While MO1 (green bars) and MO2 (orange bars) both caused these phenotypes, the combination of an equimolar mixture of both MOs (blue bars) resulted in the greatest effect. Therefore, an equimolar combination of MO1+MO2 was used in all knock-down experiments (see Figs. 4 and 5).
D: Neural crest foxd3 expression domain at st 16 is unchanged on the side injected with a combination of both MOs and MO-insensitive mRNA (MOs+mRNA); pink dots indicate lineage-labeled nuclei on the injected side. Bar graph indicates that the foxd3 expression domain is reduced in the majority of MO-injected embryos (see also Figure 4F, G), whereas the majority of embryos have a normal foxd3 expression domain with co-injection of MO-insensitive mRNA. The percentage of embryos showing reduced (dark blue), broader (medium blue) or no change (light blue) in foxd3 expression domains are significantly different between conditions (*, p<0.05, Chi-square test). Numbers above bars indicate number of embryos analyzed.
E: PPE six1 expression domain at st 17 is unchanged on the side injected with a combination of both MOs and MO-insensitive mRNA (MOs+mRNA); pink dots indicate lineage-labeled nuclei on the injected side. Bar graph indicates that the six1 expression domain is reduced in the majority of MO-injected embryos (see also Figure 4H, I), whereas the majority of embryos have a normal six1 expression domain with co-injection of MO-insensitive mRNA. The percentage of embryos showing reduced (dark blue), broader (medium blue) or no change (light blue) in six1 expression domains are significantly different between conditions (*, p<0.05, Chi-square test). Numbers above bars indicate number of embryos analyzed.
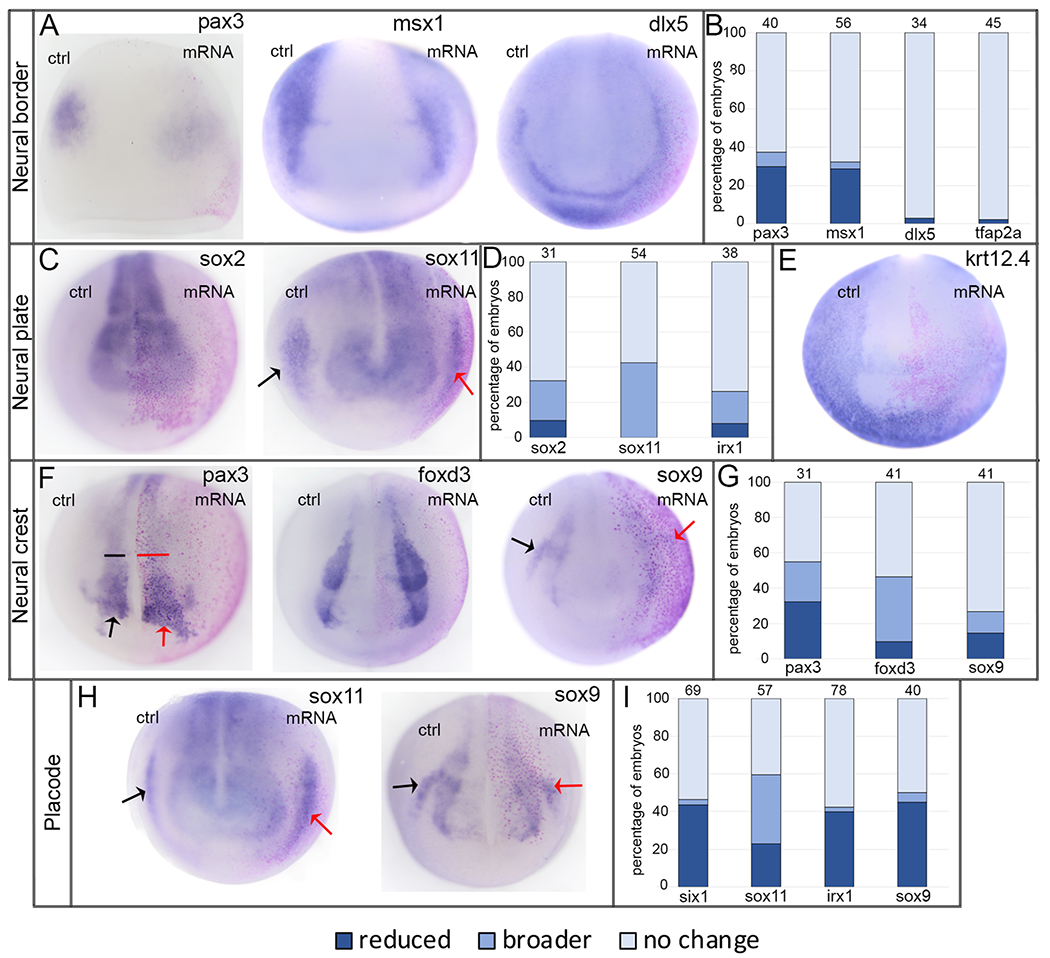
At the end of gastrulation (stage 13), a neural border zone arises between the neural and non-neural ectoderm.22 After XB5850668.L knockdown, the neural border domains of pax3 and msx1, which are mostly associated later in development with neural crest fate,23, 24 were broader in the majority of embryos (60% and 49%, respectively) but smaller in about one-fourth (22% and 25%, respectively) (Fig. 4A, B). In contrast, the neural border domains of dlx5 and tfap2α, which are mostly associated later in development with non-neural ectodermal fate,25 were predominantly reduced (59% and 64%, respectively), although a small percentage of embryos (9% and 8%, respectively) had broader domains (Fig. 4A, B).
Figure 4:
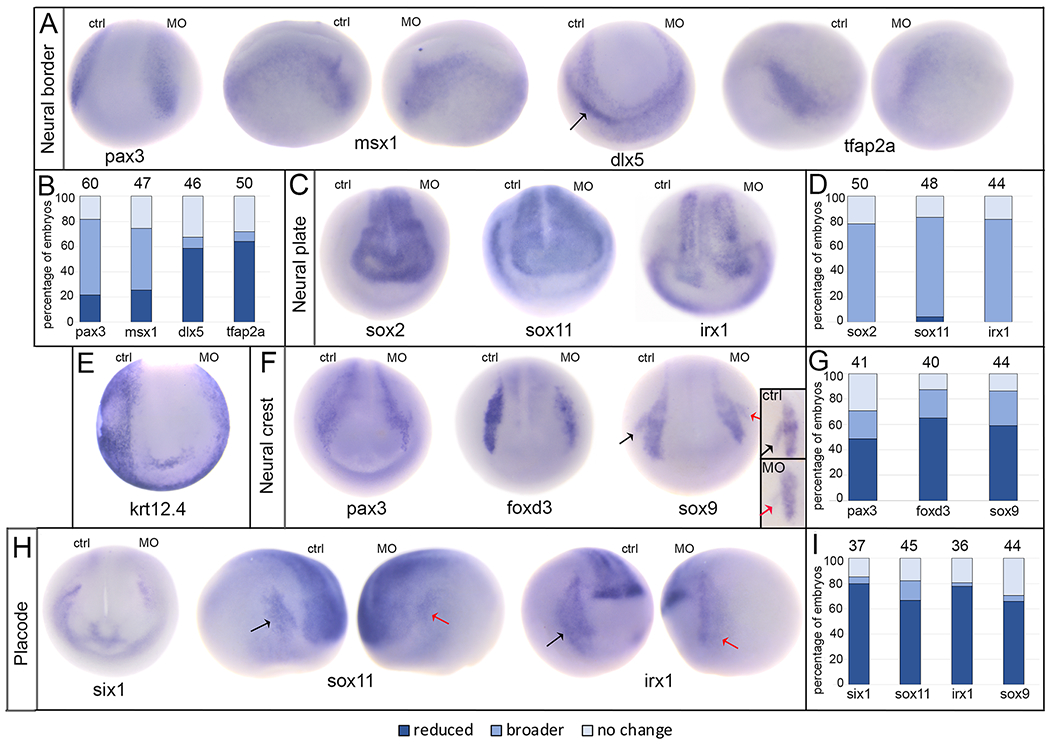
Loss of endogenous XB5850668.L alters gene expression in each ectodermal domain.
A: The expression domains of neural border genes were differentially affected by loss of XB5850668.L. Those associated later in development with the lateral neural plate and neural crest (pax3, anterior view; msx1, side views) were predominantly broader on the MO side of the embryo compared to the control side (ctrl) of the same embryo. Those associated later in development with epidermis and PPE (dlx5, anterior view; tfap2a, side views) were predominantly reduced in size and intensity on the MO side of the embryo compared to the control side (ctrl) of the same embryo. Arrow in dlx5 embryo points out the major domain (on the control side) that was analyzed. Dorsal is to the top of each image.
B: Percentage of embryos in which neural border gene expression domains were reduced in size (dark blue), broader (medium blue), or did not change (light blue) when analyzed at the end of gastrulation (st 13). The number of embryos analyzed for each gene is at the top of each bar.
C: The expression domains of three neural plate genes (sox2, sox11, irx1) were broader upon loss of XB5850668.L (MO) compared to the control side (ctrl) of the same embryo. Anterior views with dorsal to the top.
D: Percentage of embryos in which neural plate expression domains were reduced in size (dark blue), broader (medium blue) or did not change (light blue) when analyzed at neural plate stages (st 16-18). The number of embryos analyzed for each gene is at the top of each bar.
E: The expression domain of epidermis-specific keratin (krt12.4) was reduced, i.e., further from the midline, on the MO side of the embryo (95.45%, n=22). Anterior view with dorsal to the top.
F: The expression domains of three genes associated with neural crest (pax3, foxd3, sox9) were predominantly reduced in size on the XB5850668.L MO side of the embryo compared to the control side (ctrl) of the same embryo. For pax3, the most noticeable staining at this stage was in the hatching gland precursors. The black arrow on the sox9 embryo denotes the control otic placode domain and the red arrow denotes the otic placode domain on the MO side. Anterior views with dorsal to the top. Inset shows sox9 otic placode expression in a different embryo, demonstrating a reduced domain on the MO side (red arrow) compared to control side (black arrow). Lateral views with dorsal to the top.
G: Percentage of embryos in which neural crest expression domains were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at neural plate stages (st 16-18). The number of embryos analyzed for each gene is at the top of each bar.
H: The expression domains of three PPE genes (six1, anterior view; sox11, side views; irx1, side views) were reduced on the XB5850668.L MO side of the embryo compared to the control side (ctrl) of the same embryo. For sox11 and irx1, the black arrows denote the control PPE domain and the red arrows denote the PPE domain on the MO side. Dorsal is to the top of each image.
I: Percentage of embryos in which PPE expression domains were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at neural plate stages (st 16-18). The number of embryos analyzed for each gene is at the top of each bar.
We next examined gene expression at neural plate stages (stages 16-18) when the four ectodermal domains are established. Knock-down resulted in broader neural plate domains of sox2, sox11, and irx1 in the majority (>80%) of embryos (Fig. 4C, D). Consistent with these changes, nearly all embryos (95.45%, n=22) showed a reduced epidermal domain, as indicated by krt12.4 expression (Fig. 4E). At neural plate stages, pax3 expression is detected in the neural crest and hatching gland.25, 26 Both of these pax3 expression domains most frequently were reduced in size (49%), as were the neural crest domains of foxd3 and sox9 (65% and 59% respectively) (Fig. 4F, G). For each of these three genes, about one fourth of embryos had a broader domain (22%, 22%, 27%, respectively) (Fig. 4G). Similar to the neural border expression of dlx5 and tfap2α, the PPE domains of six1, sox11, and irx1, which are required for cranial placode formation,23, 27 were predominantly reduced (80%, 67%, 78%, respectively), with a minority of embryos showing broader domains (5%, 16%, 3%, respectively) (Fig. 4H, I). The otic placode domain of sox9, which is required for otic placode specification,28 also was predominantly reduced (66%) with only a few embryos (5%) showing a broader domain (Fig. 4F, I). Together, these data indicate that XB5850668.L is required for the appropriate segregation of the embryonic ectoderm into its four major domains. In its absence, the expression domains of some neural border genes (pax3, msx1) and the neural plate genes most frequently were expanded, whereas those of other neural border genes (dlx5, tfap2α) as well as neural crest, placode and epidermal genes most frequently were reduced.
Loss of XB5850668.L reduces gene expression domains in the larval otic vesicle
Since the predominant effect of XB5850668.L knockdown on the PPE was a reduced expression domain, we next monitored the expression of a number of genes that are responsible for the formation of the larval otic vesicle (OV), an important derivative of the PPE that gives rise to the inner ear. Morphant embryos were grown to early larval stages (st 28-32) when the OV has formed. Similar to effects on PPE genes, six1, irx1 and sox9 expression was primarily reduced (62%, 66%, 96%, respectively) with a few embryos (1.6%) having a larger irx1 domain (Fig. 5A, B, C, G). The expression domains of eya2, a cofactor of Six1,29 pax2, which patterns the ventromedial OV,30, 31, 32 and dlx5, which is required primarily for the vestibular OV region,33, 34 also were reduced in a high percentage of morphants (75%, 63%, 63%, respectively), with a few embryos (6%) having a larger dlx5 domain (Fig. 5D-G). Thus, the early reduction in PPE gene domains caused by knockdown persisted as reduced expression of a number of genes required for OV formation and patterning. This observation further supports the conclusion that XB5850668.L is required for cranial placode formation.
Figure 5:
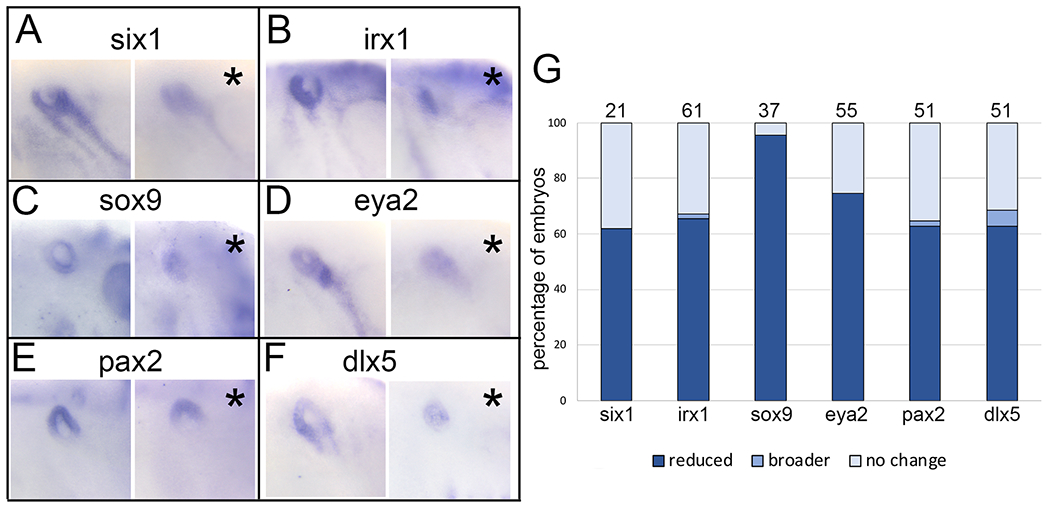
XB5850668.L is required for otic vesicle gene expression
A: Knockdown of XB5850668.L translation on one side of the larva (asterisk, right panel) results in reduced expression of six1 in the otic vesicle (OV, blue circle) compared to the control side of the same embryo (left panel).
B: Knockdown of XB5850668.L translation on one side of the larva (asterisk, right panel) results in reduced expression of irx1 in the OV (blue circle) compared to the control side of the same embryo (left panel).
C: Knockdown of XB5850668.L translation on one side of the larva (asterisk, right panel) results in reduced expression of sox9 in the OV (blue circle) compared to the control side of the same embryo (left panel).
D: Knockdown of XB5850668.L translation on one side of the larva (asterisk, right panel) results in reduced expression of eya2 in the OV (blue circle) compared to the control side of the same embryo (left panel).
E: Knockdown of XB5850668.L translation on one side of the larva (asterisk, right panel) results in reduced expression of pax2 in the OV (blue circle) compared to the control side of the same embryo (left panel).
F: Knockdown of XB5850668.L translation on one side of the larva (asterisk, right panel) results in reduced expression of dlx5 in the OV (blue circle) compared to the control side of the same embryo (left panel).
G: Percentage of embryos in which OV expression domains of genes were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at larval stages (st 28-32). The number of embryos analyzed for each gene is on top of each bar.
Increasing XB5850668.L has minor effects on embryonic ectodermal gene expression
To determine whether increased levels of XB5850668.L alters the expression of the early ectodermal genes, we injected mRNA (100 pg) encoding the wild type (wt) protein. At the completion of gastrulation, the majority of embryos showed no change in neural border gene expression. However, the intensity of staining in the pax3 and msx1 domains was reduced in about a third of embryos (30% and 29%, respectively), although a small number (7.5% and 3.6%, respectively) had darker staining (Fig. 6A, B). There were virtually no effects on the neural border expression of dlx5 or tfap2a (Fig. 6A, B). Thus, increased levels of XB5850668.L protein had an effect on the neural border domains of only pax3 and msx1.
Figure 6:
Increasing XB5850668.L has minimal effects on ectodermal gene expression domains.
A: The expression domains of neural border genes were mostly unaffected by increased XB5850668.L. However, a small percentage of those associated later in development with the lateral neural plate and neural crest (pax3, msx1, anterior views) showed reduced intensity of expression on the mRNA-injected side of the embryo (pink lineage-labeled nuclei) compared to the control side (ctrl) of the same embryo. dlx5, which later in development is associated with epidermis and PPE, was unaffected by increased XB5850668.L (anterior view); the same was found for tfap2a expression (shown in 2B). Dorsal is to the top of each image.
B: Percentage of embryos in which neural border expression domains were reduced (dark blue), broader (medium blue), or did not change (light blue) when analyzed at the end of gastrulation (st 13). The number of embryos analyzed for each gene at the top of each bar.
C: The expression domains of neural plate genes were mostly unaffected by increased XB5850668.L. However, a small percentage of embryos showed broader neural plate domains on the mRNA-injected side of the embryo (pink lineage-labeled nuclei) compared to the control side (ctrl) of the same embryo. Arrows in sox11 show its PPE expression domain on the control (black) and mRNA-injected (red) sides. In this case, increased XB5850668.L resulted in a narrower sox11 PPE domain. Anterior views with dorsal to the top.
D: Percentage of embryos in which neural plate expression domains were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at neural plate stages (st 16-18). The number of embryos analyzed for each gene is at the top of each bar.
E: The expression of epidermis-specific keratin (krt12.4) was reduced on the mRNA-injected side in only 25% of embryos; there was no change in expression in 75% of embryos (n=56). Anterior view with dorsal to the top.
F: The expression domains of neural crest genes were mostly unaffected by increased XB5850668.L (see 6H for sox9). However, small percentages of embryos showed a broader domain (pax3, foxd3) or reduced staining intensity (sox9) on the mRNA-injected side of the embryo (pink lineage-labeled nuclei) compared to the control side (ctrl) of the same embryo. For pax3, both the neural crest (bracket) and hatching gland (arrow) domains were affected. Arrows in sox9 show its otic placode domain on the control (black) and mRNA-injected (red) sides, which frequently was reduced (see also 6H, I). Anterior views with dorsal to the top.
G: Percentage of embryos in which neural crest expression domains were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at neural plate stages (st 16-18). The number of embryos analyzed for each gene is at the top of each bar.
H: The expression domains of PPE/placode genes were mostly unaffected by increased XB5850668.L. However, small percentages of embryos showed a broader domain (sox11) or a reduced domain (sox11 in 6C; sox9 and also 6F) on the mRNA-injected side of the embryo (pink lineage-labeled nuclei) compared to the control side (ctrl) of the same embryo. Arrows indicate PPE/placode domains on the control (black) and mRNA-injected (red) sides. Anterior views with dorsal to the top.
I: Percentage of embryos in which PPE expression domains were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at neural plate stages (st 16-18). The number of embryos analyzed for each gene is at the top of each bar.
At neural plate stages, most embryos showed no change in neural plate expression of sox2, sox11 or irx1 (68%, 57%, 74%, respectively). However, some embryos had a broader neural plate domain (23%, 43%, 18%, respectively), and a few had a smaller domain (9%, 0%, 8%, respectively) (Fig. 6C, D). The majority of embryos also showed no effect on the krt12.4 epidermal domain (Fig. 6E; 75%, n=56), although 25% had a smaller expression domain. Increased levels predominantly caused no effect on the pax3 neural crest and hatching gland domains or on the foxd3 and sox9 neural crest domains (45%, 54%, 73%, respectively) (Fig. 6F, G). However, some showed expanded domains (23%, 37%, 12%, respectively) and others showed reduced domains (32%, 10%, 15%, respectively) (Fig. 6F, G). Increased levels had no effect on the PPE domains of six1, irx1 and sox9 in the majority of embryos (54%, 58%, 50%, respectively), but reduced their PPE expression domains in many embryos (44%, 40%, 45%, respectively) (Fig. 6H, I). Increased levels caused pleiotropic effects on the sox11 PPE expression domain (Fig. 6I): it was not affected in 40% of embryos, broader in 37% (Fig. 6H) and reduced in 23% (Fig. 6C).
XB5850668.L gain-of-function also has minor effects on otic vesicle gene expression
Because increased levels of XB5850668.L reduced two genes expressed in the PPE that later are required for OV formation (six1, irx1) and one expressed in the otic placode (sox9), we next determined the effects of increased XB5850668.L on gene expression in the OV. For six1, irx1, sox9, pax2, eya2, dlx5, and tbx1,35, 36, 37 the predominant phenotype was either no change in gene expression (39%, 47%, 42%, 43%, 38%, 43%, 39%, respectively) or reduced gene expression (49%, 42%, 37%, 48%, 46%, 46%, 56%, respectively) (Fig. 7). Smaller numbers of embryos had enlarged gene expression domains (12%, 11%, 21%, 9%, 16%, 11%, 5%, respectively) (Fig. 7). Thus, increased levels of XB5850668.L can interfere with the expression of several of the genes required for OV formation and patterning.
Figure 7:
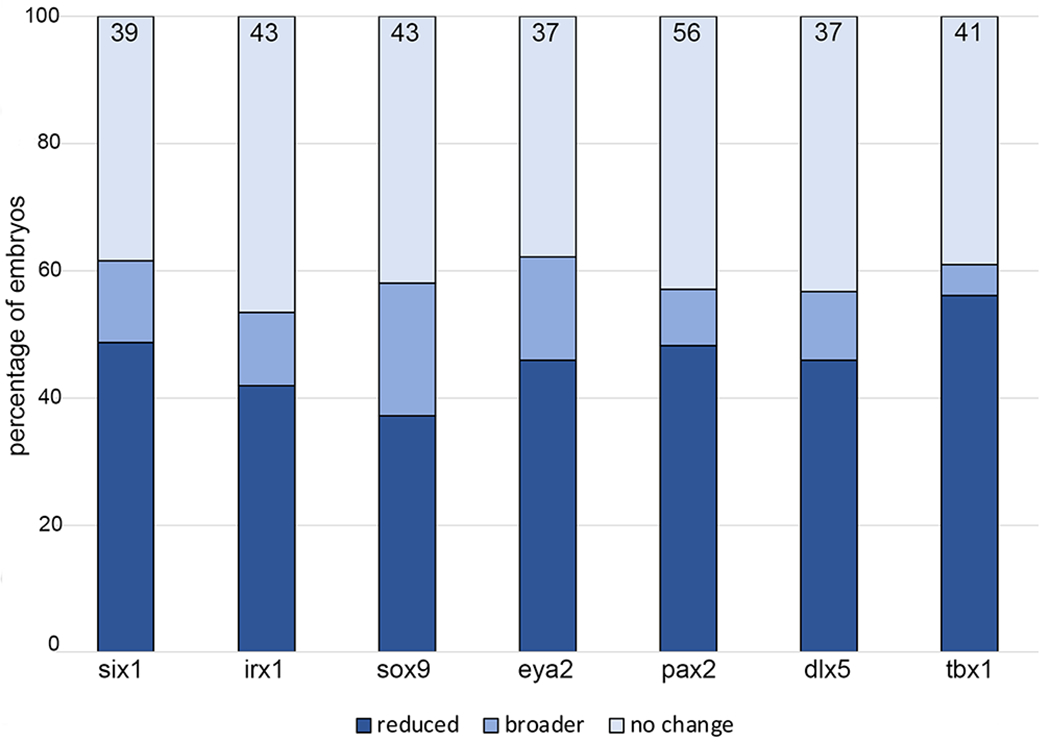
XB5850668.L gain-of-function has minimal effects on otic vesicle gene expression.
Percentage of embryos in which OV expression domains were reduced (dark blue), broader (medium blue) or did not change (light blue) when analyzed at larval stages (st 28-32). The number of embryos analyzed for each gene is inside the bars.
Two amino acids key for sulfotransferase activity may be required
Members of the SULT superfamily share considerable amino acid sequence and structural similarities.1, 4, 5 Each member contains structural features that are necessary for binding the donor, PAPS, including two key amino acids that are required for enzymatic activity: residue #40 (Lysine, K) and residue #96 (Histidine, H), which are conserved across Sult1 and Sult2 proteins, including XB5850668.L (Fig. 1). K40 lies within the highly conserved 5’ PSB loop which is required for binding the 5’phosphosulfate of PAPS; H96 is a key proton acceptor that lies within the catalytic domain and is required for enzymatic activity.5, 7, 18, 19 To determine whether these sites are necessary for the changes in gene expression in the neural plate stage and larval OV domains that occurred when wild type (wt) protein levels were increased (Figs. 6, 7), we made single (K40I; H96F) or double (K40I+H96F) amino acid substitutions that are predicted to perturb PAPS binding and/or enzymatic activity. Mutant mRNAs were injected at 100pg and compared to the results from injection of 100pg of wt XB5850668.L. For the neural plate expression of sox2 and sox11 (Fig. 8A, B) and the PPE expression of six1 and sox11 (Fig. 8F, G), the K40I mutant, H96F mutant or double mutant caused fewer effects compared to wt, suggesting the mutation caused a loss of catalytic function, either via deficient PAPS binding (K40I), enzymatic activity (H96F) or both. Consistent with these results, there were downward trends in the effects of the mutants on irx1 neural plate or sox9 otic placode expression domains (Fig. 8C, I), but these did not reach statistical significance. In contrast, the K40I mutant expanded foxd3 and sox9 neural crest domains (Fig. 8D, E), and the double mutant caused a higher frequency of reduced irx1 PPE domain (Fig. 8H) compared to wt. These results suggest that for these genes the K40I and double mutants may have caused substrates to bind in the absence of PAPS, changing the enzyme to an inactive or dominant-negative conformation, as reported for other SULTs.5
Figure 8:
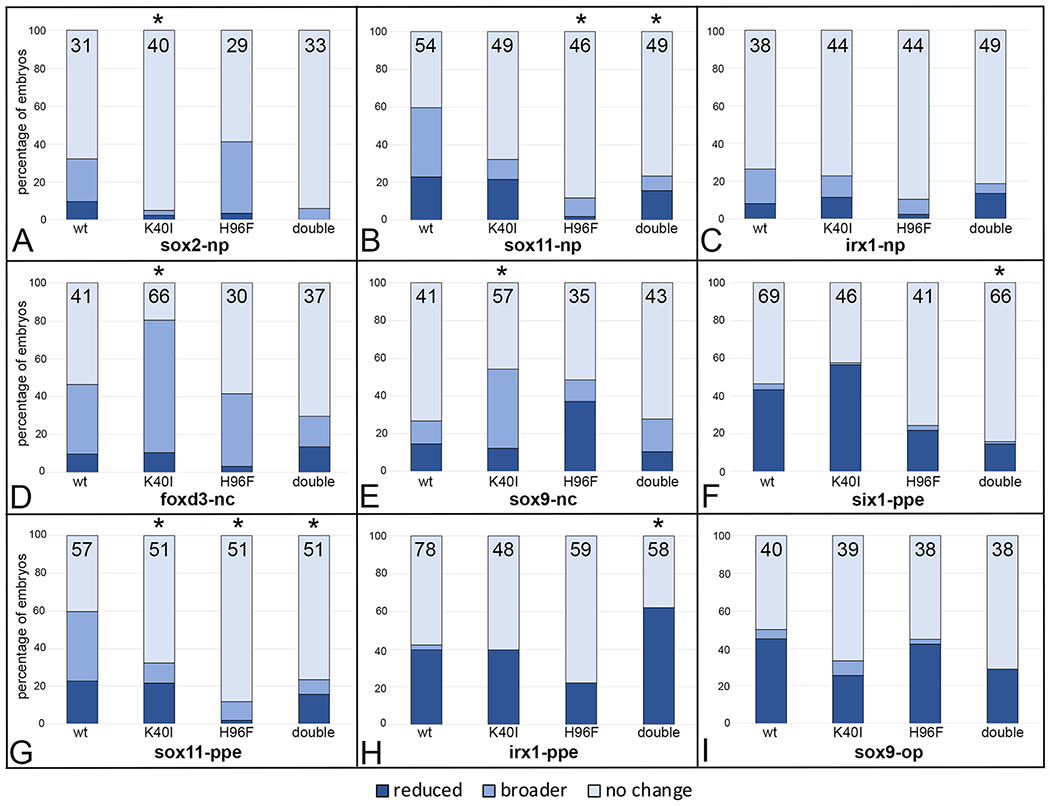
Mutation of amino acids in the PAPS binding (H40I) or catalytic (H96F) domains tend to attenuate effects of wild type XB5850668.L gain-of-function on ectodermal gene expression domains. The percentage of embryos in which the expression domain was reduced (dark blue), broader (medium blue) or did not change (light blue) after injections of mRNAs (100pg) encoding wild type (wt) or mutated versions (K40I, H96F, double = K40I+H96F) of XB5850668.L. The number of embryos analyzed is inside each bar. * indicates mutant mRNA phenotype frequencies that were significantly different from wt at the p<0.05 level (Chi square test).
A: sox2 neural plate (np) expression domain changes.
B: sox11 neural plate (np) expression domain changes.
C: irx1 neural plate (np) expression domain changes.
D: foxd3 neural crest (nc) expression domain changes.
E: sox9 neural crest (nc) expression domain changes.
F: six1 preplacodal (ppe) expression domain changes.
G: sox11 preplacodal (ppe) expression domain changes.
H: irx1 preplacodal (ppe) expression domain changes.
I: sox9 otic placode (op) expression domain changes.
For most OV genes analyzed, the double mutant caused fewer effects compared to wt, but these only reached statistical significance for sox9, eya2, pax2 and tbx1 (Fig. 9C, D, E, G). The K40I mutant also showed less frequent effects for irx1, pax2 and tbx1, but only reached significance for the latter (Fig. 9G). Likewise, the H96F mutant caused less frequent effects for six1, sox9, pax2 and tbx1, but only reached significance for the latter (Fig. 9G). Together, these results indicate that for these genes the mutations caused a loss of catalytic function. However, for sox9 and eya2, the reduced OV expression phenotype caused by wt was significantly enhanced by the K40I mutant (Fig. 9C, D), suggesting an effect that perhaps results from an inactive or dominant-negative conformation.
Figure 9:
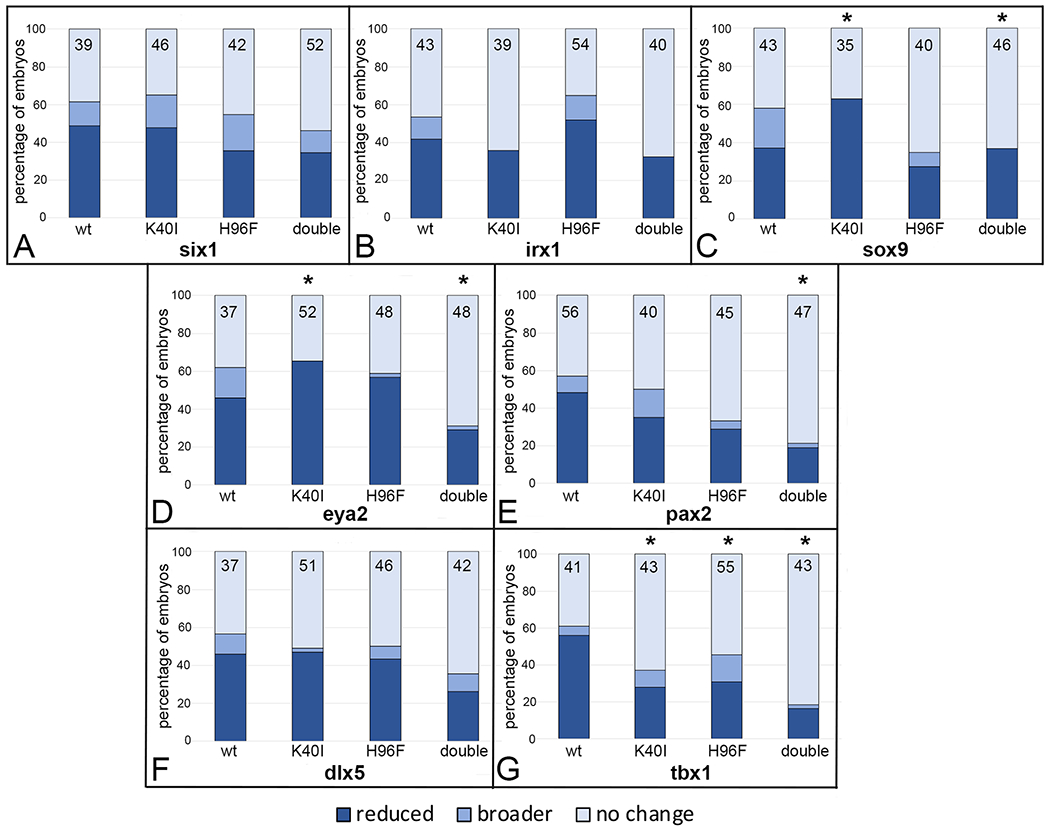
Mutation of amino acids in the PAPS binding (H40I) or catalytic (H96F) domains tend to attenuate effects of wild type XB5850668.L gain-of-function on otic vesicle gene domains.
Percentage of embryos in which an OV gene expression domain was reduced (dark blue), broader (medium blue) or did not change (light blue) after injections of mRNAs (100 pg) encoding wild type (wt) or mutated versions (K40I, H96F, double = K40I+H96F) of XB5850668.L. The number of embryos analyzed for each gene is inside the bars. * indicates mutant mRNA phenotype frequencies that were significantly different from wt at the p<0.05 level (Chi square test).
A: six1 OV expression domain changes.
B: irx1 OV expression domain changes.
C: sox9 OV expression domain changes.
D: eya2 OV expression domain changes.
E: pax2 OV expression domain changes.
F: dlx5 OV expression domain changes.
G: tbx1 OV expression domain changes.
Overall, these data indicate that the two key amino acids that are required for sulfotransferase catalytic activity are likely necessary for the effects of increased XB5850668.L on the relative sizes of many of the ectodermal domain and OV genes, but the K40I mutation in particular may affect the expression of some of these genes independent of its ability to bind PAPS or enzymatic activity.
DISCUSSION
The SULT superfamily is comprised of a large number of proteins, classified by sequence into several families, that sulfonate a wide range of substrates with overlapping specificities3, 7, 38 In humans, there are five families: twelve genes belong to the SULT1 family, three to the SULT2 family, and single genes each to the SULT3, SULT4 and SULT6 families.6, 39, 40 In mouse, there are seven families (Sult1-Sult7), each of which contains several subfamilies.4
XB5850668.L is an uncharacterized sulfotransferase that to date had not been assigned to a specific SULT family. Three protein structure databases suggest XB5850668.L is a member of the SULT1 family ([ISP-LINK MISSING]uniprot.org/uniprotkb/A1L3M6/entry; alphafold.ebi.ac.uk/entry/A1L3M6; modbase.compbio.ucsf.edu), however it was not identified in a phylogenetic analysis of the Xenopus Sult1 family.41 Instead, our phylogenetic analysis firmly places it in the SULT2 family, and in a subfamily distinct from either Sult2a or Sult2b. The SULT2 phylogenetic tree and the Xenopus Sult2 genomic arrangement on chromosome 7 suggest there have been multiple events of gene duplication and subsequent deletion in SULT2 loci during vertebrate evolution. In humans, three genes comprise the SULT2 family.6 SULT2A1 protein plays an important role in androgen and bile salt metabolism, SULT2A2P is a pseudogene, and SULT2B is expressed as two splice variants that sulfonate either a steroid hormone precursor or cholesterol. Numerous SNPs have been reported in human SULT2A1 and SULT2B that affect the efficacy of enzymatic activity and proclivity to disease.6 In zebrafish, there also are three members of the SULT2 family; these also play roles in steroid hormone and cholesterol metabolism.4 Our phylogenetic analysis indicates that the SULT2 family of the diploid Xenopus tropicalis and the allotetraploid Xenopus laevis also are comprised of three subfamilies: 1) SULT2A1 and its related genes (XB986081 in X. tropicalis and LOC108695915 and XB986081 in X. laevis); 2) SULT2B1; and 3) an intermediate class including LOC100498051 and XB5850668 in both X. tropicalis and X. laevis. While to our knowledge none of these have been functionally tested for sulfotransferase activity or had their sulfate donor or substrates identified, conservation of function with human and fish homologs would suggest potential roles in steroid hormone and cholesterol metabolism.
The roles of sulfonation in developmental processes
The addition and removal of sulfate-containing moieties on bioactive compounds play important roles in modulating their solubility, activity and efficacy.39 The regulated removal of sulfate-containing moieties by sulfatases (SULFs) are known to have many important functions during development. For example, Sulf2a, which selectively removes 6-O groups from heparin sulfate, modulates the activity of heparan sulfate proteoglycans by altering the binding sites for several signaling molecules, including Fgf, Hh, Gdnf and Wnt ligands. 42, 43, 44 In Xenopus, Sulf1 and Sulf2 enhance the axis-inducing activity of Wnt11, restrict Bmp and Fgf signaling during cell movement and differentiation, and are required for appropriate cranial neural crest migration. 45, 46, 47, 48
In contrast, although the addition of sulfuryl groups to bioactive compounds by SULTs is well recognized to modulate the activity of hormones, neurotransmitters, metabolic and xenobiotic compounds in adult tissues, there has been little insight into the roles they might play in developmental processes. It has been proposed that in humans SULT proteins protect the fetus from excessive hormonal activity as well as blood-borne maternal environmental toxins,8 but this is yet to be experimentally tested. Previously, we suggested that XB5850668.L has a role in developmental processes because XB5850668.L mRNAs were detected in the neural border zone, branchial arch-derived neural crest, cranial placodes, neural tube, somites and nephric mesoderm.16 Additionally, whole embryo RNAseq detected abundant transcripts through gastrula stages (xenbase.org based on 12, 49), and semi-quantitative RT-PCR detected the highest levels of transcripts between gastrula and neural plate stages.16 Proteomic data also indicate that it is available to participate in developmental processes such as those investigated herein (xenbase.org based on 49, 50).
Our previous work showed that XB5850668.L is upregulated by Six1 in ectodermal explants.16 Likewise, Riddiford and Schlosser reported several sulfotransferases as potential Six1/Eya1 targets in the PPE using RNAseq assays.51 These data indicate that sulfonation may play an important role in developmental processes regulated by Six1. Therefore, we performed gain- and loss-of-function studies to determine whether XB5850668.L functions during the period when Six1 has a key role in establishing and patterning the embryonic ectodermal domains - neural plate, neural crest, PPE and epidermis.27 The experiments described herein demonstrate that it is required for the appropriate segregation of the embryonic ectoderm into its four major domains. In its absence, most genes expressed in the neural border and neural plate had expanded domains, whereas in the majority of embryos the genes expressed in the neural crest, PPE and epidermis domains were reduced. These results suggest that the processes that apportion cells to these different ectodermal domains depend upon factors that require sulfonation.
In contrast, increasing the levels of XB5850668.L protein by targeted mRNA injections had minimal effects on gene expression patterns in most embryos. This result is not because the mRNA dose was too small because there was no statistical difference in phenotype frequencies when the mRNA dose was increased to 200pg (data not shown). Those embryos injected with either dose that did show a phenotype had either a moderately expanded neural plate or neural crest gene domain, or reduced epidermis or PPE domains. This is consistent with reports that when neural crest domains are expanded, PPE domains are reduced, suggesting that there is a competition between the two fates that are derived in common from the neural border zone.15, 52, 53 Both the loss and gain of XB5850668.L had long lasting effects in the PPE, as gene expression domains in the larval otic vesicle were similarly affected. It is interesting that the effects of XB5850668.L knockdown were more pronounced than the effects of increased expression. We speculate that if XB5850668.L is proved to function as a sulfotransferase, this difference might be explained by the need for a basal level of sulfonation that would be reduced by its knockdown, whereas excess enzyme might not increase sulfonation above the level produced by endogenous enzymes on a stable reservoir of available substrates.
Is sulfotranferase activity required for these effects?
Based on sequence alignment with the human SULT proteins, amino acids #40 (Lysine, K) and #96 (Histidine, H) are within the sulfotransferase catalytic domain and their corresponding amino acids in mammalian SULTs are required for enzymatic activity.5, 7, 18, 19 To determine whether these residues contributed to the phenotypes caused by increased levels of XB5850668.L reported above, we made amino acid substitutions at both of these sites. For most genes assessed, we found that the effects of mutant mRNA on gene expression domains were reduced compared to wild type, indicating that they lack wild-type function. Although not all changes reached statistical significance (Figs. 8, 9), the downward trend for most genes suggests that changing these amino acids diminishes protein function. However, it remains to be biochemically confirmed whether these effects can be attributed to loss of sulfonating activity. It also will be important to establish that the level of expression of the mutant mRNAs was comparable to the wt mRNA once an antibody to specific XB5850668.L becomes available.
The K40I substitution is predicted to prevent PAPS binding, which is required not only for enzymatic activity, but also for the formation of loops in the protein that facilitate substrate binding.1, 5, 6 In addition, aspects of protein function other than catalytic activity may be affected by lack of PAPS binding. For example, there are endogenous compounds that bind to SULTS in the absence of PAPS to prevent substrate binding, and amino acid substitutions can cause conformational changes that allow substrates with inhibitory functions to bind.1, 5, 6 Thus, the K40I mutant may affect gene expression through either reduction or loss of sulfonating activity or as a dominant-negative protein that prevents appropriate substrate binding. In addition, some SULTs may have non-sulfonating functions. For example, SULT4a1 contains all the conserved signatures for PAPS binding but does not bind PAPS and shows no sulfonating activity.5 Therefore, an important next step for interpreting the changes in gene expression reported herein is to biochemically define the sulfonation activity of XB5850668.L and identify its specific sulfate donor and substrates.
Interestingly, XB5850668.L expression patterns are similar to those of a few sulfatases, suggesting that both adding and removing sulfate moieties within the same tissue is important. For example, in mouse and Xenopus embryos, the cranial expression of Sulf2a includes the neural tube, neural crest-derived branchial arch mesenchyme and cranial sensory placodes.47, 54, 55, 56, 57 Since there is evidence from adult tissues that there are substrates that inhibit PAPS binding to Sults and thus catalytic activity,5 it will be interesting to determine putative substrates of XB5850668.L and compare them to those of SULFs to determine if they have coordinated roles during the developmental processes. For example, there is a rich literature establishing that signaling molecules involved in establishing the neural and non-neural ectodermal domains, including Bmps, Noggin, Wnts and Fgfs, require modulation of the sulfonated state of heparin sulfate proteoglycans for efficient binding and activity (e.g.,58, 59). Since these signaling molecules are important regulators of the relative sizes of the embryonic ectodermal domains,27, 60, 61, 62, 63, 64, 65 it will be important to determine whether cytoplasmic and/or membrane-bound SULTs, and specifically XB5850668.L, are involved in adding sulfate-containing moieties to biomolecules that regulate these same processes.
EXPERIMENTAL PROCEDURES
Phylogenetic analysis
Based on the amino acid sequence of X. laevis XB5850668.L, the cDNA and genomic databases in NCBI (http://www.ncbi.nlm.nih.gov) and Xenbase (http://www.xenbase.org) were searched using BLASTP. SULT2 amino acid sequences were collected from the following animals in addition to X. laevis (Xl): teleost (zebrafish Danio rerio, Dr), amphibian (X. tropicalis,, Xt), reptilian (green anole Anolis carolinensis, Ac), avian (chicken Gallus gallus, Gg) and mammalian (house mouse Mus musculusm Mm, and human Homo sapien, Hs) species (Table 1). We also collected D. rerio SULT1 gene family sequences as a SULT family out-group. Alignment and phylogenetic reconstructions were performed using the function “build” of ETE3 v3.1.1,66 (as implemented on GenomeNet (https://www.genome.jp/tools/ete/). A maximum likelihood (ML) phylogenetic tree was inferred using PhyML v20160115 run with model and parameters: --pinv e --alpha e --nclasses 4 -o tlr -f m --bootstrap −2.67 Branch supports are the Chi square-based parametric values return by the approximate likelihood ratio test.
Plasmid constructs:
A full-length Xenopus laevis LOC100037047 plasmid was purchased from Open Biosystems (BG885936). Sequencing identified it as a minor variant of LOC100037047 that had been deposited in GenBank (BC130199.1) as part of the NICHD Xenopus Gene Collection initiative.20 According to the Xenopus laevis gene assembly (v10.1; xenbase.org) the provisional gene symbol for LOC100037047 is XB5850668.L. The BG885936 ORF was subcloned into Stu1 and Xho1 sites of pCS2+ (pCS2+-XB5850668.L) using the Clone EZ PCR cloning kit (GenScript). Two HA-tagged versions of this plasmid were generated using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent): pCS2+-5’UTR-XB5850668.L-3’HA, which contains a portion of the 5’UTR rendering it sensitive to antisense morpholino oligonucleotides (MO1 and MO2; (Fig. 3A) and pCS2+-XB5850668.L −3’HA, which does not contain the 5’UTR rendering it insensitive to MO#1 (Fig. 3A). The same kit was used to introduce single nucleotide substitutions into pCS2+-XB5850668.L at sites predicted to be required for sulfotransferase activity: K40 and H96. For single amino acid changes, aa 40 was changed from lysine to isoleucine (pCS2+-XB5850668.L-K40I [AAG to AAT] and aa 96 was changed from histidine to phenylalanine (pCS2+-XB5850668.L-H96F [CAT to TTT]. To change both K40 and H96 (pCS2+-XB5850668.L-K40I+H96F, AAG was changed to ATC and CAT was changed to TTT) (pCS2+-XB5850668.L-K40I+H96F). All plasmids were confirmed by full-length sequencing in both directions.
In vitro synthesis of mRNAs and antisense RNA probes
mRNAs encoding wild type (wt) Xenopus laevis XB5850668.L, the mutated versions of XB5850668.L (K40I; H96F; K40I+H96F) and a nuclear-localized β-galactosidase (nβgal) were synthesized in vitro (mMessage mMachine kit, Thermo Fisher). Dig-labeled, antisense RNA probes for in situ hybridization (ISH) were synthesized in vitro (MEGAscript kit; Thermo Fisher) as previously described.68
Antisense morpholino oligonucleotides
To knock-down the endogenous level of XB5850668.L protein, two lissamine-labeled translation-blocking antisense morpholino oligonucleotides that bind to Xenopus laevis XB5850668.L mRNA were purchased (Gene Tools, LLC). MO2 binds in the 5’UTR and MO1 binds in the coding region at the translation start site of both BG885936 and XB5850668.L (Fig. 3A). These MOs will not bind to two other variants of XB5850668.L (variant X1, XM_018224208.2; variant X2, XM_041568315.1). To verify the ability of the MOs to block XB5850668.L translation, Xenopus stage VI oocytes were injected with 9ng of an equimolar mixture of MO1+MO2, and subsequently injected with 2ng of either 5’UTR-XB5850668.L-3’HA (MO-sensitive), or XB5850668.L-3’HA (MO2-insensitive) mRNA. Oocytes were cultured overnight at 18°C. Lysates were prepared and Western blotting performed with rabbit anti-HA antibody (C29F4, Cell Signaling) as previously described.69 Western blots of 1-egg equivalents per lane demonstrate that in the presence of both MOs, XB5850668.L-3’HA mRNA was translated (Fig. 3B), albeit at a lower level than in the absence of the MOs likely due to MO1 binding (Fig. 3A). In contrast, although 5’UTR-XB5850668.L-3’HA mRNA was translated in the absence of the MOs, in their presence its translation was blocked (Fig. 3B). To determine whether the MOs were specific to XB5850668.L, we tested whether MO2-insensitive mRNA could recue the gene expression effects of the MOs. Embryos were microinjected on one side at the 4-cell stage with 9ng of the MO mixture followed by microinjection of 100 pg of MO2-insensitive mRNA into the daughter 8-cell blastomeres. At stage 16-18, embryos were processed for foxd3 or six1 expression by ISH (Fig. 3D, E), as described below. Co-injection of MO-insensitive mRNA prevented the reduction in gene expression in a significant percent of embryos (p<0.05, Chi-square test), demonstrating specificity of the MOs.
Obtaining embryos and microinjections
Fertilized Xenopus laevis eggs were obtained by gonadotropin-induced natural mating of wild type, outbred adults as previously described.70 Embryos were selected at the 2-cell stage if the first cleavage furrow bisected the lightly pigmented region of the animal hemisphere to accurately identify the dorsal-ventral axis.71 When these selected embryos reached the 8-cell stage, the dorsal animal and ventral animal blastomeres that are the major contributors to the cranial neural crest and pre-placodal ectoderm,21 were each microinjected on one side of the embryo with 1 nl of mRNA (100 pg) or an equimolar mixture of MO1 and MO2 (9 ng), according to standard methods.72 Injections were done on only one side of the embryo and the uninjected side used as an internal control of gene expression. Embryos were cultured in a diluted series of Steinberg’s solution until fixation.
Fixation, histochemistry and in situ hybridization (ISH)
Embryos were cultured to neural border (st 13), neural plate (st 16-18) or otic vesicle (st 28-32) stages,73 fixed in 4% paraformaldehyde (in 0.1M MOPS, 2mM EGTA Magnesium, 1mM MgSO4, pH 7.4), stained for β-Gal histochemistry if injected with mRNAs, and processed for in situ hybridization (ISH) as previously described.68 Embryos in which the nβGal (mRNA injections) or lissamine (MO injections) lineage tracer was located in the appropriate tissue domain were processed by ISH.
Gene expression analysis
To assess whether altered levels of XB5850668.L affect the size of the expression domains of genes of interest, we compared the expression intensity and domain size between the injected, lineage-labeled side and the control, uninjected side of the same embryo. This direct intra-embryo comparison minimizes inter-embryo variation that might occur due to differences in developmental stages or experimental batches. Embryos were scored independently by at least two authors, and the values reported are means of their independent scores. Embryos that showed a more intense and/or larger expression domain on the injected side compared to control side of the same embryo were scored as “broader”. Embryos that showed reduced staining intensity or smaller expression domain on the injected side compared to control side of the same embryo were scored as “reduced”. The percentages of embryos that showed “broader” expression, “reduced” expression, or “no change” between injected and control sides of the same embryo were calculated for each gene assayed. To determine if the level of exogenous protein had an effect, two different doses of wild type (wt) XB5850668.L mRNA (100 pg vs. 200 pg) were injected and the frequencies of each phenotype compared by the Chi-square test (p<0.05); there were no significance differences in frequencies between the two mRNA doses. Likewise, to determine if the amino acid changes reduced activity, the frequencies of each phenotype for samples injected with either wt or mutated XB5850668.L mRNA (100pg each) were compared by the Chi-square test (p<0.05). Embryos assessed for each gene were derived from a minimum of three independent experiments from different sets of outbred parents to attain genetic diversity.
Bullet points:
XB5850668.L is an uncharacterized sulfotransferase
It is a member of the SULT2 family
It is required for proportioning the embryonic ectoderm
Some of its effects require sulfotransferase activity
ACKOWLEDGEMENTS
The National Xenopus Resource (RRID:SCR_013731,74) and from Xenbase (RRID:SCR_003280,49) provided invaluable information for carrying out this work. This work was funded by grants from the National Institutes of Health (DE022065 to SAM; DE026434 to SAM and KMN), a GWU Wilbur V. Harlan Research Fellowship (AM) and a Luther Rice Undergraduate Research Fellowship (AM).
Grant Sponsors:
NIH R01 DE022065 to SAM; NIH R01 DE026434 to SAM and KMN; GWU Wilbur V. Harlan Research Fellowship (AM); ; Luther Rice Undergraduate Research Fellowship (AM).
REFERENCES
- 1.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: structure, mechanism, biological activity, inhibition and synthetic utility. Angew Chemie Int Ed. 2004;43:3526–3548. [DOI] [PubMed] [Google Scholar]
- 2.Nimmagadda D, Cherala G, Ghatta S. Cytosolic sulfotransferases. Indian J Exp Biol. 2006;44:171–182. [PubMed] [Google Scholar]
- 3.Coughtrie MWH. Function and organization of the human cytosolic sulfotransferase (SULT) family. Chem Biol Interact. 2016;259(Pt A):2–7. [DOI] [PubMed] [Google Scholar]
- 4.Suiko M, Kurogi K, Hashiguchi T, Sakakibara Y, Liu MC. Updated perspectives on the cytosolic sulfotransferases (SULTs) and SULT-mediated sulfation. Biosci Biotechnol Biochem. 2017;81:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thonton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith AH. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurogi K, Rasool MI, Alherz FA, El Daibani AA, Bairam AF, Abunnaja M, Yasuda S, Wilson L, Hui Y, Liu MC. SULT genetic polymorphisms: physiological, pharmacological and clinical implications. Expert Opin Drug Metab Toxicol. 2021;17:767–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinshilboum RM, Otterness DM, Aksoy I, Wood TC, Her C, Raftogianis RB. Sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11:3–14. [PubMed] [Google Scholar]
- 8.Richard K, Hume R, Kaptein E, Stanley EL, Visser TJ, Coughtrie MWH. Sulfation of thyroid hormone and dopamine during human development: ontogeny of phenol sulfotransferase and Arylsulfatase in liver, lung and brain. J Clin Endocrin Metab. 2001;86:2734–2742. [DOI] [PubMed] [Google Scholar]
- 9.Hattori K, Inoue M, Inoue T, Arai H, Tamura HO. A novel sulfotransferase abundantly expressed in the dauer larvae of Caenorhabditis elegans. J Biochem. 2006;139:355–362. [DOI] [PubMed] [Google Scholar]
- 10.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahmy K, Baumgartner S. Expression analysis of a family of developmentally regulated cytosolic sulfotransferases (SULTs) in Drosophila. Hereditas. 2013;150:44–48. [DOI] [PubMed] [Google Scholar]
- 12.Session AM, Uno Y, Kwon T, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moody SA, LaMantia AS. Transcriptional regulation of cranial sensory placode development. Curr Topics Dev Biol. 2015;111:301–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlosser G Vertebrate Cranial Placodes, Volume 1: Development of Sensory and Neurosecretory Cell Types. Boca Raton, FL: CRC Press; 2021:43–71. [Google Scholar]
- 16.Yan B, Neilson KM, Ranganathan R, Streit A, Moody SA. Microarray identification of novel genes downstream of Six1, a critical factor in cranial placode, somite and kidney development. Dev Dyn. 2015;244:181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. 2000;96:253–257. [DOI] [PubMed] [Google Scholar]
- 18.Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC. Crystal structure of estrogen sulphotransferase. Nature Struct Biol. 1997;4:904–908. [DOI] [PubMed] [Google Scholar]
- 19.Kakuta Y, Pedersen LG, Pedersen LC, Negishi M. Conserved structural motifs in the sulfotransferase family. Trends Biochem Sci. 1998;23:129–130. [DOI] [PubMed] [Google Scholar]
- 20.Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. [DOI] [PubMed] [Google Scholar]
- 21.Moody SA, Kline MJ. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat Embryol. 1990;182:347–362. [DOI] [PubMed] [Google Scholar]
- 22.Groves AK, LaBonne C. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev Biol. 2014;389:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Dev Biol. 2012;370:3–23. [DOI] [PubMed] [Google Scholar]
- 24.Hovland AS, Rothstein M, Simoes-Costa M. Network architecture and regulatory logic in neural crest development. Wiley Interdiscip Rev Syst Biol Med. 2020;12:e1468. 10.1002/wsbm.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plouhinec J-L, Medina-Ruiz S, Borday C, Bernard E, Vert J-P, Eisen MB, Harland RM, Monsoro-Burq A-H. A molecular atlas of the developing ectoderm defines neural, neural crest, placode and nonneural progenitor identity in vertebrates. PLoS Biol. 2017;15:e2004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong C-S, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brugmann SA, Pandur PD. Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. [DOI] [PubMed] [Google Scholar]
- 28.Saint-Germain N, Lee YH, Zhang Y, Sargent TD, Saint-Jeannet JP. Specification of the otic placode depends on Sox9 function in Xenopus. Development. 2004;131:1755–1763. [DOI] [PubMed] [Google Scholar]
- 29.Neilson KM, Pignoni F, Yan B, Moody SA. Developmental expression patterns of candidate co-factors for vertebrate Six family transcription factors. Dev Dyn. 2010;239:3446–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton Q, Cole LK, Mulheisen M, Chang W, Wu DK. The role of pax2 in mouse inner ear development. Dev Biol. 2004;272:161–175. [DOI] [PubMed] [Google Scholar]
- 31.Heller N, Brandli AW. Xenopus Pax-2/5/8 orthologues: novel insights into pax gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric lineages. Dev Genet. 1999;24:208–219. [DOI] [PubMed] [Google Scholar]
- 32.Torres M, Gomez-Pardo E, Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development. 1996;122:3381–3391. [DOI] [PubMed] [Google Scholar]
- 33.Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. [DOI] [PubMed] [Google Scholar]
- 34.Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen R, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. [DOI] [PubMed] [Google Scholar]
- 35.Arnold JS, Braunstein EM, Ohyama T, Groves AK, Adams JC, Brown MC, Morrow BE. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Hum Mol Genet. 2006;15:1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moraes F, Novoa A, Jerome-Majewska LA, Papioannou VE, Mallo M. Tbx1 is required for proper neural crest migration and to stabilize spatial patterns during middle and inner ear development. Mech Dev. 2005;122:199–212. [DOI] [PubMed] [Google Scholar]
- 37.Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–1812. [DOI] [PubMed] [Google Scholar]
- 38.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogen. 2004;14:199–211. [DOI] [PubMed] [Google Scholar]
- 39.Coughtrie MWH, Sharp S, Maxwell K, Innes NP. Biology and function of the reversible sulfation pathway catalysed by human sulfotransferases and sulfatases. Chem Biol Interact. 1998;109:3–27. [DOI] [PubMed] [Google Scholar]
- 40.Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogen. 2004;J4:54–65. [DOI] [PubMed] [Google Scholar]
- 41.Yamauchi K, Katsumata S, Ozaki M. A prototype of the mammalian sulfotransferase 1 (SULT1) family in Xenopus laevis: molecular and enzymatic properties of XlSULT1B.S. Genes Genet Syst. 2019;94:207–217. [DOI] [PubMed] [Google Scholar]
- 42.Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Vankataramen G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. [DOI] [PubMed] [Google Scholar]
- 43.Pempe EH, Burch TC, Law CJ, Liu J. Substrate specificity of 6-O-endosulfatase (Suf-2) and its implications in synthesizing anticoagulant heparan sulfate. Glycobiol. 2012;22:1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang YH, Beck CW. Distinct patterns of endosulfatase gene expression during Xenopus laevis limb development and regeneration. Regen. 2015;2:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isaacs HV, Tannahill D, Slack JM. Expression of a novel FGF in the Xenopus embryo. A new candidate inducing factor for mesoderm formation and anteroposterior specification, Development. 1992;114:711–720. [DOI] [PubMed] [Google Scholar]
- 46.Hongo I, Kengaku M, Okamoto H. FGF signaling and the anterior neural induction in Xenopus. Dev Biol. 1999;216:561–581. [DOI] [PubMed] [Google Scholar]
- 47.Freeman SD, Moore WM, Guiral EC, Holme A, Turnbull JE, Pownall ME. Extracellular regulation of developmental cell signaling by XtSulf1. Dev Biol. 2008;320:436–445. [DOI] [PubMed] [Google Scholar]
- 48.Guiral EC, Faas L, Pownall ME. Neural crest migration requires the activity of the extracellular sulphatases XtSulf1 and XtSulf2. Dev Biol. 2010;341:375–388. [DOI] [PubMed] [Google Scholar]
- 49.Fortriede JD, Pells TJ, Chu S, Chaturvedi P, Wang DZ, Fisher ME, James-Zorn C, Wang Y, Nenni MJ, Burns KA, Lotay VS, Ponferrada VG, Karimi K, Zorn AM, Vize PD. Xenbase, deep integration of GEO and SRA RNA-seq and ChIP-seq data in a model organism database. Nucleic Acids Res. 2020;48:D776–D782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peshkin L, Gupta M, Ryazanova L, Wuhr M. Bayesian confidence intervals for multiplexed proteomics integrate ion-statistics with peptide quantification concordance. Mol Cell Proteomics. 2019;18:2108–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riddiford N, Schlosser G. Dissecting the pre-placodal transcriptome to reveal presumptive direct targets of Six1 and Eya1 in cranial placodes. Elife. 2016;5:e17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pieper M, Ahrens K, Rink E, Peter A, Schlosser G. Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development. 2012;139:1175–1187. [DOI] [PubMed] [Google Scholar]
- 53.Klein SL, Tavares ALP, Peterson M, Sullivan CH, Moody SA. Repressive interactions between transcription factors separate different embryonic ectodermal domains. Front Cell Dev Biol. 2022;10:786052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Powles N, Marshall H, Economou A, Chiang C, Murakami A, Dickson C, Krumlauf R, and Maconochie M. Regulatory analysis of the mouse Fgf3 gene: control of embryonic expression patterns and dependence upon sonic hedgehog (Shh) signaling. Dev Dyn. 2004;230:44–56. [DOI] [PubMed] [Google Scholar]
- 55.Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, Vortkamp A. Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev Dyn. 2008;237:339–353. [DOI] [PubMed] [Google Scholar]
- 56.Parain K, Mazurier N, Bronchain O, Borday C, Cabochette P Chesneau A, Colozza G, El Yakoubi W, Hamdache J, Locker M, Gilchrist MJ, Pollet N, Perron M. A large scale screen for neural stem cell markers in Xenopus retina. Dev Neurobiol. 2012;72:491–506. [DOI] [PubMed] [Google Scholar]
- 57.Winterbottom EF, Pownall ME. Complementary expression of HSPG 6-O-endosulfatases and 6-O-sulfotransferase in the hindbrain of Xenopus laevis. Gene Expr Patterns. 2009;9:166–172. [DOI] [PubMed] [Google Scholar]
- 58.Kamimura K, Rhodes JM, Ueda R, McNeeley M, Shukla D, Kimata K, Spear PG, Shworak NW, Nakato H. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J Cell Biol. 2004;166:1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem. 2004;279:5604–5611. [DOI] [PubMed] [Google Scholar]
- 60.Gómez-Skarmeta J, de La Calle-Mustienes E, Modolell J. The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and downregulates Bmp4. Development. 2001;128:551–560. [DOI] [PubMed] [Google Scholar]
- 61.Glavic A, Maris Honoré S, Gloria Feijóo C, Bastidas F, Allende ML, Mayor R. Role of BMP signaling and the homeoprotein Iroquois in the specification of the cranial placodal field. Dev Biol. 2004;272:89–103. [DOI] [PubMed] [Google Scholar]
- 62.Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. [DOI] [PubMed] [Google Scholar]
- 63.Hintze M, Prajapati RS, Tambalo M, Christophorou NAD, Anwar M, Grocott T, Streit A. Cell interactions, signals and transcriptional hierarchy governing placode progenitor induction. Development. 2017;144:2810–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maharana SK, Schlosser G. A gene regulatory network underlying the formation of pre-placodal ectoderm in Xenopus laevis. BMC Biol. 2018;16:79. 10.1186/s12915-018-0540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trevers KE, Prajapati RS, Hintze M, Stower MJ, Strobl AC, Tambalo M, Ranganathan R, Moncaut N, Khan MAF, Stern CD, Streit A. Neural induction by the node and placode induction by head mesoderm share an initial state resembling neural plate border and ES cells. Proc Natl Acad Sci USA. 2018;115:355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. [DOI] [PubMed] [Google Scholar]
- 68.Yan B, Neilson KM, Moody SA. FoxD5 plays a critical upstream role in regulating neural fate and onset of differentiation. Dev Biol. 2009;329:80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neilson KM, Abbruzzesse G, Kenyon KL, Bartolo V, Krohn P, Alfandari D, Moody SA. Pa2G4 is a novel Six1 co-factor that is required for neural crest and otic development. Dev Biol. 2017;421:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moody SA. Cell lineage analysis in Xenopus embryos. In: Tuan RS, Lo CW, eds. Meth Molec Biol: Dev Biol Protocols. 135. Totowa NJ: Humana Press; 2000:1–17. [DOI] [PubMed] [Google Scholar]
- 71.Klein SL. The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev Biol. 1987;120:299–304. [DOI] [PubMed] [Google Scholar]
- 72.Moody SA. Microinjection of mRNAs and oligonucleotides. Cold Spring Harbor Protocols. 2018; prot097261. 10.1101/pdb.prot097261. [DOI] [PubMed] [Google Scholar]
- 73.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin). New York City, NY: Garland Science. 1994. [Google Scholar]
- 74.Pearl EJ, Grainger RM, Guille M, Horb ME. Development of Xenopus resource centers: the National Xenopus Resource and the European Xenopus Resource Center. genesis. 2012;50:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]


