Abstract
Anopheles gambiae, the major vector of human malaria parasite, is an important insect model to study vector–parasite interactions. Here, we developed a simple in vivo double-stranded RNA (dsRNA) knockout approach to determine the function of the mosquito antimicrobial peptide gene Defensin. We injected dsRNA into adults and observed efficient and reproducible silencing of Defensin. Analysis of the knockdown phenotype revealed that this peptide is required for the mosquito antimicrobial defense against Gram-positive bacteria. In contrast, in mosquitoes infected by Plasmodium berghei, no loss of mosquito viability and no significant effect on the development and morphology of the parasite midgut stages were observed in the absence of Defensin. We conclude that this peptide is not a major antiparasitic factor in A. gambiae in vivo. Our results open new perspectives for the study of mosquito gene function in vivo and provide a basis for genome-scale systematic functional screens by targeted gene silencing.
INTRODUCTION
Anopheles gambiae is the most important vector for Plasmodium falciparum malaria in sub-tropical Africa and thus a critical link in the transmission cycle of one of the most serious infectious diseases of humanity (Greenwood and Mutabingwa, 2002). In recent years, this mosquito has been studied intensively by the methods of molecular and cell biology, with a special emphasis on innate immune mechanisms possibly implicated in limiting the load of parasite transmission. Such studies promise to advance rapidly when the completion of the A. gambiae genome sequencing reveals the universe of genes upon which mosquito immunity resides (Hoffman et al., 2002). Despite the existence of a good microsatellite-based genetic map (Zheng et al., 1996) and robust techniques for germ-line transgenesis (Catteruccia et al., 2000; Grossman et al., 2001), inherent limitations of mosquito stock maintenance hinder the traditional methods for gene function analysis, such as large mutagenesis screens, fine-scale gene mapping and transgenic analysis. The double-stranded RNA (dsRNA) interference is potentially adaptable to systematic reverse genetic screens (Gonczy et al., 2000), and we have shown recently that it can be used to assess gene function in cultured mosquito cells (Levashina et al., 2001). Here, we demonstrate that dsRNA can also be used to disrupt essential gene function in the whole mosquito.
When dsRNA is taken up by cells, it is cleaved into small interfering fragments that can trigger specific degradation of the endogenous target mRNA (Zamore et al., 2000). Caenorhabditis elegans is the only multicellular organism where the direct injection of dsRNA in the animal has been demonstrated to result in gene silencing throughout development (Fire et al., 1998). In Drosophila, dsRNA injection in embryos is widely used for assessing gene function in early development, but in adult flies RNA interference is routinely mediated by the expression of hairpin dsRNA in transgenic strains (St Johnston, 2002), a technique that would be very difficult to apply systematically in mosquitoes. Instead, we have opted for a rapid and direct approach, intrathoracic injection of dsRNA in adult A. gambiae, to bring about efficient and reproducible silencing of the gene encoding the antimicrobial peptide, Defensin, which is encoded by a single gene in the A. gambiae genome.
So far, three antimicrobial peptides have been characterized in A. gambiae, Defensin, Cecropin and Gambicin, which are produced by the fat body and hemocytes and secreted into hemolymph upon immune challenge (Richman et al., 1996; Vizioli et al., 2000, 2001a). These polypeptides exhibit bactericidal and/or fungicidal activities in vitro and are thought to constitute the first line of defense against microbial infections (reviewed in Dimopoulos et al., 2001; Hoffmann and Reichhart, 2002). Related peptide families exist in vertebrates and are mostly expressed by epithelia and leukocytes, acting locally to limit bacterial infection (reviewed in Lehrer and Ganz, 2002). A knockout mouse lacking the Defensin-like peptide Cathelicidin has been shown recently to be susceptible to necrotic skin infections caused by Gram-positive Group A streptococci (Nizet et al., 2001). In Drosophila, the inactivation of two major regulatory signalling pathways, Toll and Imd, has been used to turn off large groups of immune genes, including the antimicrobial peptide genes, and to associate them collectively with in vivo antifungal and antibacterial functions, respectively (Lemaitre et al., 1996). However, no loss-of-function mutants for individual antimicrobial peptide genes have been reported as yet.
The expression of Defensin is predominantly induced in the mosquito fat body shortly after bacterial challenge. It is also induced locally in the midgut and salivary gland epithelia upon invasion by malaria parasites, suggesting that Defensin may have a broad role in the defense against both microbes and parasites (Richman et al., 1996, 1997). This presumption was supported by in vitro tests of antiparasitic activity and by injection studies in Aedes mosquitoes infected by avian malaria (Shahabuddin et al., 1998). However, rigorous conclusions about Defensin function in vivo require analysis by a loss-of-function approach in the intact mosquito.
RESULTS AND DISCUSSION
To knock down the Defensin gene expression, we injected 1- to 2-day-old females with dsRNA corresponding to the genes for either Defensin (dsDEF ) or green fluorescent protein (dsGFP ) as a control and allowed the mosquitoes to recover for 4 days. The dsRNA-treated mosquitoes were then challenged with Escherichia coli, and the presence/absence of the Defensin transcripts was monitored from day 1 to 8 by RNA blotting and RT–PCR (Figure 1). In six independent experiments, the injection of either Gram-negative E. coli or Gram-positive Staphylococcus aureus induced the expression of Defensin mRNA in non-treated control mosquitoes (Figure 1A; data not shown). We observed that the injection of dsGFP partially suppressed the ultimate level of Defensin induction after bacterial challenge (Figure 1A versus C), and therefore we used dsGFP mosquitoes as controls throughout this study. In the dsDEF mosquitoes, no Defensin mRNA of proper size was detected already at day 5 after dsRNA injection (Figure 1B and C, day 1 after bacterial challenge). Instead, a strong faster migrating signal was consistently present, accompanied by a faint signal migrating more slowly than Defensin mRNA. We interpret these signals as denatured and non-denatured dsRNA, respectively, as they are also exhibited by the input dsRNA (asterisks in Figure 1D). Signals corresponding to the input dsGFP were also detected using the GFP probe (bottom asterisk in Figure 1C and D), indicating that dsRNAs are stable for at least 12 days after injection in mosquitoes and therefore can provide a long-lasting inhibition of endogenous gene expression. In contrast to robust induction of Defensin in dsGFP mosquitoes (Figure 1E, days 1, 2 and 4), only traces of Defensin mRNA were detected in dsDEF mosquitoes using sensitive RT–PCR and primers corresponding to the 5′ and 3′ UTRs of the Defensin gene. We conclude that the injection of specific dsRNA successfully inhibits induction of Defensin after bacterial challenge.
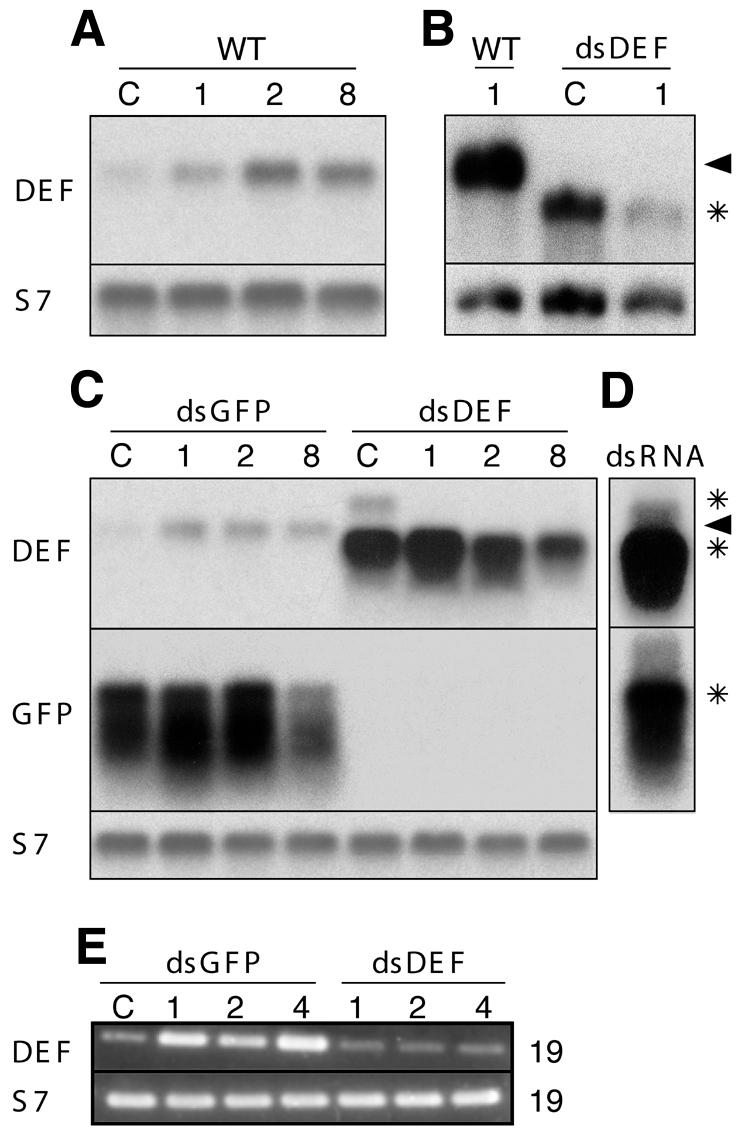
Fig. 1. RNA analysis of the Defensin gene knockout by dsRNA. RNA blots demonstrate that, over a period from 1 to 8 days (numbers above the images), E. coli challenge stably induces the expression of Defensin (DEF) mRNA (arrowheads) above the uninfected control level (C) in wild-type (WT) (A and B) and in dsGFP-treated, but not in dsDEF-treated, mosquitoes (B and C). In (D), the input dsRNAs match in size the signals (DEF and GFP) that are detected in dsRNA-injected mosquitoes (asterisks). (A), (C) and (D) were run on 1.2% agarose gels and (B) on an 1.4% agarose gel. The ribosomal protein S7 transcript was used as a loading control. GFP, green fluorescent protein. (E) RT–PCR analysis of the DEF (19 cycles) gene expression in dsGFP- and dsDEF-treated mosquitoes before (C) and after (days 1, 2 and 4) bacterial challenge. The expression of the ribosomal protein gene S7 (19 cycles) served as control.
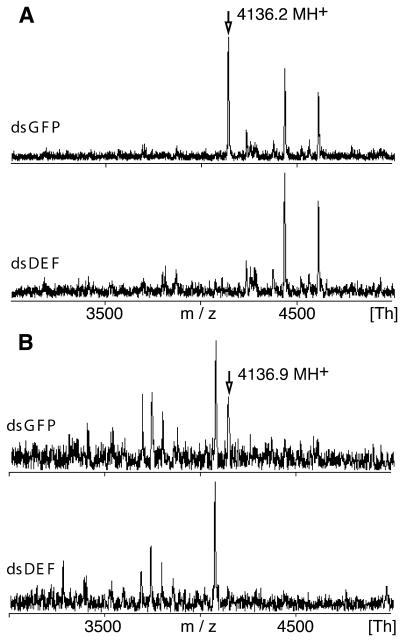
The efficacy of the Defensin knockout was further validated at the polypeptide level by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectometry (MS) of hemolymph samples (Figure 2A). Although the dsDEF and dsGFP peptide profiles were mostly comparable, the latter showed a strong peak at 4136.2 Da, which was undetectable in the dsDEF sample and corresponded to singly protonated A. gambiae Defensin bearing three disulfide bridges (calculated mass 4136.7 Da) (Vizioli et al., 2001b). This identification was confirmed by sequencing of the reduced peptide (data not shown). We also followed the Defensin knockout in the epithelium of the anterior midgut 24 h after infectious bloodmeal. The dsGFP and dsDEF midgut extracts again differed by a prominent Defensin peak at 4136.9 Da, which was present only in the midgut cells of dsGFP mosquitoes (Figure 2B). Thus, the injection of dsRNA in adult mosquitoes disrupts the expression of the targeted gene at both the RNA and polypeptide levels. Our data demonstrate that dsRNA knockout is efficient in the three different cell types tested (midgut, fat body and hemocytes), which originate from two distinct cell lineages: endoderm and mesoderm.
Fig. 2. MALDI-TOF MS analysis of the Defensin gene knockout by dsRNA. (A) Mosquitoes were challenged with a mixture of E. coli and S. aureus, and 36 h later the presence of Defensin peptide was detected in the hemolymph of control dsGFP, but not of dsDEF, mosquitoes. (B) Defensin is present in the anterior midgut 24 h after infectious bloodmeal in control dsGFP, but not in dsDEF, mosquitoes. The peaks corresponding to Defensin, and their molecular weights are indicated by arrows.
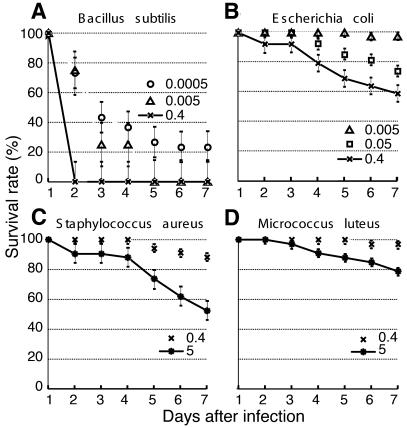
The efficacy, effectiveness and reproducibility of the knockout allowed us to determine the function and specificity of Defensin in vivo. Because of in vitro indications that Defensin is particularly potent against Gram-positive bacteria (Vizioli et al., 2001b), we focused on three members of this class, S. aureus, Micrococcus luteus and Bacillus subtilis, as well as on a Gram-negative species, E. coli. We injected measured bacterial suspensions of each species into control dsGFP A. gambiae females and followed the survival of the mosquitoes for 7 days (Figure 3). The bacteria exhibited different degrees of pathogenicity that were independent of Gram classification. Bacillus subtilis rapidly killed A. gambiae even at concentrations as low as 85 bacteria per mosquito (Figure 3A). The next most efficient pathogen, E. coli, caused comparable mosquito lethality only when injected in 10-fold higher numbers; S. aureus was substantially less pathogenic, and M. luteus was the least effective (Figure 3B–D). Differential susceptibility to bacteria has also been reported in Drosophila melanogaster, where, surprisingly, S. aureus, as well as B. subtilis, cause rapid death, while M. luteus is again only weakly pathogenic (Tzou et al., 2002). Comparative analysis of bacterial pathogenicity may be fruitful in pinpointing specificities of the immune response in related dipteran species.
Fig. 3. Lethality of control dsGFP-treated mosquitoes after infection with different doses of bacteria. Survival rates (%) are presented for mosquitoes infected with (B) the Gram-negative bacterium E. coli or the Gram-positive bacteria (A) B. subtilis, (C) S. aureus and (D) M. luteus. The bacterial concentrations are expressed as optical densities (OD) of suspensions at 600 nm: OD600 = 0.0005 (open circles), OD600 = 0.005 (open triangles), OD600 = 0.05 (open squares), OD600 = 0.4 (crosses) and OD600 = 5 (asterisks). Each experiment was performed with 50 mosquitoes for each bacterial species, and the results shown are representative of three independent experiments. Standard errors are indicated by bars.
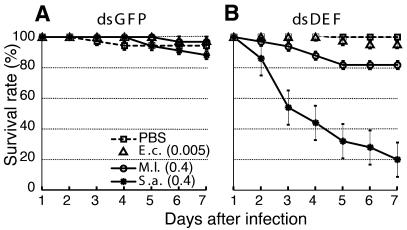
These experiments allowed us to define the sublethal concentrations for infection with the three bacterial species other than B. subtilis. We injected the selected concentrations of bacteria in both dsDEF and control dsGFP females and followed their respective survival rates over a period of 7 days (Figure 4). The profiles did not differ significantly in the case of E. coli- and mock-injected mosquitoes. In contrast, the dsDEF mosquitoes were modestly susceptible to M. luteus (20% lethality) and highly susceptible to S. aureus (80% lethality). This is the first demonstration in vivo that a single endogenous immune peptide, Defensin, is necessary for the resistance of a mosquito to two Gram-positive bacteria and not to a Gram-negative species of bacteria. The plateau in mortality that is seen in dsDEF mosquitoes 5 days after infection by M. luteus suggests that additional immune factors or cellular processes may be implicated in the clearance of this mild pathogen.
Fig. 4. Defensin knockdown mosquitoes are susceptible to Gram-positive, but not to Gram-negative, bacteria. The survival rates (%) of (A) dsGFP-treated mosquitoes and (B) dsDEF-treated mosquitoes after injection of PBS (open squares), E. coli (OD600 = 0.005, open triangles), M. luteus (OD600 = 0.4, open circles) or S. aureus (OD600 = 0.4, asterisks) are shown. Each experiment was performed with 50 mosquitoes for each bacterial species, and the results shown are representative of three independent experiments. Standard errors are indicated by bars.
We next examined the potential role of Defensin in the immune response of A. gambiae to the rodent malaria parasite, Plasmodium berghei. Defensin is constitutively expressed in the anterior midgut epithelium and is further induced by malaria infection (Richman et al., 1997; Vizioli et al., 2001b). In Aedes aegypti, the injection of high doses of related Defensins from a dragonfly and a fleshfly at specific time points after an infectious bloodmeal interfered with the development of the midgut stages of the parasite (Shahabuddin et al., 1998). These studies prompted us to monitor parasite numbers during P. berghei infections in dsGFP and dsDEF mosquitoes, using as a readout the number of oocysts per midgut 10–12 days after infection. We reasoned that, if endogenous Defensin acts antiparasitically, its absence during development of the Plasmodium midgut stages would remove a constraint and result in higher parasite loads. We observed that the mean number of oocysts per midgut was unchanged or even slightly lower in dsDEF than in dsGFP mosquitoes (Table I). Moreover, parasite-infected dsDEF and dsGFP mosquitoes showed no significant differences in mosquito viability, ookinete/oocyst morphology or the frequency distribution of oocyst numbers. We conclude that malaria-induced endogenous A. gambiae Defensin does not act as a significant antiparasitic factor in vivo.
Table I. Survival of P. berghei oocysts in the dsGFP- and dsDEF-treated mosquitoes.
| Experiment | dsRNA | Number of midguts | Oocyst number per midgut | Mean ± SEa(oocysts per midgut) | ||
|---|---|---|---|---|---|---|
| 0 | 1–29 | >30 | ||||
| 1 | dsGFP | 19 | 5 | 9 | 5 | 16.74 ± 3.7 |
| dsDEF | 24 | 7 | 14 | 3 | 10.75 ± 3.1 | |
| 2 | dsGFP | 13 | 4 | 9 | 0 | 2.77 ± 0.6 |
| dsDEF | 25 | 7 | 18 | 0 | 2.48 ± 0.4 | |
aSE, standard error.
In conclusion, the simple and convenient dsRNA technique that we describe here efficiently disrupts gene function in distinct tissues of adult mosquitoes. As a proof of principle, we have used this method to delimit phenotypically the in vivo function of a single immune peptide against different types of infections, showing it to play an important role in the resistance to Gram-positive bacteria. Evidently, antimicrobial peptides that would be unaffected by dsDEF injection cannot substitute for Defensin. Recently, we have successfully applied dsRNA to knock out 20 additional immune genes in A. gambiae (data not shown), thus confirming the general validity of this method in the study of immune responses in the mosquito. With a reverse genetics method now in hand, it should be possible to conduct systematic functional genomic analysis in this major vector of human malaria.
METHODS
Mosquito colony. Anopheles gambiae strain G3 was reared as described previously (Richman et al., 1996).
Double-stranded RNA preparation and injection in mosquitoes. dsRNAs were produced as described previously using the plasmids pLL6ds for control dsGFP (Levashina et al., 2001) and pLL80 for dsDEF. pLL80 was constructed in two steps. Defensin cDNA of 404 bp was PCR-amplified using dfn a and dfn b primers (Richman et al., 1996) and cloned into pCR2.1-TOPO (Invitrogen), resulting in pLL79. The 515 bp HindIII–XbaI fragment of pLL79 was then subcloned between the two T7 promoters of pLL10. Sense and antisense RNAs were synthesized using the T7 Ambion kit, annealed in water and stored as dsRNAs at –80°C until use. A nano-injector (Nanoject, Drummond) was used to introduce 69 nl of dsRNAs (1 mg/ml) in water in the thorax of CO2-anesthetized mosquito females, which were then allowed to recover for 4 days.
RNA analysis. Total RNA was extracted from 15 mosquitoes with TRIzol Reagent (Invitrogen) and separated by electrophoresis. Two different conditions were used: to ascertain the absence of endogenous Defensin transcripts, the electrophoresis were performed for 6 h using 1.2% agarose gels (Figure 1B); clear signals corresponding to input dsRNAs were detected using 1.4% agarose gels and 4 h migration time (Figure 1A, C and D). Separated total RNAs were transferred to a nylon membrane (Hybond-N+, Amersham Pharmacia Biotech). Blots were hybridized sequentially with the radioactively labeled probes [Ready-To-Go DNA labeling beads (dCTP), Amersham Pharmacia Biotech]: Defensin (pLL79 insert), S7 (Salazar et al., 1993) and GFP (pLL6 insert).
For RT–PCR analysis, Defensin-specific primers were selected that do not overlap with the dsRNA used for the knockout: 5′-UTR primer, 5′-AAC TCC AGC CAA GCT AAA GC-3′; and 3′-UTR primer, 5′-GAA TTA AGC CTG TGT TGT AAA C-3′. Total RNA was extracted as above from whole mosquitoes at the indicated time points. S7-specific primers and the RT–PCR conditions were as described previously (Richman et al., 1996). For amplification of both S7 and Defensin, 19 PCR cycles were used.
Mass spectrometry. The hemolymph of 40 mosquitoes was collected by centrifugation of decapitated females (3000 r.p.m., 5 min) and used in 1/10 dilution in acidified water. Five dissected anterior midguts were homogenized in 0.5% trifluoracetic acid, 50% acetonitrile; and extracts were cleared by centrifugation. The samples were analyzed without further purification in a modified thin-layer preparation (Vorm et al., 1994). MALDI-TOF MS analysis was performed on a Bruker Biflex (Bremen, Germany) mass spectrometer in linear positive mode using delayed extraction. Tandem MS with a nano-electrospray source mounted on a Micromass QTOF1 (Manchester, UK) mass spectrometer was used for peptide sequencing.
Bacterial challenge and mosquito survival. GFP-expressing E. coli OP-50 was a gift from J.J. Ewbank (INSERM, Marseille-Luminy, France). Bacillus subtilis, M. luteus and S. aureus were kind gifts from P. Bulet (IBMC, Strasbourg, France). Bacteria were cultured to OD600 = 0.4, pelleted, washed and resuspended in phosphate-buffered saline (PBS) to indicated concentrations. The number of bacteria injected was estimated by the plating of appropriate aliquots of bacterial suspension on LB plates. Mosquitoes were anesthetized with CO2, injected into the thorax with 69 nl of the bacterial suspension or PBS for controls and allowed to recover. Mosquitoes that died within 24 h of injection were not considered in the analysis. Dead mosquitoes were daily counted and removed over a period of 7 days. The results shown here are representative of at least three independent experiments, each carried out with 50 mosquitoes per tested group.
Parasite infections and oocyst counting. Parasite infections were performed essentially as described previously (Richman et al., 1997). Briefly, P. berghei parasites were passaged in CD1 mice, and parasitemia was determined from Giemsa-stained blood films. For each experiment, dsGFP and dsDEF mosquitoes were fed on the same infected mouse. Mosquito midguts were dissected 10–12 days later, fixed and DAPI stained. Morphology was examined, and the numbers of oocysts were counted using a UV-light fluorescent microscope (Zeiss).
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Doherty for help with the mosquito colony and J.J. Ewbank and P. Bulet for bacterial strains. This work was supported by EMBL, the National Institutes of Health and the European Commission.
REFERENCES
- Catteruccia F., Nolan, T., Loukeris, T.G., Blass, C., Savakis, C., Kafatos, F.C. and Crisanti, A. (2000) Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature, 405, 959–962. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G., Müller, H.M., Levashina, E.A. and Kafatos, F.C. (2001) Innate immune defense against malaria infection in the mosquito. Curr. Opin. Immunol., 13, 79–88. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gonczy P. et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- Greenwood B. and Mutabingwa, T. (2002) Malaria in 2002. Nature, 415, 670–672. [DOI] [PubMed] [Google Scholar]
- Grossman G.L., Rafferty, C.S., Clayton, J.R., Stevens, T.K., Mukabayire, O. and Benedict, M.Q. (2001) Germline transformation of the malaria vector, Anopheles gambiae, with the piggyBac transposable element. Insect Mol. Biol., 10, 597–604. [DOI] [PubMed] [Google Scholar]
- Hoffman S.L., Subramanian, G.M., Collins, F.H. and Venter, J.C. (2002) Plasmodium, human and Anopheles genomics and malaria. Nature, 415, 702–709. [DOI] [PubMed] [Google Scholar]
- Hoffmann J.A. and Reichhart, J.M. (2002) Drosophila innate immunity: an evolutionary perspective. Nat. Immunol., 3, 121–126. [DOI] [PubMed] [Google Scholar]
- Lehrer R.I. and Ganz, T. (2002) Defensins of vertebrate animals. Curr. Opin. Immunol., 14, 96–102. [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Nicolas, E., Michaut, L., Reichhart, J.M. and Hoffmann, J.A. (1996) The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell, 86, 973–983. [DOI] [PubMed] [Google Scholar]
- Levashina E.A., Moita, L.F., Blandin, S., Vriend, G., Lagueux, M. and Kafatos, F.C. (2001) Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell, 104, 709–718. [DOI] [PubMed] [Google Scholar]
- Nizet V. et al. (2001) Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature, 414, 454–457. [DOI] [PubMed] [Google Scholar]
- Richman A.M., Bulet, P., Hetru, C., Barillas-Mury, C., Hoffmann, J.A. and Kafalos, F.C. (1996) Inducible immune factors of the vector mosquito Anopheles gambiae: biochemical purification of a defensin antibacterial peptide and molecular cloning of preprodefensin cDNA. Insect Mol. Biol., 5, 203–210. [DOI] [PubMed] [Google Scholar]
- Richman A.M., Dimopoulos, G., Seeley, D. and Kafatos, F.C. (1997) Plasmodium activates the innate immune response of Anopheles gambiae mosquitoes. EMBO J., 16, 6114–6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar C.E., Mills-Hamm, D., Kumar, V. and Collins, F.H. (1993) Sequence of a cDNA from the mosquito Anopheles gambiae encoding a homologue of human ribosomal protein S7. Nucleic Acids Res., 21, 4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahabuddin M., Fields, I., Bulet, P., Hoffmann, J.A. and Miller, L.H. (1998) Plasmodium gallinaceum: differential killing of some mosquito stages of the parasite by insect defensin. Exp. Parasitol., 89, 103–112. [DOI] [PubMed] [Google Scholar]
- St Johnston R. (2002) The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet., 3, 176–188. [DOI] [PubMed] [Google Scholar]
- Tzou P., Reichhart, J.M. and Lemaitre, B. (2002) Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl Acad. Sci. USA, 99, 2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizioli J. et al. (2000) Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol., 9, 75–84. [DOI] [PubMed] [Google Scholar]
- Vizioli J., Bulet, P., Hoffmann, J.A., Kafatos, F.C., Muller, H.M. and Dimopoulos, G. (2001a) Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proc. Natl Acad. Sci. USA, 98, 12630–12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizioli J., Richman, A.M., Uttenweiler-Joseph, S., Blass, C. and Bulet, P. (2001b) The defensin peptide of the malaria vector mosquito Anopheles gambiae: antimicrobial activities and expression in adult mosquitoes. Insect Biochem. Mol. Biol., 31, 241–248. [DOI] [PubMed] [Google Scholar]
- Vorm O., Roepstorff, P. and Mann, M. (1994) Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal. Chem., 66, 3281–3287. [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A. and Bartel, D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- Zheng L., Benedict, M.Q., Cornel, A.J., Collins, F.H. and Kafatos, F.C. (1996) An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae. Genetics, 143, 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]